Back to Journals » Cancer Management and Research » Volume 13
RUNX2 as a promising therapeutic target for malignant tumors
Authors Zhao W, Yang H, Chai J , Xing L
Received 15 January 2021
Accepted for publication 27 February 2021
Published 16 March 2021 Volume 2021:13 Pages 2539—2548
DOI https://doi.org/10.2147/CMAR.S302173
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Weizhu Zhao,1,2 Haiying Yang,3 Jie Chai,4 Ligang Xing1
1Department of Radiology, Cancer Hospital Affiliated to Shandong First Medical University, Shandong Cancer Hospital and Institute, Jinan, 250117, People’s Republic of China; 2Department of Oncology, Binzhou People’s Hospital, Binzhou, 256610, People’s Republic of China; 3Department of Nursing, Binzhou People’s Hospital, Binzhou, 256610, People’s Republic of China; 4Department of Gastrointestinal Surgery, Cancer Hospital Affiliated to Shandong First Medical University, Shandong Cancer Hospital and Institute, Jinan, 250117, People’s Republic of China
Correspondence: Jie Chai
Department of Gastrointestinal Surgery, Cancer Hospital Affiliated to Shandong First Medical University, Shandong Cancer Hospital and Institute, No. 440, Jiyan Road, Jinan, 250117, People’s Republic of China
Tel +86 18678867800
Email [email protected]
Ligang Xing
Department of Radiology, Cancer Hospital Affiliated to Shandong First Medical University, Shandong Cancer Hospital and Institute, No. 440 Jiyan Road, Jinan, 250117, People’s Republic of China
Tel +86 15553181309
Email [email protected]
Abstract: The transcription factor runt-related protein 2 (RUNX2) has an important impact on the transformation of bone marrow mesenchymal stem cells to osteoblasts. Further studies have shown that RUNX2 plays a key role in the invasion and metastasis of cancers. RUNX2 is a “key” molecule in the regulatory network comprised of multiple signaling pathways upstream and its target downstream molecules. Due to the complex regulatory mechanisms of RUNX2, the specific mechanism underlying the occurrence, development and prognosis of malignant tumors has not been fully understood. Currently, RUNX2 as a promising therapeutic target for cancers has become a research hotspot. Herein, we reviewed the current literature on the modulatory functions and mechanisms of RUNX2 in the development of malignant tumors, aiming to explore its potential clinical application in the diagnosis, prognosis and treatment of tumors.
Keywords: RUNX2, malignant tumor;, research progress
Introduction
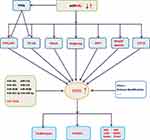
In mammals, RUNX represents a family of three transcription factors, which share a common DNA binding domain-Runt domain, homologous to the Drosophila Runt gene. RUNX family members (including RUNX1, RUNX2 and RUNX3) combine with core-binding factor β to form heterodimers that enhance their ability to bind to DNA, and participate in subsequent transcriptional regulation.1,2 The transcription factor RUNX2 is closely related to the differentiation of human osteoblasts and the maturation of chondrocytes by regulating multiple signaling pathways and transcriptional activation of a series of downstream molecules.3,4 (figure 1, figure 2, figure 3, table 1Current researches have confirmed that RUNX2 is closely related to the proliferation, invasion and bone metastasis of multiple cancers such as osteosarcoma, breast cancer (BC), prostate cancer, gastric cancer and colorectal cancer. This article summarizes the research progress of RUNX2 in malignant tumors, which focuses on the involvement of signaling pathways miRNAs regulations, histone modification and so on.
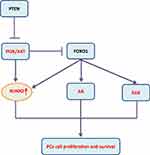 |
Figure 2 PI3K/AKT-dominated signaling pathway regulates RUNX2 in PCa. |
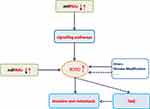 |
Figure 3 Simulation diagram of RUNX2ʹ s regulation mechanism in malignant tumors. The dotted line indicates that further research is required. |
 |
Table 1 Regulation of RUNX2 Expression by miRNAs and lncRNAs in Osteosarcoma |
The Transcription Factor RUNX2 and Malignant Tumors
RUNX2 and Breast Cancer
BC is one of the common malignant tumors in women. RUNX2 plays a vital role in BC development. RUNX2 and estradiol have opposite effects on BC.5 Previous studies have found that estrogen can trigger BC progression in situ, while presents anti-metastatic properties. Its mode of action is opposite to RUNX2. Overexpression of RUNX2 in BC cell line MCF7 induces epithelial-mesenchymal transition (EMT), which relies on signaling pathways of TGF β and Wnt.6 The high nuclear expression level of RUNX2 is related to the state of human epidermal growth factor receptor type 2 (HER2) in BC cells, and the poor prognosis is correlated with high RUNX2 expression level and negative HER2 expression in BC patients.7
Micro ribonucleic acid (miRNAs) can control gene expressions at the post-transcriptional level, and act as oncogene or tumor suppressor in BC. miRNAs directly8 or indirectly affect the expression of RUNX2 in BC cells by controlling different signaling pathways, such as PI3K/AKT, NF-κB, Wnt/β-catenin, TGF-β, BMP, Notch and Hedgehog (Figure 1).9–17 miRNAs can regulate these signaling pathways by targeting their molecular signaling components. Reduced expression levels of tumor-suppressive miRNAs and increased expression levels of oncogenic miRNAs can activate the signal transduction pathways, subsequently up-regulating the expression level of RUNX2. The enhanced expression level of RUNX2 stimulates the expressions of metastatic marker genes such as vascular endothelial growth factor (VEGF), metalloproteinase 2 (MMP-2), osteopontin (OPN), bisulfite sequencing PCR (BSP), parathyroid hormone-related peptide (PTHrP) and receptor activator of nuclear factor κB ligand (RANKL), thus promoting bone metastasis, increasing BC cell proliferation, and ultimately leading to poor prognosis of BC.18
