Back to Journals » International Journal of Nanomedicine » Volume 11
Self-assembling peptide nanofibers containing phenylalanine for the controlled release of 5-fluorouracil
Authors Ashwanikumar N, Kumar NA, Saneesh Babu PS, Sivakumar KC, Vadakkan MV, Nair P, Hema Saranya I, Asha Nair S, Vinod Kumar GS
Received 20 January 2016
Accepted for publication 2 April 2016
Published 25 October 2016 Volume 2016:11 Pages 5583—5594
DOI https://doi.org/10.2147/IJN.S104707
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Narayanan Ashwanikumar,1 Nisha Asok Kumar,2 Padma S Saneesh Babu,2 Krishnankutty C Sivakumar,3 Mithun Varghese Vadakkan,1 Parvathi Nair,1 Ilamathi Hema Saranya,1 Sivakumari Asha Nair,2 Gopalakrishnapillai S Vinod Kumar1
1Chemical Biology, Nano Drug Delivery Systems, Bio-Innovation Center, 2Cancer Research Programme, 3Distributed Information Sub-Centre (Bioinformatics Centre), Rajiv Gandhi Centre for Biotechnology, Poojappura, Thiruvananthapuram, Kerala, India
Abstract: The study shows that RADA-F6 peptide with pH-responsive self-assembling nature can be effectively used as a drug delivery system for the sustained release of a potent anticancer drug 5-fluorouracil (5-FU) at basic pH. As 5-FU contains the aromatic pyrimidine ring, RADA-F6 system is suitable for entrapping an aromatic drug due to effective π–π stacking with phenylalanine and be able to show better controlled release behavior. The stability and controlled release nature of RADA-F6 in different conditions followed by 5-FU entrapment at in silico conditions was confirmed by molecular dynamics simulation taking RADA-16 as control. Cytotoxicity of the drug-loaded RADA-F6 was measured by MTT assay and cellular uptake by confocal microscopy. Physicochemical characterization and further Western blot analysis and flow cytometric studies confirm that RADA-F6 can be successfully used as an efficient vector for pH-sensitive, controlled 5-FU delivery system.
Keywords: scaffold, drug delivery, nanofibrous, aromatic
Introduction
An autonomous, ordered organization of materials leading to the formation of specific pattern or structure can be termed as self-assembly. At nanoscale level, the molecular self-assembly depicts the information encoded in individual objects such as shape, size charge, etc.1 Molecular self-assembly utilizes noncovalent interactions such as ionic, hydrogen, hydrophobic, van der Waals, and coordination bonds to a greater extent. These contribute to the inherent features of the materials, which can be fine tuned by altering the interactions.
Self-assembling peptide-based biomaterials were able to uphold their application in various fields such as tissue engineering, drug delivery, etc, in recent times.2–4 They put forward an attractive platform for designing and synthesizing biomaterials consisting of hierarchical nanoarchitectures. Versatile chemical functionalities available through side chain of amino acids, in combination with each other, at specific conditions, open up new possibilities for the construction of nanomaterials with ultimate desired structure and chemical properties. The molecular structure, available charged groups, and concentration of peptide lead to the effective formation of self-assembling peptide nanofibrous scaffold (SAPNS).5,6 Tailoring the amino acid sequence, concentration of the peptide nanofiber, the effective loading and controlled release of drugs from the SAPNS can be achieved successfully. Depending upon the specific binding affinities between aromatic groups, the aromatic ring containing scaffold would serve as an ideal release system for the incorporated functional biomolecules.7,8 Increasing therapeutic efficacy of such controlled drug delivery system (CDDS) can be achieved by incorporating suitable peptide architecture along with proper drug molecule.
Bioinspired and de novo designed self-assembling peptides have been reported in literature earlier.9–12 RADA-16 [CH3-CO-RADARADARADARADA-CO-NH2] is one of the SAPNS, which was proved to be effective in variety of applications such as encapsulation of proteins,13 growth factors,14,15 wound healing,16 scaffold for neural stem cell differentiation,17 in vivo bone regeneration, etc.18 Like a value addition to scientific traditional knowledge of RADA-16, we explored the property of pH-dependent self-assembly and controlled release property of an in silico designed peptide RADA-F6 [CH3-CO-RADARFDARADARADA-CO-NH2] by introducing phenylalanine at sixth position. RADA-F6 displays the property of aromatic π–π interaction between the sequences followed by other noncovalent interactions, which will aid in self-assembly. RADA-F6 was studied further as a CDDS for 5-fluorouracil (5-FU), a major drug used in the treatment of colon cancer. Colon cancer continues to be the major malignancy, which is responsible for higher mortality rate globally.19 The treatment of 5-FU shows several shortcomings such as short biological half-life, poor absorption due to dihydropyrimidine dehydrogenase enzyme, and nonselective action against healthy cells of gastrointestinal tract and bone marrow.20,21 To overcome these limitations, drug needs to be administered in nanoformulations through a biodegradable, biocompatible CDDS.
