Back to Journals » International Journal of Nanomedicine » Volume 12
Sensitive detection of microRNAs based on the conversion of colorimetric assay into electrochemical analysis with duplex-specific nuclease-assisted signal amplification
Authors Xia N, Liu K, Zhou Y, Li Y, Yi X
Received 2 April 2017
Accepted for publication 13 June 2017
Published 13 July 2017 Volume 2017:12 Pages 5013—5022
DOI https://doi.org/10.2147/IJN.S138656
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Ning Xia,1,2 Ke Liu,1 Yingying Zhou,1 Yuanyuan Li,1 Xinyao Yi2
1Key Laboratory of New Optoelectronic Functional Materials, College of Chemistry and Chemical Engineering, Anyang Normal University, Anyang, 2College of Chemistry and Chemical Engineering, Central South University, Changsha, China
Abstract: miRNAs have emerged as new biomarkers for the detection of a wide variety of cancers. By employing duplex-specific nuclease for signal amplification and gold nanoparticles (AuNPs) as the carriers of detection probes, a novel electrochemical assay of miRNAs was performed. The method is based on conversion of the well-known colorimetric assay into electrochemical analysis with enhanced sensitivity. DNA capture probes immobilized on the electrode surface and ferrocene (Fc)-labeled DNA detection probes (denoted “Fc-DNA-Fc”) presented in the solution induced the assembly of positively charged AuNPs on the electrode surface through the electrostatic interaction. As a result, a large number of Fc-DNA-Fc molecules were attached on the electrode surface, thus amplifying the electrochemical signal. When duplex-specific nuclease was added to recycle the process of miRNA-initiated digestion of the immobilized DNA probes, Fc-DNA-Fc-induced assembly of AuNPs on the electrode surface could not occur. This resulted in a significant fall in the oxidation current of Fc. The current was found to be inversely proportional to the concentration of miRNAs in the range of 0–25 fM, and a detection limit of 0.1 fM was achieved. Moreover, this work presents a new method for converting colorimetric assays into sensitive electrochemical analyses, and thus would be valuable for design of novel chemical/biosensors.
Keywords: microRNA, duplex-specific nuclease, gold nanoparticle, signal amplification, electrochemical biosensor, colorimetric assay
Introduction
miRNAs are a class of single-stranded, endogenous nonprotein-coding RNAs 17-25 nucleotides long, and are posttranscriptional regulators for gene expression in a broad range of animals, plants, and viruses.1–3 Expression of miRNAs has been associated with the regulation and progression of numerous cancers.4–6 Therefore, the expression profiles of miRNAs have been increasingly used as effective biomarkers for the diagnosis, classification, progression, and treatment response of cancers.7,8 However, due to the smallness, low expression level, and high sequence similarity of miRNAs,9,10 it is still a challenge to develop a simple, sensitive, and selective method for their assay.
Duplex-specific nuclease (DSN) has the capability of cleaving DNA in double-stranded DNA or DNA–RNA heteroduplexes. More importantly, this enzyme can clearly discriminate among mismatched duplexes, even a one-base mismatch.11,12 Therefore, analytic specificity has been significantly improved with DSN-mediated analytical strategies, including colorimetric assays,13–15 electrochemistry,16–24 fluorescence,25–37 chemiluminescence,38 surface-plasmon resonance,39 and electrochemiluminescence.40,41 In combination with nanomaterials, such as gold nanoparticles (AuNPs),27,42 graphene,32,43 magnetic microparticles,17,35,44,45 and quantum dots,33 ultralow detection limits could be achieved with DSN-assisted signal amplification.