With the increase of whole-genome acetylation level, histone deacetylase inhibitor (HDACi) promotes delocalization of bromodomain-containing protein 4 (BRD4) from active Enhancers (ENHs) to other sites and reduces the expression of highly expressed genes; JQ1 inhibits RUNX2 by blocking the recruitment of BRD4 to the RUNX2 promoter and ENHs; It is also speculated that HDACi and bromodomain and extraterminal inhibitors (BETi) synergistically inhibit RUNX2 and other cancer driver genes, supporting the rational combined use of these drugs in cancer treatment.19,20
RUNX2 has an important impact on osteogenesis and BC-mediated bone metastasis. It also plays a critical role in osteolytic lesions. BC cells with a lack of RUNX2 expression suppress osteoblasts differentiation and increase osteoclast differentiation.21 RANKL expression is positively correlated with connective tissue growth factor (CTGF) in BC tissues and the expression levels of RANKL and CTGF are higher in bone metastasis tissues than in other sites. CTGF also promotes the recruitment of RUNX2 to the RANKL promoter, thereby increasing the production of RANKL in tumor cells, and subsequently stimulating osteoclastogenesis.22 In BC cells, RUNX2 is implicated in an adhesion-dependent mechanism of bone tropism and bone colonization, which is a potential target for predicting and treating bone metastasis of BC.23 In the process of BC metastasis, cancer cells promote survival under nutrient starvation by autophagy. In addition, RUNX2 promotes autophagy through α-tubulin acetylation and autophagic vesicle transport. Therefore, the levels of LC3B and RUNX2 can be used to predict metastasis of Bethesda categories (BCs).24
The mechanism of RUNX2 underlying BC is relatively thorough, which provides reference for the study of other malignant tumors.
RUNX2 and Osteosarcoma
Osteosarcoma is a malignant bone tumor, which occurs more commonly in adolescents or children under the age of 20. RUNX2 has the potential to regulate osteoblast differentiation and carcinogenesis, and it exerts an important impact on the development and progression of osteosarcoma. RUNX2 is overexpressed in human osteosarcoma tissues, especially in tumors that respond poorly to chemotherapy.25 Increasing number of studies have confirmed that miRNAs and long non-coding RNAs (lncRNAs) are aberrantly expressed in osteosarcoma and can directly or indirectly participate in the regulation of RUNX2 expression. The potential diagnostic and therapeutic values of the differentially expressed miRNAs and lncRNAs by targeting RUNX2 are extremely important in the clinic (Table 1).26–34 For example, miR-150 is suggested as the therapeutic target of osteosarcoma due to its anti-tumor function in promoting chemotherapy sensitivity and inhibiting tumor cell proliferation via RUNX2. In details, the results of luciferase reporter assay demonstrated that RUNX2 is a direct target of miR-150 and miR-150-RUNX2 axis affects the chemical sensitivity of osteosarcoma cells by regulating the expression levels of apoptosis proteins such as increasing the expression levels of cleaved caspase-3 and cleaved caspase-8 while reducing the expression levels of cleaved caspase-3 and cleaved caspase-8.27 With the deepening of research, the regulatory signaling pathways, potential targets and corresponding drugs related to RUNX2 are emerging research hotspot against osteosarcoma.35–37
RUNX2 and Prostate Cancer
Prostate cancer (PCa) is a heterogeneous disease at both the genetic and clinical levels. At present, it is divided into different genotypes and treatment methods were applied based on a large amount of clinical data. The molecular mechanism remains to be clarified.38 Through RUNX2 immunohistochemical staining analysis on PCa tissues, it was found that patients with the positive RUNX2 staining had higher PSA levels, higher Gleason grades, and stronger metastatic ability than the negative staining ones.39 Overexpression of RUNX2 is related to the up-regulation of matrix metalloproteinases, bone resorption factors and the enhanced metastasis of PCa cells to bone.40 The increased RUNX2 protein level is related to the decrease of phosphatase and tensin homolog (PTEN) protein expression. The expression of RUNX2 is down-regulated in PCa cells by interacting with Forkhead Box O1 (FOXO1). Correspondingly, FOXO1/PTEN expression is decreased in patients with bone metastasis, while RUNX2 expression is increased, suggesting the potential value of FOXO1 in the metastasis of PCa to bone.41 Further studies have confirmed that PTEN deletion promotes the activation of the AKT-RUNX2-OCN-GPRC6A-CREB signal axis and further induces the expressions of cytochrome P450 family 1 subfamily A1 (CYP1A1) and CYP17A1 in PCa cells and intratumoral androgen synthesis (IAS).42 An important target of the PI3K/AKT signaling pathway is FOXO1 protein that can be phosphorylated directly by AKT leading to translocation of FOXO1 from the cytoplasm to the nucleus. This not only impairs FOXO1 activities on transactivation of downstream target genes, but also abolishes its transcriptional activity-independent inhibitory effect on other targets such as androgen receptor (AR), extracellular regulated protein kinases (ERK) and RUNX2 (Figure 2). Additionally, miR-466 inhibits tumor growth and bone metastasis in PCa by direct regulation of RUNX2 expression.43
Furthermore, RUNX2 S319 phosphorylation plays an important role in the development of PCa; P-S319-Runx2 is a marker for more aggressive metastatic disease in a patient population, which has an exclusively nuclear localization and is regulated by both RAS/MAPK and PI3K/AKT signaling pathways.44 Therefore, based on the conventional signaling pathways research, the research of histone modification function of RUNX2 should be paid more attention, and it may be used as a potential target for the diagnosis and treatment of prostate cancer.
