Back to Journals » International Journal of Nanomedicine » Volume 15
Protein–Nanoparticle Interaction: Corona Formation and Conformational Changes in Proteins on Nanoparticles
Authors Park SJ
Received 20 March 2020
Accepted for publication 21 May 2020
Published 6 August 2020 Volume 2020:15 Pages 5783—5802
DOI https://doi.org/10.2147/IJN.S254808
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Thomas Webster
Sung Jean Park
College of Pharmacy and Gachon Institute of Pharmaceutical Sciences, Gachon University, Incheon 21936, Korea
Correspondence: Sung Jean Park
College of Pharmacy and Gachon Institute of Pharmaceutical Sciences, Gachon University, 191 Hambakmoero, Yeonsu-gu, Incheon 21936, Korea
Email [email protected]
Abstract: Nanoparticles (NPs) are highly potent tools for the diagnosis of diseases and specific delivery of therapeutic agents. Their development and application are scientifically and industrially important. The engineering of NPs and the modulation of their in vivo behavior have been extensively studied, and significant achievements have been made in the past decades. However, in vivo applications of NPs are often limited by several difficulties, including inflammatory responses and cellular toxicity, unexpected distribution and clearance from the body, and insufficient delivery to a specific target. These unfavorable phenomena may largely be related to the in vivo protein–NP interaction, termed “protein corona.” The layer of adsorbed proteins on the surface of NPs affects the biological behavior of NPs and changes their functionality, occasionally resulting in loss-of-function or gain-of-function. The formation of a protein corona is an intricate process involving complex kinetics and dynamics between the two interacting entities. Structural changes in corona proteins have been reported in many cases after their adsorption on the surfaces of NPs that strongly influence the functions of NPs. Thus, understanding of the conformational changes and unfolding process of proteins is very important to accelerate the biomedical applications of NPs. Here, we describe several protein corona characteristics and specifically focus on the conformational fluctuations in corona proteins induced by NPs.
Keywords: nanoparticle, protein corona, structure, surface characteristic, conformational change
Introduction
Understanding the interaction between nanoparticles (NPs) and proteins is imperative to the application of NPs as nanomedicines or nanocarriers and to control NP-associated environmental biohazards.1–3 Various types of NPs are currently being used in clinical research.4 Aside from metal and inorganic NPs, different organic NPs have been successfully used in the medical field. The largest economic value-added sectors are magnetic resonance imaging (MRI) and drug delivery, and several NPs have received FDA approval or are currently in clinical trials.5–7
Drug delivery systems are generally designed to control the release and absorption of the carrier drug or to deliver it to a specific part of the body.8,9 Drug delivery systems reduce the side effects of the drug while maximizing its efficacy by delivering effective amounts to the target site for a specific period of time. NPs have been actively studied as effective drug delivery systems.8 They have a large surface area to volume ratio, allowing excellent drug adsorption and controlled drug release kinetics according to the adsorption method.10 NPs have been developed as carriers for the delivery of various molecules such as growth factors, genes, antibacterial agents, anti-inflammatory agents, and antibodies.2,11,12
The use of nanomaterials and carbon nanotubes (CNTs) as drug carriers has increased greatly but their use faces some technical limitations. Nanomaterials may cause immune reactions in vivo and side-effects such as inflammation.13,14 Toxicity is reported after their in vivo administration and long-term retention.15 Further, problems such as biological signal disturbances caused by their unexpected interactions and binding with various biomolecules such as proteins in vivo have been reported. Therefore, to use these particles as general-purpose drug carriers, the biophysical and biochemical characteristics of the starting material must be investigated.16