In the present study, we have designed a sequence RADA-F6 [CH3-CO-RADARFDARADARADA-CO-NH2], which exhibits the property of forming SAPNS. The introduction of phenylalanine leads to effective π–π stacking.7 The self-assembling behavior, pH stability, and 5-FU release from RADA-F6 were analyzed by molecular dynamics (MD) studies taking RADA-16 as control. As 5-FU contains the aromatic pyrimidine ring,22 RADA-F6 system is suitable for entrapping an aromatic drug and shows better controlled release. This is the first report explaining physicochemical characterization and biological evaluation studies of a self-assembling peptide for the release of 5-FU as an anticancer therapeutic. The drug-entrapped hydrogel can be given in oral route through pH-sensitive polymer-coated capsules.23
Materials and methods
Synthesis of peptide
RADA-F6 peptide was synthesized manually through solid-phase peptide synthesis. Fmoc Rink linker was utilized in order to get the peptide C-terminal as amide (Figure S1).
Preparation of hydrogel
RADA-F6 peptide (5 mg/mL) was dissolved in Milli-Q water and Tris–HCl buffer of pH 7.4 and observed for gelation behavior. After placing at room temperature for few minutes, a well-defined hydrogel was formed via the supramolecular self-assembling of the peptide (Figure 1). The 5-FU-loaded RADA-F6 was prepared by dissolving 5-FU in buffer during the formation of SAPNS. For in vitro cytotoxicity experiments, different concentrations of 5-FU-loaded RADA-F6 were prepared from a stock solution of 5-FU (5 mg/mL of Tris–HCl buffer, pH 7.4) in a serial dilution manner (peptide:5-FU; 20:1). Similarly, 5-FU-loaded RADA-16 hydrogel was prepared to compare the entrapment efficiency. Fluorescein isothiocyanate (FITC) (1% wt/v)-loaded RADA-F6 hydrogel was also prepared for cellular uptake experiments.
TEM analysis
Morphological studies of RADA-F6 hydrogel were monitored by a transmission electron microscope (TEM) (JEOL, Tokyo, Japan). Five microliters of peptide hydrogel was taken on a copper-coated formvar microscopy grid and pictures were taken using a Digital Micrograph and Soft Imaging Viewer software.
Fluorescence spectroscopy
Measurements of fluorescence were carried out using a FP-777 spectrofluorometer (JASCO International, Tokyo, Japan). Analysis was done at ambient temperature using a quartz cell with a 10 mm path length. In this study, 8-anilino-1-naphthalenesulfonic acid (ANS) was used to investigate the self-assembly peptide fibers as reported in literature.24 Emission spectra of ANS in aqueous solutions of RADA-F6 at different concentrations were recorded and excited at λex=356 nm.
Circular dichroism measurement
Circular dichroism (CD) spectra were obtained using a JASCO J-820K spectropolarimeter. Wavelength scans were recorded at 0.5 nm intervals from 260 to 190 nm, using a 0.1 cm path length quartz cuvette.25
Molecular dynamics simulation studies
To understand how acidic, neutral, and basic pH affects peptides conformation and to have a bird view of 5-FU release, we employed molecular dynamics simulations. Initial structure of the peptide was build using an external tool such as the PyMol builder. The chains were positioned manually in the vicinity of the nearby chain to form a bilayer sheet with four chains in each layer. Karlsberg+26 web interface was used to protonate all the titratable amino acid residues in order to have effective pKa values at different pH. MD simulations were conducted using GROMACS 4.5.4.27 After protonation, the peptides were solvated with the four-point transferable intermolecular potential charged molecules in a box of dimension (5.712×5.352×3.802 nm). Na and Cl were added at a physiological concentration of 0.15 M to provide charge neutrality. For each pH systems, initially energy was minimized with steepest descent method for 1,000 steps. After minimization, a 50 ps of MD simulation was performed with position restraints on the peptide to allow the relaxation of the solvent molecules.