RUNX2 and Colorectal Cancer
Colorectal cancer (CRC) is a common malignant tumor in the digestive system. Most CRC-related deaths are attributed to liver metastases. Researches have shown that RUNX2 is closely related to Duke staging, liver metastasis and ERβ, and is an independent factor in the prognosis of colon cancer patients.45 OPN in CT26 CRC cells is regulated by RUNX2 and ETS-1. Therefore, inhibiting these transcription factors could result in a significant down-regulation of the osteopontin transfer proteins.46 RUNX2, OPN and MMP-7 are highly expressed in CC531 colon cancer cells metastases explanted from the liver, while the expressions are reduced and/or disappeared in cell culture in vitro. The opposite expression profiles of Hoxc8, OPN and RUNX2 indicate that these genes may be regulated in a feedback loop way. Transforming Growth Factor β-1 (TGFβ-1) induces the overexpression of OPN and RUNX2 in hepatocytes, but does not show the same effect on hepatocytes co-cultured with CC531 cells.47 Small nucleolar RNA host gene 3 (SNHG3) binding to miR-539 up-regulates RUNX2 expression, which promotes the growth and metastasis of CRC. SNHG3 may be a potential target for CRC treatment.48 RNA-metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) binding to miR-15 family members affects the expression of LRP6, thereby enhancing β-catenin signaling and leading to an increase in the transcription level of the downstream target gene RUNX2. Moreover, MALAT1 can also bind to the splicing factor proline/glutamine-rich (SFPQ) protein, contributing to dissociation of the SFPQ/polypyrimidine tract binding protein-2 (PTBP2) dimer, the release of PTBP2 and the increase of RUNX2 expression by interplaying with the IRES domain of the corresponding mRNA 5ʹUTR region.49
Our previous studies indicate that miR-455 inhibits the progression of CRC via RAF. Plasmacytoma variant translocation 1 (PVT1) silencing can inhibit the progression of CRC by miR-455 in vivo. RUNX2 increases the expression level of PVT1 in CRC, while miR-455 inhibits RUNX2 expression, which forms a feedback loop between RUNX2/PVT1/miR-455. In conclusion, the RUNX2/PVT1/miR-455/RAF-1 axis has been considered a potential target for CRC treatment.50,51
RUNX2 and Gastric Cancer
Gastric cancer (GC) remains to be one of the world’s leading malignant tumors of the digestive system. Half of the new affected cases occurred in East Asia, including China, Japan and South Korea. RUNX2 in GC tissue is related to the degree of differentiation, depth of invasion, and lymph node metastasis. RUNX2 is one of the independent prognostic factors for patients with GC. RUNX2 enhances transcription and promotes the progression of GC by directly binding to CXCR4.52 In GC patients, RUNX2 plays a regulatory role by interacting with lncRNAs under certain conditions. The expression levels of lncRNA EPEL and RUNX2 are up-regulated in GC. Overexpression of lncRNA EPEL leads to the up-regulation of RUNX2 expression, while overexpression of RUNX2 does not affect EPEL expression. Thence, lncRNA EPEL may be regulated by the interaction with RUNX2.53 MiR-539 as a tumor suppressor inhibits GC progression by targeting RUNX2.54 JQ1 can inhibit the progression of GC by down-regulating chromatin accessibility and inactivating the RUNX2/Nidogen 1 (NID1) signaling pathway. In addition, NID1 may be a new therapeutic target for GC.55
RUNX2 and Lung Cancer
Non-small cell lung cancer (NSCLC) accounts for nearly 80% of lung cancers. The expression level of RUNX2 in NSCLC is significantly correlated with tumor size, stage and lymph node metastasis. RUNX2 is an independent risk factor in NSCLC.56 Besides, the co-expression network of RUNX2 in lung squamous cell carcinoma (LUSC) reveals that the complex interactions between RUNX2 and 45 co-expressed genes, which are involved in extracellular matrix-receptor interactions, local adhesion, protein digestion and absorption, human papillomavirus infection and PI3K/AKT signaling pathway, etc.57 In lung adenocarcinoma, RUNX2 transcriptionally regulated its potential target genes are involved in cell cycle-related proteins and MAPK signaling pathways, providing a new perspective for targeted drug development and drug resistance research.58 WW domain-containing oxidoreductase (WWOX) is a tumor suppressor gene.59 Overexpression of WWOX in highly aggressive H1299 lung cancer cells suppresses cell motility and invasiveness, and inhibits the expressions of RUNX2 and its target gene MMP-9.60 BMP-2 induces lung cancer migration through increasing the activation of ERK and p38 and up-regulating Runx2 and Snail expressions. ERK-RUNX2-Snail and p38-RUNX2-Snail can induce lung cancer cell migration and EMT. It is also shown that RUNX2 can increase the expression of Snail to regulate the early metastatic events of lung cancer.61 In addition, miR-218 and lncRNA H19 directly or indirectly regulate RUNX2 in lung cancer.62,63
RUNX2 and Other Cancers