In addition to their use as small drug carriers, NPs have recently been used as immunomodulators, enzyme inhibitors, and transporters of therapeutic proteins. These new applications are associated with the protein–nanoparticle interaction, a phenomenon termed as “protein corona.” The layer of proteins adsorbed on NPs functions as a biological module in vivo and is difficult to understand at the level of a single protein. The process underlying corona formation is complicated and involves complex kinetics and dynamics. Thus, the in vivo behavior of NPs relies on protein corona formation, which defines the biological activity or toxicity of NPs. Protein corona formation is highly diverse depending on the sources and/or production methods of NP systems, including metals, metal oxides, polymers, composites, and CNTs. For example, gold nanoparticles affected albumin in the circulating system,17 while those produced by enzymatic method showed binding to cellular RNAs and proteins.18 Casein-coated gold NP can translocate across the blood-brain barrier (BBB), resulting in corona formation with Abeta42.19 Magnesium oxide (MgO) NPs spontaneously form an albumin complex upon introduction,20 while MgO NPs produced through green synthesis can also interact with cellular proteins such as superoxide dismutase 1 (Sod1) and p53 after internalization into cells.21
Regarding protein corona formation, the structural changes in proteins on NPs should be noted and studied to enhance the in vivo availability of NPs. The correct structure of the protein is necessary for its proper function. Various biological systems, including chaperons, proteasome, autophagy, and ER-associated degradation monitor the quality of the protein structure and tightly regulate the fate of proteins in cells depending on their conformational state.22 Uncontrolled disruption of the folding pathway can lead to disease onset. For example, the unfolding or misfolding of proteins is directly related to amyloid-associated diseases, such as Alzheimer’s disease,23–25 Parkinson’s disease,26,27 type 2 diabetes,28 and familial amyloidosis.29,30 The immunoglobulin light-chain in the blood stream occasionally forms aggregates, accumulates at various sites, and causes a significant problem in the body, which may be due to protein unfolding.31 Thus, the unfolding or structural disruption of corona proteins by NPs could be linked to unexpected abnormalities in vivo. A recent study highlighted that quantum dots composed of CdSe/ZnS could disrupt the secondary structure of insulin and induce aggregation and fibrillation, depending on the specific size and surface charge of NPs.32
To overcome the potential problems of NP application, recent studies have focused on how the protein corona can be used to produce beneficial effects in vivo. Pre-incubation or artificial coating of NPs with a corona protein can enhance the NPs function.33 The designed corona formation with NPs can be helpful for evading the host immunity.34 To obtain successful results with these approaches, a better understanding of the structural changes in corona proteins on NPs may be helpful. In this regard, we initially described the general aspects of protein coronas, including physicochemical factors affecting corona formation, and explored the structural details of corona proteins in various NPs.
Factors Influencing Protein Binding and Corona Formation
The physical characteristics of NPs, such as their size and surface properties, serve as the basic determinants of their biochemical, physiological, and pharmacological applications. The details of the basic properties of NPs affecting corona formation have been well reviewed in other articles.35 Thus we briefly summarized the influential parameters. Protein adsorption and binding are governed by several factors such as the size, shape, surface area, and surface charge of NPs. This binding is dependent on the source of nanomaterials such as metals,36–38 organic polymers,39,40 and CNTs.41,42
Surface Area
In general, NPs have a large surface area, which may be advantageous for their nonspecific interactions with serum proteins.42, 43 Platelet aggregation was recently shown to be dependent on the surface area of NPs and not on NP size.44 Further, the high ratio of the total available surface area of NPs and the protein concentration does not always result in high adsorption and binding of plasma proteins. For instance, silica NPs bound more plasma proteins than polystyrene NPs of the same size in the presence of low concentrations of plasma proteins, but their binding capacity was lower in the presence of high concentrations of plasma proteins. The polystyrene NPs showed the opposite behavior.40 Moreover, the protein species bound to silica and polystyrene NPs were quite different,40 indicating that not only the density but also the species of NP is important for selective protein binding. The solvent-accessible surface area of a protein is a counterpart determinant of selective corona formation. Plasma proteins such as bovine fibrinogen, gamma globulin, and serum albumin were found to be adsorbed onto CNTs through the π-π stacking of the exposed aromatic residues (Trp, Phe, Tyr) of proteins.45 The amount of protein adsorbed onto CNTs was proportional to the exposed surface area of the CNTs.45
Radius of Curvature