During MD simulations, the peptide and rest of the system were coupled separately to the temperature bath. The temperature was maintained close to the intended values (300 K) by weak coupling to an external temperature bath using a coupling time τT=0.1 ps. The pressure was kept constant at P0=1 by weak coupling to a bath of constant pressure with a coupling time τP=0.5 ps.28 Simulations were performed under constant pressure and temperature, using periodic boundary conditions and particle mesh Ewald electrostatics with short range interactions cutoff of 0.9 nm. LINCS algorithm29 was used to constrain all bond lengths. For the water molecules, the SETTLE algorithm30 was used. A relative dielectric permittivity, εr=1, and a time step of 2 fs were used. A twin-range cutoff was used for the calculation of nonbonded interactions. The short-range cutoff radius was set to 0.8 nm and the long-range cutoff radius to 1.4 nm for both Coulombic and Lennard-Jones interactions. No reaction field corrections beyond the long-range cutoff were included in the cutoff simulations. Interactions within the short-range cutoff were updated every time step, whereas interactions within the long-range cutoff were updated every five time steps together with the pair-list. All atoms were given an initial velocity obtained from a Maxwellian distribution at the desired initial temperature. MD simulations were run for 1 ns long in case of blank RADA-F6 and RADA-16. Then 5-FU-entrapped system of RADA-F6 and RADA-16 was simulated for 200 ns. Four molecules of 5-FU were kept inside the scaffold to monitor for controlled release.
In vitro drug release studies
In vitro release kinetics study of the drug was performed to assess the release behavior and stability of the peptide hydrogel in different pH conditions. Freshly prepared phosphate-buffered saline (PBS) of two different pH values, namely acidic (pH 2) and basic (pH 7.8), was opted for the experiment. Drug-loaded peptide hydrogel was taken in a dialysis membrane (molecular weight cutoff 500 kDa) and subjected to shaking at 50 rpm on a rotary shaker at 37°C. One milliliter of the reaction mixture was withdrawn at different time intervals. The amount of 5-FU released was estimated using a UV–Visible Spectrophotometer (Perkin-Elmer, Lambda 25, Waltham, MA, USA) at an absorbance wavelength of 266 nm.
Cell culture
HCT-116 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in RPMI supplemented with 10% FBS at 37°C under humidified atmosphere containing 5% carbon dioxide. Until reaching 70% confluence in tissue culture flasks, the cells were trypsinized with PBS solution containing 0.25% trypsin and 0.03% ethylenediaminetetraacetic acid (EDTA). The cells were seeded into culture plate as desired and allowed to attach for 24 hours. RADA-F6 hydrogel containing 5-FU in cell culture medium was then diluted to specific concentrations and added into the cells. After incubation for predetermined durations, cells were subsequently analyzed for cellular uptake, cytotoxicity, apoptosis assay, and cell cycle analysis.
Cell uptake studies
To study cellular uptake, HCT-116 colon cancer cells were cultured in a 12-well plate containing a sterile glass cover slip in complete growth media at a density of 8×103 cells for 24 hours. FITC-tagged RADA-F6 was added to the culture media at a concentration of 75 μM. After 3 hours and 45 minutes of incubation at 37°C, the cells were counterstained with Hoechst for 15 minutes. After PBS washing, the plasma membrane of cells was stained with Cell Mask™ (Thermo Fisher Scientific, Waltham, MA, USA) at 1 μM concentration for 2–4 minutes. Then they were washed twice with PBS and fixed with 4% paraformaldehyde for 30 minutes at room temperature. The cells were again washed twice with PBS and mounted using Fluoromount®. The slides were analyzed under a confocal laser scanning microscope (Nikon Eclipse Ti (AIR), Tokyo, Japan) at a wavelength of 405, 488, and 640 nm to view the blue, green, and red fluorescence, respectively. Images were captured at a magnification of 63× and a scan speed of 400 Hz.
Cytotoxicity assay
MTT reduction assay was performed to assess cell viability. Briefly, cells were seeded (5×103 cells/well) in a 96-well culture plate and incubated for 24 hours. Cells were treated with formulations of 5-FU (10–100 μM) for 24 and 48 hours. After the required duration, media were aspirated and 100 μL fresh media was added. Ten microliters (10% v/v) of MTT (5 mg/mL in PBS) was added to each well, and the plates were further incubated for 4 hours at 37°C. The assay determines cell viability based on the ability of mitochondrial dehydrogenase enzyme present in the metabolically active cells to convert water soluble yellow compound MTT into water-insoluble purple formazan crystals. Four hours after incubation, the medium was completely removed and crystals formed were dissolved in a 100 μL isopropyl alcohol. Therefore, the color yield is proportional to the number of metabolically active cells. The optical density was measured at 570 nm. Percentage of cell inhibition was calculated by the formula: [Absorbance of control – Absorbance of test]/[Absorbance of control] ×100. Control cells are those cells treated with media alone.