Malignant melanoma is highly aggressive and resistant to chemotherapy. RUNX2 is overexpressed in melanoma cells, and its expression is related to the process of proliferation and migration.64 The RUNT domain in RUNX2 is responsible for the proliferation and migration of melanoma.65 The RUNT domain increases the angiogenic properties of melanoma cells. Proteomic analysis allows us to point out that the RUNT domain is involved in the process of neovascularization.66 RUNT domain promotes bone metastasis of melanoma cells through the complex interactions with genes involved in bone remodeling through the ERK/p-ERK and AKT/p-AKT pathways.67 All these findings indicate that the RUNT domain is involved in melanoma metastasis and cell migration. RUNX2 can reactivate the MAPK and PI3K/AKT pathways, thereby endowing melanoma cells with high metastatic potential.68
In patients with thyroid cancer, the expression level of RUNX2 was higher in patients with microcalcification, compared with those without microcalcification.69 RUNX2 promotes bone homing and bone metastasis by interacting with its target genes such as Stromal Sell-Derived Factor 1 (SDF1), CXCR7 and BSP.70 Thyroid hormone receptor β, (TRβ) modulating RUNX2 expression is a signal axis shared by thyroid cancer and BC. TRβ directly interacts with the proximal promoter of RUNX2 through the thyroid hormone response element, resulting in the reduced RUNX2 promoter activity.71 As a cis-regulatory element, RAIN promotes carcinogenic characteristics in thyroid and BC cells, promoting the expression of RUNX2 through two patterns, including binding to WD repeat domain 5 (WDR5) and facilitating its positioning on RUNX2 promoter; and changing the transcription status of the RUNX2 locus and promoting transcription initiation. RAIN acts as a bait for the negative elongation factor (NELF) complex to inhibit its inhibitory effect on transcription elongation.72 Ectopic expression of miR-218 inhibits the development of papillary thyroid cancer by targeting RUNX2 to inactivate the PTEN/PI3K/AKT pathway.73
Transcription Factor RUNX2 and Cancer Stem Cells in Some Cancers
The theory of cancer stem cells (CSCs) suggests that there is a group of “stem” cell populations with self-renewal, multidirectional differentiation, multi-drug resistance to radiotherapy and chemotherapy, and invasion and metastasis.74 CSCs were first found in hematological malignant tumors, and then many CSCs or tumor stem cell-like cells have been identified and isolated from solid tumors, such as BC, PCa, colon cancer, etc.75–77 CSCs are usually hidden in the cancer nest, in a static state, in which DNA replication is not active, cells can escape DNA damage induced by chemotherapeutic drugs, enhance the ability to repair, and maintain the stable inheritance of genes. CD44 is one of the most commonly used markers for identifying CSCs. Essentially, cell-cell adhesion proteins have an important impact on tumor invasion and metastasis.78 These biological characteristics of CSCs are controlled by complex intracellular and extracellular regulatory networks. RUNX2 is closely related to the biological behavior of malignant tumors, and tumor stem cells are the root causes of the malignant behavior of tumors. There are a few related studies. BMP-2 can induce bone formation and restrain CSCs in the human osteosarcoma OS99-1 cell line, accompanied by the accumulation of RUNX2 and Osx.79 The function of RUNX2 begins with breast stem cells that differentiated into progenitor cells which develop into luminal and basal breast lineages. RUNX2 can promote the activity of CD44+/CD24−/low BC stem cells and regulate the malignant phenotype of BC.80 The prostate cancer stem cells are characterized by high expression of CD49f and RUNX2, low expression of CD44, CD133 and Androgen Receptor.81
Outlook
In recent years, RUNX2 as a specific transcription factor in malignant tumors has attracted much attention, and its related molecular pathways have been put forward as research hotspots. Current research mainly focuses on its upstream signaling pathways and the regulation of miRNAs, and the researches of its downstream mechanisms are relatively few (Figure 3). Its research on and histone modifications has also begun to take shape and has excellent research value. At the same time, combined with the regulatory characteristics of the RUNX family in malignant tumors, both RUNX1 and RUNX3 can regulate the differentiation and activity of immune cells. Combined with rare reports in previous studies, we believe that RUNX2 has a unique mechanism in regulating the tumor microenvironment (TME), which can not only reshape the microenvironment also regulate the activity of immune cells, leading to tumor immune escape. With the in-depth study of RUNX2-related regulatory mechanisms, RUNX2 is expected to become a new therapeutic target and contribute to the development of new drugs and the improvement of clinical efficacy.