Among various methods for nucleic acid detection, AuNP-based liquid-phase colorimetric assays have received considerable attention since they require very simple sample handling and enable color visualization without specific instruments. In particular, the unmodified AuNP-based colorimetric assay is more convenient and cost-effective for its absence of the elaborate and expensive synthesis of ligand-modified AuNPs.46 Usually, there are two categories of unmodified AuNP-based colorimetric assays with DNA as the probe. In the first strategy, DNA absorbs onto the unmodified negatively charged AuNPs through the interaction between gold and the nitrogen-containing base, thus stabilizing them against salt-induced aggregation.47,48 In the other strategy, the single-strand DNA probe adsorbs onto the positively charged AuNPs via electrostatic interaction to induce their aggregation.49–52 Upon binding to the target or being digested by the nuclease, the DNA probe loses its ability to inhibit salt-induced aggregation of negatively charged AuNPs or to trigger the assembly of positively charged AuNPs. The combination of DSN and AuNPs has also attracted lots of attention for colorimetric assays of miRNAs. For example, Shen et al reported a real-time sensitive colorimetric detection of target miRNAs by inducing DSN into DNA-modified AuNP networks.13 Shi et al demonstrated that the DNA probe protected the NaCl-induced aggregation of AuNPs. However, cleavage of DNA in the DNA–miRNA hybrid by DSN into small fragments allowed for the NaCl-induced aggregation of AuNPs.14 Based on the target-mediated probe digestion by DSN and the probe-triggered aggregation of DNA-modified AuNPs as a switch for signal output, Wang et al designed a colorimetric method for miRNA detection.15 In the present work, we found that the DNA probe triggered the aggregation of positively charged AuNPs. Digestion of DNA by DSN in the presence of target miRNAs interfered with aggregation. However, although the colorimetric assay with the combination of unmodified AuNPs and DSN provides an easy and rapid way for miRNA detection, it has some unavoidable limitations, such as poor sensitivity and anti-interference capability. Therefore, the colorimetric example is expected to recreate an existing platform with improved sensitivity and specificity.
Electrochemical biosensors have been widely used in the detection of biomarkers, because of their low cost, portability, rapid response, and high sensitivity.17,21,22,53–56 Furthermore, we demonstrated that the DSN-assisted colorimetric assay can be converted into an electrochemical analysis with improved sensitivity and specificity. Specifically, the DNA probe immobilized on the electrode surface and that labeled with ferrocene (Fc) (denoted “Fc-DNA-Fc”) in the solution triggered the in situ assembly of positively charged AuNPs through electrostatic interaction. The electrochemical signal of Fc-DNA-Fc AuNP network architecture can be measured by recording the oxidation current of Fc. Once the sensing electrode was incubated with the mixture of DSN and miRNAs, DSN cleaved the DNA strand in the miRNA–DNA heteroduplexes and released the target miRNAs for recycling. Cleavage of DNA anchored on the electrode surface prevented the in situ formation of Fc-DNA-Fc AuNPs, thus depressing the oxidation current of Fc.
Materials and methods
6-Mercapto-1-hexanol (MCH), tris(carboxyethyl)phosphine, HAuCl4, NaBH4, tetraoctylammonium bromide, and 4-dimethylaminopyridine (DMAP) were acquired from Sigma-Aldrich (St Louis, MO, US). DSN was provided by Evrogen Joint Stock Company (Moscow, Russia). Diethyl pyrocarbonate and the thiolated and Fc-labeled DNA with sequences of TCAACATCAGTCTGATAAGCTATTT-(CH2)6-SH, Fc-TCAACATCAGTCTGATAAGCTA (Fc-DNA), and Fc-TCAACATCAGTCTGATAAGCTA-Fc (Fc-DNA-Fc) were supplied by Sangon Biotech Co Ltd (Shanghai, China). The miRNAs were synthesized and purified by GenePharma Co Ltd (Shanghai, China). Their sequences were 5′-UAGCUUAUCAGACUGAUGUUGA-3′ (miRNA-21), 5′-UAGCUUAUCGGACUGAUGUUGA-3′ (single-base mismatch), 5′-UUGCUUAUCGGACUGAUCUUGA-3′ (three-base mismatch), and 5′-GUAAGGCAUCUGACCGAAGGCA-3′ (noncomplementary). Serum samples were obtained from Anyang Maternal and Child Heath Care Hospital (Anyang, China). All solutions were prepared with diethyl pyrocarbonate-treated deionized water in an RNase-free environment. The DNA probes and the target miRNAs were incubated in 20 mM Tris buffer (pH 7.4) containing 200 mM NaCl and 50 mM MgCl2.