Western blot analysis
Cells (1×106) were seeded in 60 mm culture dishes and treated with 5-FU (50 μM) for 24 hours. Cells were lysed and the total protein content was measured using Bradford’s reagent. Sixty micrograms of total protein was loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblotting was carried out using antibodies specific for poly(ADP-ribose) polymerase (PARP) and detected using the enhanced chemiluminescence method.
Cell cycle analysis
For flow cytometric analysis, 1×106 cells were seeded in 60 mm culture dishes for 24 hours. 5-FU treatment (50 μM) was given for 24 hours. Trypsinized cell suspensions and floated cells were fixed in 70% ethanol. The samples were then placed at 4°C until cell cycle analysis. RNA was removed by RNAase treatment and stained with propidium iodide, and samples were analyzed for DNA content using a laser flow cytometer (Becton Dickinson FACS ARIA I; BD Biosciences, San Jose, CA, USA) system and results were analyzed using a DIVA software.
Statistics
All the measurements were done in triplicate, and results are expressed as arithmetic mean ± standard error of the mean.
Results
Preparation of self-assembled nanofibrous hydrogel of RADA-F6
Molecular weight of the peptide RADA-F6 (Figure S2) was determined by MALDI TOF and purity was checked by high-performance liquid chromatography (HPLC) (Figure S3). The experimental value (1,789.2) agreed very well with the calculated value (1,789.2). Hydrogel formation was observed by adjusting the pH of Tris–HCl buffer at 7.4. Self-supporting elastic hydrogel at different conditions consisting of interconnecting fibrillar network was visualized in Figure 1. The loading of guest molecules (5-FU, FITC) was not found to alter the property of gelation of the synthesized peptide hydrogel. Three-dimensional (3D) structure of RADA-F6 also shows the protrusion of the phenyl ring outside the single RADA-F6 molecule, which was assumed to play a role in loading of aromatic guest molecules inside the nanofibrous matrix (Figure 1). Optimization of the gelation behavior with 5-FU further lead to the confirmation that SAPNS of RADA-F6 can be used for drug delivery applications.
Nanostructure of the hydrogels
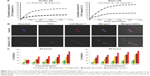
Interior nanofibrillar morphology leading to the scaffold formation was vividly visualized using TEM. Images of self-assembling hydrogel with and without 5-FU of RADA-F6 and RADA-16 were shown in Figure 2A(a–d). But π stacking from F residue contributes some dense network in RADA-F6 compared with RADA-16. Both consist of crosslinked network of nanostructures, suggesting that the obtained hydrogel was constructed by interconnecting nanofibers. The nanofibers organize in a form of self-supporting, supramolecular, and stable scaffolds. The intensity of the nanofibers was more with 5-FU loading compared to the free RADA-F6 at pH 7.4. The individual nanofibers formed were of 5–10 nm thickness and 50–100 nm in length initially and leads to micrometers in size. After complete gelation, it is observed that the hydrogel matrix consists of dense fibrillar network of peptide, which leads to the formation of self-supporting hydrogel.
ANS fluorescent probe was used for the study, to ascertain the amphiphilic nature leading to the self-assembly of the peptide. Fluorescence spectra of RADA-F6 nanofibers in ANS solutions exhibited an intense peak at 479 nm, which showed blue shift compared to spectrum of ANS solution, and the intensity was also increased. ANS shows emission when binds to a self-assembled peptide moiety at a hydrophobic region as described in literature. It is also evident from the spectrofluorimetric analysis that intensity of fluorescence is increased with increase in concentration. It can therefore speculate that specific hydrophobic pockets are essential for an ordered supramolecular self-assembly, which arises due to phenylalanine in the peptide sequence7 (Figure 2Ba).
CD measurements revealed the formation of a well-arranged antiparallel beta sheet of self-assembled peptides through the peaks of 196 nm and 215 nm (Figure 3Aa). The beta sheet arrangement was disruptive at higher basic pH namely 12.5, whereas it was stable at other two pH conditions. This substantiates the use of RADA-F6 as an efficient DDS at physiological pH. But the drug-entrapped system was not showing any significant change in the CD pattern. So it can be stated that the drug entrapment does not cause any significant change in the geometry of the peptide and no reaction takes place between 5-FU and RADA-F6 (Figure 3Ab).