Abbreviations
RUNX2, transcription factor runt-related protein 2; BC, Breast cancer; HER2, human epidermal growth factor receptor type 2; EMT, epithelial-mesenchymal transition; VEGF, metastatic marker genes vascular endothelial growth factor; MMP-2, metalloproteinase 2; OPN, osteopontin; BSP, bisulfite sequencing PCR,; PTHrP, parathyroid hormone-related peptide; RANKL, receptor activator of nuclear factor κB ligand; miRNAs, micro ribonucleic acid; HDACi, histone deacetylase inhibitor; BRD4, bromodomain-containing protein 4; ENHs, Enhancers; BETi, bromodomain and extraterminal inhibitors; CTGF, connective tissue growth factor; BCs, Bethesda categories; lncRNAs, long non-coding RNAs; Pca, Prostate cancer; PTEN, phosphatase and tensin homolog; FOXO1, Forkhead Box O1; CYP1A1, cytochrome P450 family 1 subfamily A1; IAS, intratumoral androgen synthesis; AR, androgen receptor; ERK, extracellular regulated protein kinases; CRC, Colorectal cancer; TGFβ-1, Transforming Growth Factor β-1; SNHG3, small nucleolar RNA host gene 3, MALAT1, RNA-metastasis-associated lung adenocarcinoma transcript 1; SFPQ, splicing factor proline/glutamine rich; PTBP2, polypyrimidine tract binding protein-2; PVT1, Plasmacytoma variant translocation 1; NID1, Nidogen 1; EMT, epithelial-mesenchymal transition; WWOX, WW domain-containing oxidoreductase; NSCLC, Non-small cell lung cancer; LUSC, lung squamous cell carcinoma; SDF1, Stromal Sell-Derived Factor 1; TRβ, Thyroid hormone receptor β; WDR5, WD repeat domain 5; NELF, negative elongation factor; CSCs, cancer stem cells; TME, tumor microenvironment.
Consent for Publication
All authors have agreed to the publication of this manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Key Research and Development Project of Shandong Province of China (NO.2019JZZY011008).
Disclosure
The authors declare no conflicts of interest for this work and that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
1. Blyth K, Vaillant F, Jenkins A, et al. Runx2 in normal tissues and cancer cells: a developing story. Blood Cells Mol Dis. 2010;45(2):117–123. doi:10.1016/j.bcmd.2010.05.007
2. Chimge N, Frenkel BJ. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32(17):2121–2130. doi:10.1038/onc.2012.328
3. Hill T, Später D, Taketo M, et al. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8(5):727–738. doi:10.1016/j.devcel.2005.02.013
4. Zhang Y, Yasui N, Ito K, et al. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A. 2000;97(19):10549–10554. doi:10.1073/pnas.180309597
5. Ferrari N, McDonald L, Morris J, et al. RUNX2 in mammary gland development and breast cancer. J Cell Physiol. 2013;228(6):1137–1142. doi:10.1002/jcp.24285
6. Chimge N, Baniwal S, Luo J, et al. Opposing effects of Runx2 and estradiol on breast cancer cell proliferation: in vitro identification of reciprocally regulated gene signature related to clinical letrozole responsiveness. Clin Cancer Res. 2012;18(3):901–911. doi:10.1158/1078-0432.Ccr-11-1530
7. Onodera Y, Miki Y, Suzuki T, et al. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Sci. 2010;101(12):2670–2675. doi:10.1111/j.1349-7006.2010.01742.x
8. Iorio M, Ferracin M, Liu C, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi:10.1158/0008-5472.Can-05-1783
9. Miao Y, Zheng W, Li N, et al. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci Rep. 2017;7:41942. doi:10.1038/srep41942
10. Wang F, Li L, Chen Z, et al. MicroRNA-214 acts as a potential oncogene in breast cancer by targeting the PTEN-PI3K/Akt signaling pathway. Int J Mol Med. 2016;37(5):1421–1428. doi:10.3892/ijmm.2016.2518
11. Tong L, Yuan Y, Wu S. Therapeutic microRNAs targeting the NF-kappa B signaling circuits of cancers. Adv Drug Deliv Rev. 2015;81:1–15. doi:10.1016/j.addr.2014.09.004
12. Sweeney K, Cameron E, Blyth KJM, et al. Complex interplay between the RUNX transcription factors and wnt/β-catenin pathway in cancer: a tango in the night. Mol Cells. 2020;43(2):188–197. doi:10.14348/molcells.2019.0310
13. Han M, Wang F, Gu Y, et al. MicroRNA-21 induces breast cancer cell invasion and migration by suppressing smad7 via EGF and TGF-β pathways. Oncol Rep. 2016;35(1):73–80. doi:10.3892/or.2015.4360
14. Taipaleenmäki H, Browne G, Akech J, et al. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75(7):1433–1444. doi:10.1158/0008-5472.Can-14-1026