Preparation of AuNPs
The positively charged AuNPs were synthesized as previously reported by the spontaneous phase-transfer reaction.57 In brief, 7.4 mL of 1% HAuCl4 aqueous solution was added to 16 mL of 25 mM tetraoctylammonium bromide in toluene solution. Then, 5 mL of 0.4 M freshly prepared NaBH4 solution was added to the stirred mixture. After being stirred for 30 minutes, the toluene phase was collected, subsequently washed with 0.1 M H2SO4, 0.1 M NaOH, and deionized water (three times), and then dried with anhydrous Na2SO4. For the phase transfer, 2 mL of 0.1 M DMAP solution was added to 2 mL of the as-prepared NP suspension. The AuNPs were stable in a wide pH range and under high salt concentration. To remove the free DMAP, the NPs were centrifuged and redispersed in 20 mM Tris buffer. The AuNPs were characterized by observed by spectrophotometry (Cary 60; Agilent Technologies, Santa Clara, CA, US) and transmission electron microscopy (TEM; Tecnai G2 T20; Thermo Fisher Scientific, Waltham, MA, US). The average size of the AuNPs was determined to be 6.5±1.2 nm using a Nano ZS laser-scattering particle-size analyzer (Malvern Instruments, Malvern, UK). The concentration of AuNPs was calculated according to NP size and Au-atom concentration.
Colorimetric assay
A DNA/DSN solution (25 μL) was first mixed with 25 μL of miRNAs at a given concentration for 1 hour. Then, 450 μL of AuNP suspension was added to the mixture for incubation of 5 minutes. The absorption change was monitored by the ultraviolet-visible spectrophotometry.
Electrochemical detection
The gold electrode (diameter 2 mm) was polished with 0.3 μm alumina slurry on a polishing cloth and then cleaned by sonication in ethanol and water. The cleaned electrode was first immersed into 100 μL of 1 μM thiolated DNA probe for over 12 hours. This was followed by washing the electrode thoroughly with tris buffer and deionized water and drying by N2. The DNA-covered electrode was then incubated in 0.1 mM MCH for 1 hour to block the unreacted gold sites. After being washed with ethanol/water, the DNA/MCH-covered electrode was incubated with the desired concentration of miRNAs in the presence of DSN at 37°C for 1 hour. Then, the electrode was rinsed and exposed to 20 μL of AuNP suspension in a homemade plastic cell, followed by addition of 20 μL of Fc-DNA-Fc to incubate for 30 minutes. After being washed with water again, the electrode was placed in 0.1 M KClO4 for differential pulse voltammetry (DPV) measurement on a 660E electrochemical workstation (CH Instruments, Shanghai, China). A platinum wire and Ag/AgCl electrode were used as the counter and reference electrodes, respectively.
Results and discussion
Principle and feasibility of AuNP-based colorimetric assay
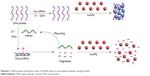
A schematic illustration of the DSN-assisted colorimetric assay of miRNA based on unmodified positively charged AuNPs is shown in Scheme 1. In the absence of miRNA, DSN is inactive to single-stranded DNA. As such, the negatively charged DNA molecules can absorb onto the surface of the positively charged AuNPs to trigger their aggregation.50 In the presence of miRNA targets, the complementary DNA probes form the miRNA–DNA heteroduplexes by hybridization, thus activating the DSN. When DNA in the DNA–miRNA heteroduplex was cleaved into small fragments, the miRNA was released to initiate another round of hybridization and DSN digestion. As a result, the resulting small DNA fragments cannot induce AuNP aggregation.50
  | Scheme 1 DSN-assisted colorimetric assay of miRNA based on unmodified positively charged AuNPs. |
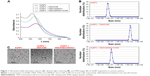
The feasibility of the method was first characterized by ultraviolet-visible spectroscopy. As shown in Figure 1A, the dispersed AuNPs displayed a characteristic absorbance peak at 520 nm (A520; black curve). Upon addition of DNA and DSN, the AuNP suspension exhibited a decrease in A520 with a bathochromic shift to ~560 nm, demonstrating that DNA caused aggregation of the positively charged AuNPs (red curve). With the addition of miRNA21 to the DNA solution in the absence of DSN, the absorbance peak was the same as the red curve, indicating that a small amount of miRNA21 showed no impact on the DNA-triggered AuNP aggregates (blue curve). Interestingly, in the presence of DSN, introducing the same concentration of miRNA21 into the DNA solution caused a significant increase in the absorption intensity of AuNPs (green curve). This is indicative of the cleavage of DNA in the DNA–miRNA heteroduplex by DSN. The result was further confirmed by dynamic light scattering. As shown in Figure 1B, the dispersed AuNP suspension showed an average diameter of ~6.5 nm. Upon addition of DNA, the AuNPs aggregated to form larger bodes (~1,281 nm). After the DNA probes were hybridized with miRNA21 and follow-up cleaved by DSN into small fragments, the AuNPs were almost dispersed, with an average diameter of ~15.7 nm. The aggregation and dispersion of AuNPs were also monitored by TEM (Figure 1C). The dispersed AuNPs assembled into larger aggregates in the presence of DNA/DSN, but introduction of miRNA21 prevented DNA-triggered AuNP assembly. The TEM results were good in agreement with those observed by dynamic light scattering, clearly verifying that the AuNP-based colorimetric assay is viable for miRNA detection with DSN-assisted signal amplification.