Elucidation of mechanism of self-assembly and controlled release in silico
MD simulation studies were able to alleviate in elucidating the mechanism of self-assembly process of the peptides. The newly formed system RADA-F6 at basic pH is at a lower energy level of ~10 kJ/mol from acidic pH (pH 2) conditions and hence higher stability as illustrated in Figure 3B. At basic pH (pH 7.8), RADA-F6 system was quite stable compared to acidic and neutral environment. The introduction of phenylalanine leads to the formation of a stable system which is having a significant energy difference of ~200 kJ/mol with that of RADA-16 at three different pH conditions (Figures 3B and S4). The presence of phenylalanine in RADA-F6 leads to the formation of well-ordered network of peptides with an additive π–π interaction. This is an important observation in terms of the stability of the designed system in experimental condition.
Solvent accessible surface area (SASA) of the peptide was measured. At basic pH, it was low up to 500 ps and found to be increasing, an added advantage for better controlled release property at basic pH (Figures 3B and S4). The earlier observation substantiates the fact that drug can be effectively loaded inside the peptide scaffold and a sustained release pattern of the drug can be observed from the nanofibrous scaffold. The orientation of the scaffolds at different pH and time interval suggests less hindered stable geometry at basic pH condition (Figure S5).
Further, the sustained release of 5-FU from RADA-F6 was monitored from the simulation data. From Figure 4A, after 200 ns, the 5-FU molecules were started to diffuse from the scaffold matrix of RADA-F6. Of the four 5-FU molecules placed inside the scaffold, three come to the surface of the peptide scaffold (Figure 4A). But in RADA-16, the initial drug molecules are still buried inside the scaffold. The energy variation graph during the simulation time was illustrated in Figure 4B. This says that RADA-F6 shows an energy difference of 13 kJ/mol at the starting point of simulation and at the end difference reaches up to 30 kJ/mol from the control. The difference still expected to increase as time passes by looking at the nature of the graph. The earlier observations suggest that RADA-F6 is a suitable candidate for controlled delivery of 5-FU. The Phe-5-FU interaction is sufficient to hold the drug in the peptide matrix and to deliver in a sustained manner. As controlled release occurs, the DDS attain stability during the course of time. A schematic representation of the 5-FU loading is shown in Figure S6. These observations were further confirmed by in vitro data of drug release.
Effective loading of 5-FU inside peptide nanofibrous scaffold of RADA-F6
In situ preparation of 5-FU-loaded RADA-F6 was accomplished by 5-FU encapsulation within the hydrogel network along with peptide self-assembly. 5-FU is soluble in water and organic solvents such as DMSO, ethanol, methanol, or acetone. We used water to solubilize 5-FU. The drug-entrapped peptide hydrogels were further used for cell studies. The drug entrapment efficiency was 94.34%±0.86% for RADA-F6, whereas for RADA-16 it was 78.57%±1.2%, which indicates the effective loading of 5-FU inside the nanofibrous RADA-F6 is due to Phe-5-FU interaction.
To assess the release pattern of 5-FU from RADA-F6, in vitro kinetics studies were performed and compared to RADA-16. Major aim of the in vitro drug release studies was to illustrate the pH-sensitive drug release properties of the peptide hydrogel as given in Figure 5A. In acidic pH, low concentration of drug was released supporting the fact that the stability of the RADA-F6 was considerable for the controlled release. The drug release from RADA-F6 increased as the pH of the buffer was increased. Kinetics studies clearly indicated that the hydrogel matrix of RADA-F6 shows pH-sensitive drug release behavior than that of RADA-16. At pH 7.8, it was observed that the peptide hydrogel showed sustained release pattern over a period of time (Figure 5A).
Investigation of nanofiber uptake
The cellular uptake of RADA-F6 in colon cancer cell line HCT-116 was studied using FITC-tagged RADA-F6, and results showed that the nanofibrous RADA-F6 was internalized as early as 4 hours of incubation. The confocal microscopic images depict that the FITC-loaded peptides (Green) were localized in the cytoplasm of the cell, which can be visualized clearly by counterstaining it by plasma membrane stain (Cell Mask™-Red) and nuclear stain (Hoechst-Blue) (Figure 5B).
Significant cytotoxicity of the 5-FU-loaded RADA-F6
MTT assay revealed that 5-FU-entrapped RADA-F6 exhibited higher cytotoxicity than free 5-FU and 5-FU-entrapped RADA-16 over a range of experimental concentrations against tumor cell line (Figure 5C). After 24 hours, 23% inhibition of HCT-116 cells was observed when the cells were treated with 25 μM of 5-FU-entrapped RADA-F6, but free 5-FU inhibits 13% and control showed 14.6%. After 48 hours of treatment with free 5-FU, a cytotoxicity of 31.1% was observed whereas the 5-FU-entrapped RADA-F6 formulation gave an inhibition of 60.8%. The significant amount of cytotoxicity of 5-FU-entrapped RADA-F6 hydrogel after 24 hours was observed for all the concentrations. This is a crucial observation with regard to a CDDS. From the dose–response curve, the IC50 value after 48 hours was less than 10 μM for RADA-F6 with 5-FU, whereas for free 5-FU it was 65.96 μM. Unmodified peptide RADA-16 with 5-FU showed 49.92 μM as IC50 value (after 48 hours) and blank peptide showed negligible amount of inhibition, which confirms the biocompatibility of RADA-F6.