15. Brabletz S, Bajdak K, Meidhof S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30(4):770–782. doi:10.1038/emboj.2010.349
16. Wang Z, Li Y, Kong D, et al. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010;292(2):141–148. doi:10.1016/j.canlet.2009.11.012
17. Liu X, Zhao T, Bai X, et al. LOC101930370/MiR-1471 axis modulates the hedgehog signaling pathway in breast cancer. Cell Physiol Biochem. 2018;48(3):1139–1150. doi:10.1159/000491980
18. Pranavkrishna S, Sanjeev G, Akshaya R, et al. Regulation of Runx2 and its signaling pathways by microRNAs in breast cancer metastasis. Curr Protein Pept Sci. 2020;21. doi:10.2174/1389203721666201116115337
19. Bhadury J, Nilsson L, Muralidharan S, et al. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc Natl Acad Sci U S A. 2014;111(26):E2721–2730. doi:10.1073/pnas.1406722111
20. Sancisi V, Manzotti G, Gugnoni M, et al. RUNX2 expression in thyroid and breast cancer requires the cooperation of three non-redundant enhancers under the control of BRD4 and c-JUN. Nucleic Acids Res. 2017;45(19):11249–11267. doi:10.1093/nar/gkx802
21. van der Deen M, Akech J, Lapointe D, et al. Genomic promoter occupancy of runt-related transcription factor RUNX2 in Osteosarcoma cells identifies genes involved in cell adhesion and motility. J Biol Chem. 2012;287(7):4503–4517. doi:10.1074/jbc.M111.287771
22. Kim B, Kim H, Jung S, et al. A CTGF-RUNX2-RANKL axis in breast and prostate cancer cells promotes tumor progression in bone. J Bone Miner Res. 2020;35(1):155–166. doi:10.1002/jbmr.3869
23. Li J, Cai S, Peng J, et al. Time dependent distribution of MicroRNA 144 after intravenous delivery. Microrna. 2016;5(1):36–49. doi:10.2174/2211536605666160322152146
24. Tandon M, Othman A, Ashok V, et al. The role of Runx2 in facilitating autophagy in metastatic breast cancer cells. J Cell Physiol. 2018;233(1):559–571. doi:10.1002/jcp.25916
25. Sadikovic B, Thorner P, Chilton-Macneill S, et al. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202. doi:10.1186/1471-2407-10-202
26. Li N, Luo D, Hu X, et al. RUNX2 and Osteosarcoma. Anticancer Agents Med Chem. 2015;15(7):881–887. doi:10.2174/1871520615666150304151228
27. Ling Z, Fan G, Yao D, et al. MicroRNA-150 functions as a tumor suppressor and sensitizes osteosarcoma to doxorubicin-induced apoptosis by targeting RUNX2. Exp Ther Med. 2020;19(1):481–488. doi:10.3892/etm.2019.8231
28. Sheng K, Li YJE, Medicine T. LncRNA TUG1 promotes the development of osteosarcoma through RUNX2. Exp Ther Med. 2019;18(4):3002–3008. doi:10.3892/etm.2019.7880
29. Jia F, Zhang Z, Zhang X. MicroRNA-338-3p inhibits tumor growth and metastasis in osteosarcoma cells by targeting RUNX2/CDK4 and inhibition of MAPK pathway. J Cell Biochem. 2019;120(4):6420–6430. doi:10.1002/jcb.27929
30. Wang W, Luo P, Guo W, et al. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. 2018;503(3):1927–1933. doi:10.1016/j.bbrc.2018.07.137
31. Pan B, Wu L, Pan L, et al. Up-regulation of microRNA-340 promotes osteosarcoma cell apoptosis while suppressing proliferation, migration, and invasion by inactivating the CTNNB1-mediated Notch signaling pathway. Biosci Rep. 2018;38(4). doi:10.1042/bsr20171615
32. Xie Y, Sun W, Deng Z, et al. MiR-302b suppresses osteosarcoma cell migration and invasion by targeting Runx2. Sci Rep. 2017;7(1):13388. doi:10.1038/s41598-017-13353-9
33. Lin W, Zhu X, Yang S, et al. MicroRNA-203 inhibits proliferation and invasion, and promotes apoptosis of osteosarcoma cells by targeting Runt-related transcription factor 2. Biomed Pharmacother. 2017;91:1075–1084. doi:10.1016/j.biopha.2017.05.034
34. Zhang C, Long F, Wan J, et al. MicroRNA-205 acts as a tumor suppressor in osteosarcoma via targeting RUNX2. Oncol Rep. 2016;35(6):3275–3284. doi:10.3892/or.2016.4700
35. Alegre F, Ormonde A, Godinez D, et al. The interaction between RUNX2 and core binding factor beta as a potential therapeutic target in canine osteosarcoma. Vet Comp Oncol. 2020;18(1):52–63. doi:10.1111/vco.12526
36. Wang X, Qin G, Liang X, et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat Commun. 2020;11(1):1141. doi:10.1038/s41467-020-14870-4
37. Montazeri-Najafabady N, Dabbaghmanesh M, Chatrabnous N, et al. The effects of astaxanthin on proliferation and differentiation of MG-63 osteosarcoma cells via Aryl Hydrocarbon Receptor (AhR) pathway: a comparison with AhR endogenous ligand. Nutr Cancer. 2020;72(8):1400–1410. doi:10.1080/01635581.2019.1679199