Optimized experimental conditions and sensitivity of the colorimetric assay
We optimized the experimental conditions for the miRNA assay, ie, DNA concentration, DSN concentration, and hybridization/incubation time. As shown in Figure 2A, A520 was decreased upon addition of increasing concentrations of DNA and began to level off beyond 2 μM. In the following assays, a compromise concentration of 1 μM was used for further experiments. DSN concentration was then investigated. A520 increased with increasing concentrations of DSN (Figure 2B). When the concentration increased to 0.2 U, no enhancement of A520 was observed. Furthermore, we found the increase in A520 was dependent upon hybridization/incubation time below 60 minutes (Figure 2C). With these results, 0.2 U and 60 minutes were chosen as the optimal DSN concentration and reaction time, respectively. Under the optimal experimental conditions, miRNA21 at various concentrations was determined with the colorimetric assay. As depicted in Figure 2D, A520 increased with increased miRNA21 concentration, ranging from 1 to 2,000 pM. The inset shows the linear portion of the calibration curve between 1 to 1,000 pM. The detection limit was estimated to be 1 pM. Note that the concentration of miRNA21 was normally as low as dozens of femtomoles or lower; therefore, such a detection limit was not sufficient for miRNA detection in clinical samples.
Principle of the electrochemical assay
To achieve a low detection limit, the proposed colorimetric assay was converted to electrochemical analysis, in view of the high sensitivity of electrochemical biosensors. The electrochemical method based on DSN-assisted signal amplification employs the Fc-DNA-Fc/AuNPs network architecture as the signal indicators. Its design principle is the same as that of the colorimetric assay, as shown in Scheme 2. The polished gold electrode was first modified with thiolated DNA probes, which was followed by blocking the empty sites on the electrode with MCH to prevent aspecific adsorption. When the DNA/MCH-covered electrode was immersed in the AuNP suspension (path I), the positively charged AuNPs were captured by DNA through the electrostatic interaction. After addition of DNA labeled with Fc tags in both the 5′ and 3′ terminals (Fc-DNA-Fc), the negatively charged Fc-DNA-Fc absorbed onto the positively charged AuNPs and induced the in situ assembly of AuNPs on the electrode surface. The aggregated AuNPs incorporated numerous Fc-DNA-Fc molecules in a network, thus leading to an amplified detectable electrochemical signal. When miRNA21 hybridized with the DNA probe to form a DNA–miRNA21 heteroduplex (path II), the DNA probe was able to be cleaved into small fragments by DSN. The miRNA21 was then released and hybridized with the other DNA probe on the electrode surface. In this manner, each miRNA21 can trigger thousands of hybridization/cleavage-cycle events to remove the DNA probes from the electrode surface. In this case, introducing the mixture of Fc-DNA-Fc and AuNPs did not cause the formation of a network of Fc-DNA-Fc AuNPs on the electrode surface. Therefore, a poor or undetectable electrochemical signal would be observed.
  | Scheme 2 DSN-assisted electrochemical assay of miRNA21 based on in situ formation of AuNP aggregates on electrode surface. |
Feasibility of the electrochemical assay
Figure 3 shows the DPV responses of the sensing electrodes after incubation with various solutions. After incubation of the DNA/MCH-covered electrode with the mixture of Fc-DNA-Fc AuNPs, a much larger DPV signal with an oxidation potential of ~0.47 V was observed (curve a). Note that no electrochemical signal was observed when the sensing electrode was incubated with the mixture of DNA AuNPs (curve d), demonstrating the oxidation peak in the curve a was due to the oxidation of Fc tags. We also found that the current with dual-labeled DNA (Fc-DNA-Fc) as the detection probe (curve a) was higher than that with the single-labeled detection probe Fc-DNA (curve b). Considering the concentration of miRNAs was quite low, Fc-DNA-Fc was chosen as the detection probe because of the strong electrochemical signal. As expected, incubation of the sensing electrode with miRNA21 and DSN and follow-up incubation with the mixture of Fc-DNA-Fc AuNPs caused a poor electrochemical signal (curve c). The result indicated that cleavage of the anchored DNA probes diminished the assembly of Fc-DNA-Fc AuNPs on the electrode surface.