Western blotting to detect apoptosis
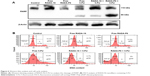
To understand the mechanism in induction of cell death, PARP cleavage assay was carried out. Immunoblotting was performed to detect the cleavage of PARP (116 kDa), a DNA repairing protein. PARP enzyme is involved in catalyzing poly(ADP-ribosyl)ation of a variety of nuclear proteins such as lamins, histones, topoisomerases, and DNA polymerases with nicotinamide adenine dinucleotide as substrate.31 PARP activated by binding to DNA strand breaks, and was subsequently cleaved by caspase-3 into 89- and 24-kDa fragments that contain active site and the DNA-binding domain of the enzyme, respectively, during drug-induced apoptosis. This cleavage inactivates the enzyme by destroying its ability to respond to DNA strand breaks. Immunoblot analysis demonstrated the cleavage of PARP with significant amount after 24 hours of treatment with 5-FU-entrapped RADA-F6 over 5-FU-entrapped RADA-16 and free 5-FU. Since PARP cleavage is a hallmark of apoptosis, the result clearly shows the efficiency of 5-FU-entrapped RADA-F6 to induce apoptosis-mediated cell death (Figure 6A).
Effect on cell cycle progression in HCT-116 cells
The influence of 5-FU-entrapped RADA-F6 on cell cycle was analyzed by subjecting the nanoparticle-treated cells to cell cycle analysis by flow cytometry. Sub-G1 accumulation of cells after 24 hours of treatment with RADA-F6+5-FU indicates apoptosis. Here, we observed no cell cycle arrest in RADA-16 and RADA-F6. 5-FU alone demonstrated S-phase cell cycle arrest but upon entrapment with RADA-F6 enhanced sub-G1 arrest (28.2%) than RADA-16 as carrier (20.3%). This depicts the superior apoptotic activation of RADA-F6 with drug entrapment (Figure 6B). This kind of differential growth inhibition was described in different human colorectal carcinoma cell lines earlier.32
Discussion
Ease of drug loading, high loading capacity, biocompatibility, reversible character, and biodegradability makes self-assembling peptide-based nanostructures as an asset in the field of drug delivery.1,33,34 Unlike the polymer-based DDS, degradation products consist of polymer fragments with heterogeneous chain lengths which may present complex toxicity profiles. These peptide-based DDS give rise to amino acids and drug on degradation. The convenient design and synthesis, ease of drug loading, and felicitous physical and biological evaluation make RADA-F6 an admirable system for the pH-sensitive delivery of 5-FU. Unlike other aromatic amino acids (Tyr, Trp, and His), phenylalanine proves to be an ideal choice due to its charge and structure which retains the beta sheet geometry of RADA-16. Incorporation of phenylalanine at suitable position enhances the property of self-assembling behavior and is illustrated in silico and in vitro.
The peptide RADA-F6 showed considerable amount of gelation with the formation of nanofibrous scaffolds as demonstrated in TEM analysis. The initial nanofibers formed intertwine with each other in a sequential manner leading to the formation of well-ordered nanostructures (Figure 2Aa, b). Spectrofluorimetric analysis of RADA-F6 shows the formation of hydrophobic pockets inside SAPNS by blue shift in fluorescence. The presence of polar amino acids such as arginine and aspartic acid imparts hydrophilicity to DDS. Such an amphiphilicity is a desired character for the DDS to be effective.35–38 The inherent beta sheet geometry of the peptide was unaltered by the modification in sequence and drug loading, as observed in CD spectra. Superior amount of 5-FU loading inside the peptide matrix facilitated the use of RADA-F6 as an ideal tool for 5-FU delivery. The propensity about self-assembly, pH sensitivity, 5-FU loading, and sustained release from RADA-F6 was clarified by MD simulation studies. Further, the pH-sensitive drug release profile of 5-FU-entrapped RADA-F6 was able to substantiate the sustained release pattern of 5-FU (Figure 5A).