The radius of curvature of NPs is another factor that affects the binding and unfolding of proteins.46 The curvature of NPs is directly related to their size and shape. A large radius of curvature results in a planar surface, and small-sized NPs possess a small radius of curvature. The planar surface may provide a wide contact area for protein binding, resulting in stronger interactions between proteins and NPs.47,48 As many proteins are smaller than NPs such as CNTs, a bulk of proteins could be adsorbed and may accumulate on the surface of NPs. The effect of NP size on blood protein binding was well explained in a study using gold NPs;49 the smaller the gold NP size, the lower the amount of human serum albumin (HSA) bound. Consistent with the size effect, the association constant of plasma proteins, as determined by a fluorescence quenching assay, was found to increase with an increase in the size of gold NPs.49 However, high molecular weight proteins, such as immunoglobulin G (150 kDa), have molecular diameters (10–15 nm) comparable to NPs and may bind to small NPs at a 1-to-1 ratio. Figure 1 compares the sizes of several proteins to NPs.50 The binding stoichiometry between proteins and NPs may affect the conformational state and the functionality of the bound protein. Boselli et al used 2–5 nm gold NPs and found that smaller NPs did not form a conventional “protein corona” and that the layers comprised plasma proteins bound around NPs.51 Thus, the specific interaction between NPs and proteins may be regulated by controlling the size of NPs. In terms of the effect of NP size on protein binding, the influence of single-walled and multi-walled carbon nanotubes (SWCNTs and MWCNTs) on corona formation is interesting. Several reports have shown their toxicity to cells and different effects on protein structures. SWCNTs were more toxic to macrophages than MWCNTs.52 Tau protein strongly bound to SWCNTs and underwent significant structural changes, while the interaction between the MWCNT and Tau protein was negligible.53
Shape
The effects of NP shape as well as size, curvature, and surface area on protein binding cannot be excluded.54 The binding affinity of HSA to spherical-shaped gold NPs was three times higher than that of branched-shaped gold NPs of similar size (50–70 nm). Furthermore, the thickness of the protein corona adsorbed on gold NPs differed depending on their shape and was smaller for spherical-shaped gold NPs, as measured from the hydrodynamic radius of the NP and protein complex.55 Small zinc oxide (ZnO) NPs showed different activity for β-galactosidase (GAL) depending on the shape of the NPs, such as pyramids, plates, and spheres. The strongest inhibition was observed with pyramid-shaped ZnO NPs.56 These authors suggested that GAL has a long groove at the active site on the surface and that the pyramidal-shaped NPs could bind well to this region as compared to plates or spheres.
Biofluid
The main driving force for protein corona formation may be the noncovalent, nonspecific, and hydrophobic interactions between proteins and NPs. The hydrophobic aggregation of molecules is largely affected by the solution being examined. Biological systems, especially human body fluids, contain various electrolytes, small metabolites, peptide hormones, lipids, sugars, and nucleotides, all of which may significantly affect the hydrophobic strength of the interaction.40 Thus, protein binding to NPs in biological fluids may be different from that in culture media or buffers.57 Immune activation by the NP–immune protein complex in cellular systems does not always correlate with the immune activation reported under in vivo conditions.58 Gold NPs incubated in Dulbecco’s modified Eagle’s medium showed greater adsorption of proteins than those incubated in Roswell Park Memorial Institute medium.59
Affinity and Exposure Time
Many in vitro studies on the interaction between proteins and NPs have mainly focused on specific protein binding; however, the behavior of NPs in biological systems should be understood with respect to protein corona formation. NPs can aggregate with other biomolecules such as organic compounds, carbohydrates, and lipids. However, the majority of bio-coronas are protein coronas,60,61 which are dynamically formed in vivo and undergo changes in composition over time.1,15,57 Time-dependent corona formation is defined by the Vroman effect,62,63 which explains how fibrinogen at the surface of materials may be replaced by other high-affinity proteins. That is, abundant proteins are nonspecifically adsorbed on the surface of NPs immediately after their in vivo administration. Most of these proteins may have low affinity for NPs and may be released from the complex and replaced by those with higher affinity after a long exposure time.64 This weakly bound layer is called a “soft” corona, while the strongly bound layer is called a “hard” corona.64–66 The composition of soft and hard corona is affected by many factors, such as the surface characteristics, biological environment around NPs, times of exposure, and physiochemical properties of NPs.