The concentration of the DNA probe had a profound influence on the aggregation of AuNPs (Figure 2A). Herein, the optimal concentration of the Fc-DNA-Fc detection probe for the DPV signal was examined. Dependence of the DPV current (ipa) on the concentration of Fc-DNA-Fc is shown in Figure 4A. The ipa increased with the excitation of the Fc-DNA-Fc concentration and reached its maximum value at 0.8 μM for 4 nM AuNPs. The result was acceptable, since a low concentration of Fc-DNA-Fc would cause a small amount of Fc-DNA-Fc AuNP aggregates on the electrode surface. However, further increase in the concentration of Fc-DNA-Fc over 0.8 μM led to a decrease in the ipa. This was probably because free Fc-DNA-Fc molecules in solution competed with the anchored DNA capture probe to bind with AuNPs, thus hampering the in situ assembly of AuNPs on the electrode surface. Additionally, the dependence of the AuNP concentration on ipa was examined (Figure 4B). It was found that ipa increased with increasing AuNP concentrations and began to level off beyond 1.5 nM. In the following quantitative assays, the final concentrations of AuNPs and Fc-DNA-Fc were kept at 1.5 nM and 0.3 μM, respectively.
Sensitivity and selectivity
The dependence of current response upon the concentration of miRNA21 is depicted in Figure 5A. Increasing miRNA21 concentration led to a decrease in electrochemical signal, demonstrating that high miRNA21 concentration benefits the cleavage of DNA and thus leaves a small amount of DNA probes on the electrode surface to allow for the formation of Fc-DNA-Fc AuNP network. As shown in Figure 5B, the oxidation peak current was inversely proportional with the concentration of miRNA21 in the range of 0–100 fM. The insets show the linear portion of the calibration curve for 0–25 fM. The linear regression equation was expressed as ipa=0.275–0.008miRNA21 (fM); R=0.996. The detection limit was estimated to be 0.1 fM (n=11), which is fourfold lower than the aforementioned colorimetric assay. Therefore, the electrochemical method enables highly sensitive detection of miRNAs in a similar design principle to that of the colorimetric assay. Moreover, the detection limit is lower than (or comparable to) the other DSN-based colorimetric and electrochemical methods reported previously (Table 1).
The short length and high sequence similarity of miRNAs in the RNA family greatly complicate miRNA detection in a biological matrix through hybridization, because the sequence mismatch may easily produce a false-positive signal. Therefore, high specificity of the biosensor is very significant in miRNA-profiling measurement in clinical applications. The selectivity of the proposed biosensor toward miRNA21 was further investigated. The single-base and three-base mismatched and noncomplementary sequences of miRNA21 were chosen for evaluating selectivity. As depicted in Figure 6, in comparison with miRNA21, the three tested miRNAs caused negligible current changes. The results demonstrate the method is highly selective for discriminating target miRNA21 from other miRNAs with similar sequences. The good specificity should be attributed to the latent capability of DSN to discriminate the complementary DNA–RNA duplex from the mismatched ones.
  | Figure 6 Selectivity of proposed electrochemical method for 25 fM miRNAs with different sequences. |
Assays of serum samples
To demonstrate the viability of our method to measure target-miRNA levels in biological samples, we determined the concentration of miRNA21 in two serum samples. The values were found to be in the femtomolar range without spiking of target miRNAs (Table 2), indicating that the two donors were healthy.58,59 Moreover, after addition of miRNA21, recovery rates were 92%–105%, demonstrating the accuracy of the assay for biological samples.