38. Robinson D, Van Allen E, Wu Y, et al. Integrative clinical genomics of advanced prostate cancer. Cell Physiol Biochem. 2015;161(5):1215–1228. doi:10.1016/j.cell.2015.05.001
39. Akech J, Wixted J, Bedard K, et al. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29(6):811–821. doi:10.1038/onc.2009.389
40. Senbanjo L, AlJohani H, AlQranei M, et al. Identification of sequence-specific interactions of the CD44-intracellular domain with RUNX2 in the transcription of matrix metalloprotease-9 in human prostate cancer cells. Cancer Drug Resist. 2020;3(3):586–602. doi:10.20517/cdr.2020.21
41. Zhang H, Pan Y, Zheng L, et al. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer Res. 2011;71(9):3257–3267. doi:10.1158/0008-5472.Can-10-2603
42. Yang Y, Bai Y, He Y, et al. PTEN loss promotes intratumoral androgen synthesis and tumor microenvironment remodeling via aberrant activation of RUNX2 in castration-resistant prostate cancer. Clin Cancer Res. 2018;24(4):834–846. doi:10.1158/1078-0432.Ccr-17-2006
43. Colden M, Dar A, Saini S, et al. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis. 2017;8(1):e2572. doi:10.1038/cddis.2017.15
44. Ge C, Zhao G, Li Y, et al. Role of Runx2 phosphorylation in prostate cancer and association with metastatic disease. Oncogene. 2016;35(3):366–376. doi:10.1038/onc.2015.91
45. Sase T, Suzuki T, Miura K, et al. Runt-related transcription factor 2 in human colon carcinoma: a potent prognostic factor associated with estrogen receptor. Int J Cancer. 2012;131(10):2284–2293. doi:10.1002/ijc.27525
46. Wai P, Mi Z, Gao C, et al. Ets-1 and runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells. J Biol Chem. 2006;281(28):18973–18982. doi:10.1074/jbc.M511962200
47. Georges R, Adwan H, Zhivkova M, et al. Regulation of osteopontin and related proteins in rat CC531 colorectal cancer cells. Int J Oncol. 2010;37(2):249–256. doi:10.3892/ijo_00000672
48. Dacheng W, Songhe L, Weidong J, et al. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother. 2020;125:110039. doi:10.1016/j.biopha.2020.110039
49. Ji Q, Cai G, Liu X, et al. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019;10(6):378. doi:10.1038/s41419-019-1598-x
50. Chai J, Guo D, Ma W, et al. A feedback loop consisting of RUNX2/LncRNA-PVT1/miR-455 is involved in the progression of colorectal cancer. Am J Cancer Res. 2018;8(3):538–550.
51. Chai J, Wang S, Han D, et al. MicroRNA-455 inhibits proliferation and invasion of colorectal cancer by targeting RAF proto-oncogene serine/threonine-protein kinase. Tumour Biol. 2015;36(2):1313–1321. doi:10.1007/s13277-014-2766-3
52. Guo Z, Yang L, Qian F, et al. Transcription factor RUNX2 up-regulates chemokine receptor CXCR4 to promote invasive and metastatic potentials of human gastric cancer. Oncotarget. 2016;7(15):20999–21012. doi:10.18632/oncotarget.8236
53. Fu J, Zhao W, Guo D, et al. LncRNA E2F-mediated cell proliferation enhancing lncRNA regulates cancer cell behaviors and affects prognosis of gastric cancer. Dig Dis Sci. 2020;65(5):1348–1354. doi:10.1007/s10620-019-05855-5
54. Jin W, Han H, Liu D. Downregulation miR-539 is associated with poor prognosis of gastric cancer patients and aggressive progression of gastric cancer cells. Cancer Biomark. 2019;26(2):183–191. doi:10.3233/cbm-190384
55. Zhou S, Zhang S, Wang L, et al. BET protein inhibitor JQ1 downregulates chromatin accessibility and suppresses metastasis of gastric cancer via inactivating RUNX2/NID1 signaling. Oncogenesis. 2020;9(3):33. doi:10.1038/s41389-020-0218-z
56. Li H, Zhou R, Zhang G, et al. Clinical significance of RUNX2 expression in patients with nonsmall cell lung cancer: a 5-year follow-up study. Tumour Biol. 2013;34(3):1807–1812. doi:10.1007/s13277-013-0720-4
57. Yang D, Lu H, Chen G, et al. Integrated expression analysis revealed RUNX2 upregulation in lung squamous cell carcinoma tissues. IET Syst Biol. 2020;14(5):252–260. doi:10.1049/iet-syb.2020.0063
58. Yang D, Huang W, Chen G, et al. Clinical significance of transcription factor RUNX2 in lung adenocarcinoma and its latent transcriptional regulating mechanism. Comput Biol Chem. 2020;89:107383. doi:10.1016/j.compbiolchem.2020.107383
59. Bednarek A, Laflin K, Daniel R, et al. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res Treat. 2000;60(8):2140–2145. doi:10.1016/S0165-4608(99)00201-0