Our biological evaluation vividly depicts the efficacy of drug-entrapped peptide for anticancer activity. FITC-tagged peptide nanofibers were readily internalized by the HCT-116 cells as seen in the confocal laser scanning microscope analysis. The biocompatibility of RADA-F6 can be witnessed by MTT assay (Figure 5C). The significant observation of high amount of cytotoxicity by the drug-loaded peptides was essential in case of an apt DDS. The drug-entrapped RADA-F6 imparts higher amount of cytotoxicity compared to the control in a dose- and time-dependent manner. Induction of cytotoxicity by apoptosis is highly desirable for DDS. Apoptosis is a process of an essential tissue homeostasis, and regarded as the ideal way to inhibit cancer cell growth.39 The induction of apoptosis by RADA-F6+5-FU from Western blotting analysis can be viewed as a promising result for the cancer treatment through peptide-based drug delivery systems. The flow cytometric analysis confirms the mechanism of action of 5-FU at specific phases of cell cycle. The earlier observations strongly substantiate our hypothesis of RADA-F6 as an ideal DDS to 5-FU.
Conclusion
Combined physical and biological evaluation of RADA-F6 peptide suggests that it can be successfully used as a pH-responsive drug delivery system for 5-FU. The synthesized peptide showed spontaneous self-assembly providing specific pH conditions. The need of a biocompatible pH-responsive drug delivery system for 5-FU is inevitable for the treatment of disease like colon cancer. RADA-F6 has its own significance due to considerable stability, pH-dependent self-assembling nature, and as a CDDS for 5-FU. Higher stability of the peptide was noticeable from the energy graph and controlled release was evident from SASA data and MD simulation analysis. The antiparallel beta sheet peptide showed ergonomic appearance in TEM analysis with marked amphiphilicity. The controlled release of 5-FU at basic pH and higher cytotoxicity by the induction of apoptosis of drug-entrapped RADA-F6 was an added advantage to use it as an effective biocompatible pH-sensitive drug delivery system. Hence, we are proposing RADA-F6 as an oral DDS through pH-sensitive polymer-coated capsules for colon cancer treatment.
Acknowledgments
The authors are thankful to the Department of Biotechnology, New Delhi for providing the financial assistance for the research and the Council for Scientific and Industrial Research (CSIR), New Delhi for the Junior Research Fellowship to N Ashwanikumar.
Disclosure
The authors report no conflicts of interest in this work.
References
Busseron E, Ruff Y, Moulin E, Giuseppone N. Supramolecular self-assemblies as functional nanomaterials. Nanoscale. 2013;5(16):7098–7140. | ||
Zandén C, Hellström Erkenstam N, Padel T, Wittgenstein J, Liu J, Kuhn HG. Stem cell responses to plasma surface modified electrospun polyurethane scaffolds. Nanomedicine. 2014;10(5):949–958. | ||
Loo Y, Zhang S, Hauser CA. From short peptides to nanofibers to macromolecular assemblies in biomedicine. Biotechnol Adv. 2012;30(3):593–603. | ||
Wang H, Yang Z. Short-peptide-based molecular hydrogels: novel gelation strategies and applications for tissue engineering and drug delivery. Nanoscale. 2012;4(17):5259–5267. | ||
Nagai Y, Unsworth LD, Koutsopoulos S, Zhang S. Slow release of molecules in self-assembling peptide nanofiber scaffold. J Control Release. 2006;115(1):18–25. | ||
Aravinda S, Shamala N, Das C, Sriranjini A, Karle IL, Balaram P. Aromatic-aromatic interactions in crystal structures of helical peptide scaffolds containing projecting phenylalanine residues. J Am Chem Soc. 2003;125(18):5308–5315. | ||
Zhao Y, Tanaka M, Kinoshita T, Higuchi M, Tan T. Nanofibrous scaffold from self-assembly of β-sheet peptides containing phenylalanine for controlled release. J Control Release. 2010;142(3):354–360. | ||
Zhang L, Zhong J, Huang L, Wang L, Hong Y, Sha Y. Parallel-oriented fibrogenesis of a β-sheet forming peptide on supported lipid bilayers. J Phys Chem B. 2008;112(30):8950–8954. | ||
Hoffman AS. Hydrogels for biomedical applications. Adv Drug Deliv Rev. 2002;54(1):3–12. | ||
Rajagopal K, Schneider JP. Self-assembling peptides and proteins for nanotechnological applications. Curr Opin Str Biol. 2004;14(4):480–486. | ||
Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc. 2002;124(5):15030–15037. | ||
Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci U S A. 1993;90(8):3334–3338. | ||
Koutsopoulos S, Unsworth LD, Nagai Y, Zhang S. Controlled release of functional proteins through designer self-assembling peptide nanofiber hydrogel scaffold. Proc Natl Acad Sci U S A. 2009;106(12):4623–4628. | ||
Davis ME, Hsieh PC, Takahashi T, et al. Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc Natl Acad Sci U S A. 2006;103(21):8155–8160. | ||
Davis ME, Motion JM, Narmoneva DA, et al. Injectable self-assembling peptide nanofibers create intramyocardial microenvironments for endothelial cells. Circulation. 2005;111(4):442–450. | ||
Ellis-Behnke RG, Liang Y-X, Tay DK, et al. Nano hemostat solution: immediate hemostasis at the nanoscale. Nanomedicine. 2006;2(4):207–215. | ||
Cheng T-Y, Chen M-H, Chang W-H, Huang M-Y, Wang T-W. Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials. 2013;34(8):2005–2016. | ||
Misawa H, Kobayashi N, Soto-Gutierrez A, et al. PuraMatrix facilitates bone regeneration in bone defects of calvaria in mice. Cell Transplant. 2006;15(10):903–910. | ||
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Fata F, Ron IG, Kemeny N, O’Reilly E, Klimstra D, Kelsen DP. 5-Fluorouracil-induced small bowel toxicity in patients with colorectal carcinoma. Cancer. 1999;86(7):1129–1134. | ||
Di Paolo A, Danesi R, Falcone A, et al. Relationship between 5-fluorouracil disposition, toxicity and dihydropyrimidine dehydrogenase activity in cancer patients. Annal Oncol. 2001;12(9):1301–1306. | ||
Cysewski P. An ab initio study on nucleic acid bases aromaticities. J Mol Struct Theochem. 2005;714(1):29–34. | ||
Cole ET, Scott RA, Connor AL, et al. Enteric coated HPMC capsules designed to achieve intestinal targeting. Int J Pharm. 2002;231(1):83–95. | ||
Jayakumar R, Murugesan M, Asokan C, Aulice Scibioh M. Self-assembly of a peptide Boc-(Ile) 5-OMe in chloroform and N, N-dimethylformamide. Langmuir. 2000;16(4):1489–1496. | ||
Zhao Y, Yokoi H, Tanaka M, Kinoshita T, Tan T. Self-assembled pH-responsive hydrogels composed of the RATEA16 peptide. Biomacromolecules. 2008;9(6):1511–1518. | ||
Kieseritzky G, Knapp EW. Optimizing pKa computation in proteins with pH adapted conformations. Proteins Struct Funct Bioinf. 2008;71(3):1335–1348. | ||
Lindahl E, Hess B, Van Der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7(8):306–317. | ||
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. | ||
Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. | ||
Miyamoto S, Kollman PA. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–962. | ||
Naegeli H, Loetscher P, Althaus F. Poly ADP-ribosylation of proteins. Processivity of a post-translational modification. J Biol Chem. 1989;264(24):14382–14385. | ||
Tokunaga E, Oda S, Fukushima M, Maehara Y, Sugimachi K. Differential growth inhibition by 5-fluorouracil in human colorectal carcinoma cell lines. Eur J Cancer. 2000;36(15):1998–2006. | ||
Altunbas A, Lee SJ, Rajasekaran SA, Schneider JP, Pochan DJ. Encapsulation of curcumin in self-assembling peptide hydrogels as injectable drug delivery vehicles. Biomaterials. 2011;32(25):5906–5914. | ||
Xu X-D, Liang L, Chen C-S, et al. Peptide hydrogel as an intraocular drug delivery system for inhibition of postoperative scarring formation. ACS Appl Mater Interfaces. 2010;2(9):2663–2671. | ||
Elgersma RC, Meijneke T, Posthuma G, Rijkers DTS, Liskamp RMJ. Self assembly of amylin (20–29) amide-bond derivatives into helical ribbons and peptide nanotubes rather than fibrils. Chem Eur J. 2006;12(14):3714–3725. | ||
She W, Luo K, Zhang C, et al. The potential of self-assembled, pH-responsive nanoparticles of mPEGylated peptide dendron–doxorubicin conjugates for cancer therapy. Biomaterials. 2013;34(5):1613–1623. | ||
Mart RJ, Osborne RD, Stevens MM, Ulijn RV. Peptide-based stimuli-responsive biomaterials. Soft Matter. 2006;2(10):822–835. | ||
Ashwanikumar N, Kumar NA, Nair SA, Kumar GSV. Methacrylic-based nanogels for the pH-sensitive delivery of 5-fluorouracil in the colon. Int J Nanomed. 2011;7:5769–5779. | ||
Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.