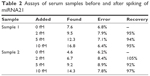  | Table 2 Assays of serum samples before and after spiking of miRNA21 |
Conclusion
By integrating the advantages of DSN for signal amplification and DNA-induced aggregation of AuNPs for signal readout, this work provides an option for miRNA detection. It also conceives a way of converting colorimetric assays into electrochemical analyses. The detection limits of the proposed methods are comparable to or even lower than those achieved by other DSN-based colorimetric and electrochemical assays reported previously. Additionally, this method requires a simple detection principle, and obviates the specific modification of nanomaterials, as well as the use of extra enzymes for molecular recognition and signal readout, thus reducing operation complexity. The excellent capability of identifying the complementary DNA–miRNA duplex by DSN ensures the selectivity and accuracy of the assay for clinical use. Our work also provides a hint for designing novel optical and electrochemical biosensors through the DNA-induced in situ formation of NP aggregates.
Acknowledgment
Financial support from the National Natural Science Foundation of China (21305004), Joint Fund for Fostering Talents of National Natural Science Foundation of China, and Henan Province (U1304205) and Science and Technology Foundation of Henan Province (17A150001) is gratefully acknowledged.
Disclosure
The authors report no conflicts of interest in this work.
References
Buchan JR, Parker R. Molecular biology: the two faces of miRNA. Science. 2007;318:1877–1878. | ||
Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. | ||
Laterza OF, Lim L, Garrett-Engele PW, et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–1983. | ||
Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. | ||
Tricoli JV, Jacobson JW. MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67:4553–4555. | ||
Paul S, Maji P. Rough sets for in silico identification of differentially expressed miRNAs. Int J Nanomedicine. 2013;8:63–74. | ||
Braicu C, Cojocneanu-Petric R, Chira S, et al. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomed. 2015;10:791–800. | ||
Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. | ||
Cissell KA, Shrestha S, Deo SK. MicroRNA detection: challenge for analytical chemists. Anal Chem. 2007;79:4754–4761. | ||
Ye DK, Zuo XL, Fan CH. DNA nanostructure-based engineering of the biosensing interface for biomolecular detection. Prog Chem. 2017;29:36–46. | ||
Li XL, Wang YC, Zhang XJ, Zhao YJ, Liu CH, Li ZP. Double strand-specific nuclease-assisted sensitive detection of microRNA. Acta Chim Sin. 2014;72:395–400. | ||
Qiu X, Zhang H, Yu H, Jiang T, Luo Y. Duplex-specific nuclease-mediated bioanalysis. Trends Biotechnol. 2015;33:180–188. | ||
Shen W, Deng H, Ren Y, Gao Z. A real-time colorimetric assay for label-free detection of microRNAs down to sub-femtomolar levels. Chem Commun (Camb). 2013;49:4959–4961. | ||
Shi HY, Yang L, Zhou XY, et al. A gold nanoparticle-based colorimetric strategy coupled to duplex-specific nuclease signal amplification for the determination of microRNA. Microchim Acta. 2016;184:525–531. | ||
Wang Q, Li RD, Yin BC, Ye BC. Colorimetric detection of sequence-specific microRNA based on duplex-specific nuclease-assisted nanoparticle amplification. Analyst. 2015;140:6306–6312. | ||
Li BC, Li X, Wang M, Yang ZQ, Yin HS, Ai AY. Photoelectrochemical biosensor for highly sensitive detection of microRNA based on duplex-specific nuclease-triggered signal amplification. J Solid State Electrochem. 2015;19:1301–1309. | ||
Li XM, Wang LL, Luo J, Wei QL. A dual-amplified electrochemical detection of mRNA based on duplex-specific nuclease and bio-bar-code conjugates. Biosens Bioelectron. 2015;65:245–250. | ||
Li Y, Tian R, Zheng X, Huang R. Amplified electrochemical detection of nucleic acid hybridization via selective preconcentration of unmodified gold nanoparticles. Anal Chim Acta. 2016;934:59–65. | ||