Protein corona formation is highly related to the functionality and activity of NPs in either a positive or negative manner.13 Which of the strongly or weakly bound proteins influences the biological functions of NPs is still unclear. Weber et al have suggested the role of hard corona proteins, such as immunoglobulin G and clusterin, in the internalization of NPs into cells.67
Synthetic Methods
Due to ethical and environmental reasons, various production methods of NPs, so called green synthesis, have been introduced in consideration of the environment and human health. In particular, there are many studies on the production of metal and metal oxide nanomaterials.68,69 Interestingly, the different synthetic methods could result in different biocompatibility and toxicity of NPs as well as different binding property to cellular proteins. The green-synthesized MgO NPs showed different mortality, biocompatibility, and toxicity in zebrafish compared to the commercially synthesized MgO NPs: authors suggested that the different toxicity may be related to different protein binding property in the ROS cascade even though the size and charge characteristics of both NP systems were similar.21 The green-synthesized gold NPs also showed reduced toxicity with reduced ROS, resulting in reduced apoptosis.18
Conformational Changes of Proteins in the Protein Corona
NPs have different chemical environments on their surfaces, which drive structural changes in the bound proteins. Physical properties such as size, shape, curvature, and surface area affect the structural characteristics of the bound proteins. The surface characteristics of proteins are important factors that induce structural changes upon binding. Structural changes in corona proteins are biologically meaningful, as the loss-of-function after structural changes may provoke the destruction of physiological homeostasis and unwanted immune responses.70 In addition, bulk conformational changes may trigger protein aggregation and amyloid fiber formation.71,72 Hydrogenated NPs promote β-sheet structures and aggregation through a reduction in zeta potential, which may enhance the collision between intact Aβ40 peptides.73 Hen egg white lysozyme was unfolded by silica NPs, resulting in the formation of β-sheet-rich fiber-like protein aggregates.74 Thus, the knowledge of the functionality of NPs demands an understanding of the conformational changes in the protein corona of each NP system (Figure 2).