60. Zheng Q, Zhou Y, You Q, et al. WWOX inhibits the invasion of lung cancer cells by downregulating RUNX2. Cancer Gene Ther. 2016;23(12):433–438. doi:10.1038/cgt.2016.59
61. Hsu Y, Huang M, Yang C, et al. Lung tumor-associated osteoblast-derived bone morphogenetic protein-2 increased epithelial-to-mesenchymal transition of cancer by Runx2/Snail signaling pathway. J Biol Chem. 2011;286(43):37335–37346. doi:10.1074/jbc.M111.256156
62. Zhou M, Zhang L, Song M, et al. MicroRNA-218 prevents lung injury in sepsis by inhibiting RUNX2. Eur Rev Med Pharmacol Sci. 2018;22(23):8438–8446. doi:10.26355/eurrev_201812_16543
63. Wu L, Sun L, Hua Y, et al. Overexpression of long non-coding RNA H19 protects lung fibroblasts from LPS-induced injury by targeting miR-181a and Runx2 via activation of Notch and JNK pathways. J Cell Biochem. 2018. doi:10.1002/jcb.26660
64. Perduca M, Dalle Carbonare L, Bovi M, et al. Runx2 downregulation, migration and proliferation inhibition in melanoma cells treated with BEL β-trefoil. Oncol Rep. 2017;37(4):2209–2214. doi:10.3892/or.2017.5493
65. Deiana M, Dalle Carbonare L, Serena M, et al. New insights into the runt domain of RUNX2 in melanoma cell proliferation and migration. Cells. 2018;7(11):220. doi:10.3390/cells7110220
66. Cecconi D, Brandi J, Manfredi M, et al. Runx2 stimulates neoangiogenesis through the Runt domain in melanoma. Sci Rep. 2019;9(1):8052. doi:10.1038/s41598-019-44552-1
67. Deiana M, Dalle Carbonare L, Serena M, et al. A potential role of RUNX2- RUNT domain in modulating the expression of genes involved in bone metastases: an in vitro study with melanoma cells. Cells. 2020;9(3):751. doi:10.3390/cells9030751
68. Valenti M, Dalle Carbonare L, Mottes MJ. RUNX2Ectopic expression of the osteogenic master gene in melanoma. World J Stem Cells. 2018;10(7):78–81. doi:10.4252/wjsc.v10.i7.78
69. Dalle Carbonare L, Frigo A, Francia G, et al. Runx2 mRNA expression in the tissue, serum, and circulating non-hematopoietic cells of patients with thyroid cancer. J Clin Endocrinol Metab. 2012;97(7):E1249–1256. doi:10.1210/jc.2011-2624
70. Baniwal S, Khalid O, Gabet Y, et al. Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer. 2010;9:258. doi:10.1186/1476-4598-9-258
71. Bolf E, Gillis N, Barnum M, et al. The thyroid hormone receptor-RUNX2 axis: a novel tumor suppressive pathway in breast cancer. Horm Cancer. 2020;11(1):34–41. doi:10.1007/s12672-019-00373-2
72. Rossi T, Pistoni M, Sancisi V, et al. RAIN is a novel enhancer-associated lncRNA that controls RUNX2 expression and promotes breast and thyroid cancer. Mol Cancer Res. 2020;18(1):140–152. doi:10.1158/1541-7786.Mcr-19-0564
73. Han M, Chen L, Wang YJO, et al. miR-218 overexpression suppresses tumorigenesis of papillary thyroid cancer via inactivation of PTEN/PI3K/AKT pathway by targeting Runx2. Onco Targets Ther. 2018;11:6305–6316. doi:10.2147/ott.S172152
74. Akbari-Birgani S, Paranjothy T, Zuse A, et al. Cancer stem cells, cancer-initiating cells and methods for their detection. Drug Discovery Today. 2016;21(5):836–842. doi:10.1016/j.drudis.2016.03.004
75. Clouston H, Rees P, Lamb R, et al. Effect of tissue factor on colorectal cancer stem cells. Anticancer Res. 2018;38(5):2635–2642. doi:10.21873/anticanres.12505
76. Pindiprolu S, Krishnamurthy P. Chintamaneni PJN-Ssaop. Pharmacological targets of breast cancer stem cells: a review. Naunyn Schmiedebergs Arch Pharmacol. 2018;391(5):463–479. doi:10.1007/s00210-018-1479-3
77. Skvortsov S, Skvortsova I, Tang D, et al. Concise Review: prostate Cancer Stem Cells: current Understanding. Stem Cells. 2018;36(10):1457–1474. doi:10.1002/stem.2859
78. Jing F, Kim H, Kim C, et al. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int J Oncol. 2015;46(4):1582–1588. doi:10.3892/ijo.2015.2844
79. Wang L, Park P, La Marca F, et al. Bone formation induced by BMP-2 in human osteosarcoma cells. Int J Oncol. 2013;43(4):1095–1102. doi:10.3892/ijo.2013.2030
80. Zhang L, Liu L, Xu X, et al. miR-205/RunX2 axis negatively regulates CD44/CD24 breast cancer stem cell activity. Am J Cancer Res. 2020;10(6):1871–1887.
81. Liao C, Adisetiyo H, Liang M, et al. Cancer-associated fibroblasts enhance the gland-forming capability of prostate cancer stem cells. Cancer Res. 2010;70(18):7294–7303. doi:10.1158/0008-5472.Can-09-3982
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.