Liu L, Gao Y, Liu H, Xia N. An ultrasensitive electrochemical miRNAs sensor based on miRNAs-initiated cleavage of DNA by duplex-specific nuclease and signal amplification of enzyme plus redox cycling reaction. Sens Actuators B Chem. 2015;208:137–142. | ||
Miao P, Tang YG, Wang BD, et al. Nuclease assisted target recycling and spherical nucleic acids gold nanoparticles recruitment for ultrasensitive detection of microRNA. Electrochim Acta. 2016;190:396–401. | ||
Ren Y, Deng H, Shen W, Gao Z. A highly sensitive and selective electrochemical biosensor for direct detection of microRNAs in serum. Anal Chem. 2013;85:4784–4789. | ||
Yang C, Dou B, Shi K, Chai Y, Xiang Y, Yuan R. Multiplexed and amplified electronic sensor for the detection of microRNAs from cancer cells. Anal Chem. 2014;86:11913–11918. | ||
Zhang J, Wu DZ, Cai SX, et al. An immobilization-free electrochemical impedance biosensor based on duplex-specific nuclease assisted target recycling for amplified detection of microRNA. Biosens Bioelectron. 2016;75:452–457. | ||
Zhang X, Wu D, Liu Z, et al. An ultrasensitive label-free electrochemical biosensor for microRNA-21 detection based on a 2′-O-methyl modified DNAzyme and duplex-specific nuclease assisted target recycling. Chem Commun (Camb). 2014;50:12375–12377. | ||
Zhou Y, Zhang J, Zhao L, et al. Visual detection of multiplex microRNAs using cationic conjugated polymer materials. ACS Appl Mater Interfaces. 2016;8:1520–1526. | ||
Yin BC, Liu YQ, Ye BC. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J Am Chem Soc. 2012;134:5064–5067. | ||
Degliangeli F, Kshirsagar P, Brunetti V, Pompa PP, Fiammengo R. Absolute and direct microRNA quantification using DNA-gold nanoparticle probes. J Am Chem Soc. 2014;136:2264–2267. | ||
Zhou H, Yang C, Chen H, Li X, Li Y, Fan X. A simple G-quadruplex molecular beacon-based biosensor for highly selective detection of microRNA. Biosens Bioelectron. 2017;87:552–557. | ||
Li Y, Zhang J, Zhao J, Zhao L, Cheng Y, Li Z. A simple molecular beacon with duplex-specific nuclease amplification for detection of microRNA. Analyst. 2016;141:1071–1076. | ||
Xu F, Dong H, Cao Y, et al. Ultrasensitive and multiple disease-related microRNA detection based on tetrahedral DNA nanostructures and duplex-specific nuclease-assisted signal amplification. ACS Appl Mater Interfaces. 2016;8:33499–33505. | ||
Kim E, Howes PD, Crowder SW, Stevens MM. Multi-amplified sensing of microRNA by a small DNA fragment-driven enzymatic cascade reaction. ACS Sens. 2016;2:111–118. | ||
Guo SA, Yang F, Zhang YL, Ning Y, Yao QF, Zhang GJ. Amplified fluorescence sensing of miRNA by combination of graphene oxide with duplex-specific nuclease. Anal Methods. 2014;6:3598–3603. | ||
Jou AF, Lu CH, Ou YC, et al. Diagnosing the miR-141 prostate cancer biomarker using nucleic acid-functionalized CdSe/ZnS QDs and telomerase. Chem Sci. 2015;6:659–665. | ||
Hu Z, Chen J, Li W, et al. Label-free fluorescence turn-on detection of microRNA based on duplex-specific nuclease and a perylene probe. Anal Chim Acta. 2015;895:89–94. | ||
Zhang J, Wu D, Chen Q, et al. Label-free microRNA detection based on terbium and duplex-specific nuclease assisted target recycling. Analyst. 2015;140:5082–5089. | ||
Tian T, Xiao H, Zhang Z, et al. Sensitive and convenient detection of microRNAs based on cascade amplification by catalytic DNAzymes. Chemistry. 2013;19:92–95. | ||
Zhang K, Wang K, Zhu X, Xu F, Xie M. Sensitive detection of microRNA in complex biological samples by using two stages DSN-assisted target recycling signal amplification method. Biosens Bioelectron. 2017;87:358–364. | ||
Bi S, Zhang J, Hao S, Ding C, Zhang S. Exponential amplification for chemiluminescence resonance energy transfer detection of microRNA in real samples based on a cross-catalyst strand-displacement network. Anal Chem. 2011;83:3696–8702. | ||
Qiu X, Liu X, Zhang W, et al. Dynamic monitoring of microRNA-DNA hybridization using DNAase-triggered signal amplification. Anal Chem. 2015;87:6303–6310. | ||