Detection Methods
Tertiary structural changes in proteins are difficult to monitor owing to methodological limitations. The most frequently used method is circular dichroism (CD) spectroscopy, which highlights secondary structural changes as well as alterations in protein melting temperatures. For instance, the serum protein fibrinogen was studied by CD, and a reduction in the α-helical content and an increase in the β-strand content of fibrinogen were reported depending on the surface characteristics and density of nanotubes.75 Immunoglobulin G was highly unfolded in MWCNTs, while polyethylene glycol (PEG)-modified nanotubes maintained its structure.76 CD spectroscopy is widely used for corona protein studies, although it suffers from low sensitivity. The combined use of CD with a highly sensitive photothermal detection system was recently described as photothermal CD, which enhanced the sensitivity of CD spectroscopy.77 Dynamic light scattering (DLS) may be used to observe alterations in particle size after protein binding. Through DLS, the thickness and aggregation of the corona on NPs can be probed. The hydrodynamic radii of bovine serum albumin (BSA) and lysozyme were measured with DLS, and the relationship between the aggregation of NPs and the radius was highlighted with anionic silica NPs.78 Electron microscopy methods such as transmission electron microscopy (TEM) and atomic force microscopy (AFM) are useful for obtaining information on a single NP. The size and shape of a single NP with/without protein corona can be determined using these techniques. A study described tau protein aggregation on the surface of titanium NPs, and suggested that the early-stage tau aggregates were unordered and amorphous.79 Nuclear magnetic resonance (NMR) is not commonly used in NP studies, as the molecular weight of NP systems is usually over the detection limitation of NMR. However, the TROSY technique80 was successfully used to quantify GB3, a small immunoglobulin-binding domain from Staphylococcus aureus adsorbed onto various sizes of gold NPs, and to describe the molecular mechanism of the interaction between them.81
Cases
Many studies on the conformational changes in proteins caused by NPs have revealed that α-helices typically decrease in number and/or β-sheet formation typically increases.47,82 However, the same protein may exhibit different changes upon binding to different NPs. Furthermore, the same NPs with different surface characteristics, such as charges or length of aliphatic branches may exert different effects on protein structures. Capomaccio et al revealed secondary structural changes in HSA following binding to gold NPs (~20 nm).83 Native HSA is an α-helical protein and its helicity decreased following interaction with gold NPs with an increase in β-structure. The decrease in helicity was proportional to the increase in the concentration of gold NPs. The molecular dynamic simulation between HSA and gold NPs (large crystalline shape) showed that the domain III (lipid-binding site) of HSA mainly interacted with the NP surface through a loop carrying Lys464, Thr504, Phe505, and Leu581.84 This study also highlighted the significant decrease in α-helicity from 68% to 45%. However, citrate-coated silver NPs failed to induce significant structural changes in HSA;85 the smallest silver NPs (~16 nm) induced only a 4% reduction in α-helicity, while an increased size of ~40 nm resulted in less than 1% reduction in helicity.
Table 1 shows several proteins studied with NPs and their effects on protein structure upon NP binding. The heterogeneous changes in the protein corona can be identified.
 |
Table 1 Representative Conformational Changes in Proteins Following Complexation with NPs |
Cellular Proteins
Besides the corona formation in the circulating system, the corona formation with cellular protein is also related to the toxic effects of NPs (Figure 2). The cellular responses by NPs appear as oxidative stress, inflammatory responses, DNA damage, apoptosis, ER stress, and etc., which were widely studied while the direct complexation of cellular proteins with NPs was less studied. The SWCNTs and MWCNTs bind to adhesive proteins such as extracellular fibronectin probably by the nonspecific manner, which subsequently results in integrin-mediated cell attachment.86 Cai et al showed the over 750 proteins from human HeLa cell lysate could bind MWCNTs by quantitative proteomics:87 among the potential corona proteins, secondary structural characteristics of 269 proteins were analyzed and the α-helical content of those proteins notably seems to be important for complexation with NPs (Figure 3). The skeletal proteins such as actin may be affected by SWCNTs through changing the filament assembly.88 The in silico docking studies described the protein-metal NPs complexes such as the transcription factor Oct4-AgNPs,89 the enzyme Sod1-AgNPs,90 the apoptotic factor p53-AgNPs,90 and the transmembrane lipoprotein VLDLR-titanium oxide NPs.38 These studies showed specific binding of NPs to the pockets in the target proteins without disruption of folded state. The silica NPs (SiO2) might interact with the nucleoplasmic proteins such as topoisomerase I clusters and induce the large aggregates in the cell nucleus.91 Various cellular proteins may be directly affected by the internalized NPs, resulting in numerous disruption of cellular events such as transcription, proliferation, signal transduction, cell cycle regulation, metabolism, apoptosis, and etc. However, structural studies on the interaction between these proteins and NPs at the atomic level are currently limited.