Feng XB, Gan N, Zhang HR, et al. Ratiometric biosensor array for multiplexed detection of microRNAs based on electrochemiluminescence coupled with cyclic voltammetry. Biosens Bioelectron. 2016;75:308–314. | ||
Zhang P, Wu X, Yuan R, Chai Y. An “off-on” electrochemiluminescent biosensor based on DNAzyme-assisted target recycling and rolling circle amplifications for ultrasensitive detection of microRNA. Anal Chem. 2015;87:3202–3207. | ||
Wang J, Yi X, Tang H, Han H, Wu M, Zhou F. Direct quantification of microRNA at low picomolar level in sera of glioma patients using a competitive hybridization followed by amplified voltammetric detection. Anal Chem. 2012;84:6400–6406. | ||
Jin B, Wang P, Mao H, et al. Multi-nanomaterial electrochemical biosensor based on label-free graphene for detecting cancer biomarkers. Biosens Bioelectron. 2014;55:464–469. | ||
Lv W, Zhao J, Situ B, et al. A target-triggered dual amplification strategy for sensitive detection of microRNA. Biosens Bioelectron. 2016;83:250–255. | ||
Tian B, Ma J, Qiu Z, et al. Optomagnetic detection of microRNA based on duplex-specific nuclease-assisted target recycling and multilayer core-satellite magnetic superstructures. ACS Nano. 2017;11:1798–1806. | ||
Sun K, Chang Y, Zhou BB, Wang XJ, Liu L. Gold nanoparticles-based electrochemical method for the detection of protein kinase with a peptide-like inhibitor as the bioreceptor. Int J Nanomedicine. 2017;12:1905–1915. | ||
Fan D, Zhai Q, Zhou W, Zhu X, Wang E, Dong S. A label-free colorimetric aptasensor for simple, sensitive and selective detection of Pt (II) based on platinum (II)-oligonucleotide coordination induced gold nanoparticles aggregation. Biosens Bioelectron. 2016;85:771–776. | ||
Xia N, Zhou B, Huang N, Jiang M, Zhang J, Liu L. Visual and fluorescent assays for selective detection of β-amyloid oligomers based on the inner filter effect of gold nanoparticles on the fluorescence of CdTe quantum dots. Biosens Bioelectron. 2016;85:625–632. | ||
Cao R, Li B. A simple and sensitive method for visual detection of heparin using positively-charged gold nanoparticles as colorimetric probes. Chem Commun (Camb). 2011;47:2865–2867. | ||
Cao R, Li B, Zhang Y, Zhang Z. Naked-eye sensitive detection of nuclease activity using positively-charged gold nanoparticles as colorimetric probes. Chem Commun (Camb). 2011;47:12301–12303. | ||
Gao QA, Zhang WY, Guo YY, Qi HL, Zhang CX. Highly sensitive impedimetric sensing of DNA hybridization based on the target DNA-induced displacement of gold nanoparticles attached to ssDNA probe. Electrochem Commun. 2011;13:335–337. | ||
Su J, Zhou W, Xiang Y, Yuan R, Chai Y. Target-induced charge reduction of aptamers for visual detection of lysozyme based on positively charged gold nanoparticles. Chem Commun (Camb). 2013;49:7659–7661. | ||
Zhao CF, Yang SF, Lin LQ, et al. Chronocoulometric biosensor for K-ras point mutation detection based on E. coli DNA ligase and AuNPs amplification effects. Sens Actuators B Chem. 2016;223:946–951. | ||
Tsai JJ, Bau IJ, Chen HT, Lin YT, Wang GJ. A novel nanostructured biosensor for the detection of the dust mite antigen Der p2. Int J Nanomedicine. 2011;6:1201–1208. | ||
Zeng Y, Zhu ZH, Du D, Lin YH. Nanomaterial-based electrochemical biosensors for food safety. J Electroanal Chem (Lausanne). 2016;781:147–154. | ||
Wen YL, Lin MH, Pei H, Lu N, Fan CH. Electrochemical-based microRNA sensors. Prog Chem. 2012;24:1656–1664. | ||
Gittins DI, Caruso F. Spontaneous phase transfer of nanoparticulate metals from organic to aqueous media. Angew Chem Int Ed Engl. 2001;40:3001–3004. | ||
Liu L, Chang Y, Xia N, et al. Simple, sensitive and label-free electrochemical detection of microRNAs based on the in situ formation of silver nanoparticles aggregates for signal amplification. Biosens Bioelectron. 2017;94:235–242. | ||
Zhuang J, Tang D, Lai W, Chen G, Yang H. Immobilization-free programmable hairpin probe for ultrasensitive electronic monitoring of nucleic acid based on a biphasic reaction mode. Anal Chem. 2014;86:8400–8407. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.