Effects of Surface Modification on Protein Conformation
The surface chemistry of NPs is inevitable for their stabilization and to prevent coagulation and agglomeration. Thus, surface modification of NPs is generally required to maintain them in a well-dispersed colloidal state. Agglomerated NPs exert different effects on protein conformation; i.e., the characteristics of protein coronas of agglomerated NPs are quite different from those of coronas formed on normal NPs.92,93 This observation may be associated with alterations in available surface area, modified exposure of chemical motifs, changes in shape, and loss of charge balance. Studies have shown that agglomeration could be controlled by the formation of protein corona on polystyrene NPs.94,95 In this case, proteins may serve as a protective shield against the accumulating NPs. Thus, it is important to properly design the surface modification of NPs to control protein corona formation and achieve NP stability for clinical applications.
Surface modification can be performed in two major ways, namely, by controlling the hydrophobicity and by controlling the charge property. Polymers such as PEG and poly ethyl ethylene phosphate (PEEP) are widely used to control hydrophobicity.96 NPs coated with polymers have extended half-lives in circulation in vivo.97 PEG has been used to recover the ability of herceptin-conjugated NPs to specifically bind to targeted receptors by “backfilling” the surface of NPs.98 In this system, protein corona formation was suppressed by PEG, resulting in the retention of specific-targeting ability. PEEP-coated polystyrene NPs also showed decreased protein corona formation, but weak and nonspecific binding was still observed at their surfaces.99 The structures of corona proteins on PEG-NPs have not been extensively studied, and whether conformational changes are highly affected by PEG is unclear. A study using different lengths of PEG reported that the effect on the secondary structure of lysozyme was not significant, but fluctuations in melting temperatures were obvious.100 It seems that the contact of PEG-coated gold NPs with lysozyme may occur within a limited range, but further studies are warranted.
The hydrophobic contact between NPs and proteins may induce large structural changes, as the inner hydrophobic core of proteins may be exposed to the polar solvent environment through hydrophobic interactions. However, it is difficult to predict the effect of hydrophobicity of NPs on protein binding and conformational changes. Chakraborti et al suggested that the conformational change in α-lactalbumin, a highly hydrophobic protein, may be mainly attributed to its hydrophobic interaction with zinc NPs.101 Neutral gold NPs were bound by only a few plasma proteins, while charged gold NPs attracted several proteins, including fibrinogen.102 Several model proteins, including BSA, transferrin, and apolipoprotein, showed high binding affinity to neutral hydrophobic polystyrene NPs, but not to charged NPs.103
Surface charge characteristics may highly influence the conformation of bound proteins104 and regulate protein adsorption, binding affinity, and structural changes, all of which may induce alterations in the protein-protein (ligands) interaction and biological functions of NPs. For instance, the positively charged model NPs prepared with polylactide showed two-fold higher uptake in HeLa cells. Interestingly, the positively charged NPs, but not the negatively charged NPs, were internalized via the clathrin pathway, which indicated a different corona protein interaction in the latter case.105 In the case of quantum dot NPs, negatively charged NPs were more efficiently transported into HEK cells via the lipid raft/caveolae pathway but not the clathrin pathway.106 Polystyrene NPs modified with carboxyl (-COOH) and amine (-NH2) groups showed differential cellular uptake. The carboxylated NPs were better internalized by macrophages, while the positively charged NPs preferred monocyte THP-1 cells.107 Positively charged residues such as lysine and arginine of lysozyme may be exposed to the solvent following binding to gold NPs, thereby facilitating NP aggregation owing to the interaction between these exposed residues of lysozyme and other proteins.108 The species of surface charge may determine the protein types bound to NPs. Positively charged polystyrene NPs showed high affinity to albumin, which has an isoelectric point (PI) below 5.5, but negative NPs preferred to form corona with IgG with a PI above 5.5.109 In addition to charged species, charge density may also affect corona formation. The increase in the surface charge density of polystyrene NPs was accompanied by an increase in the adsorbed amount of proteins without any change in protein species.110
To compare the effect of charge on corona formation and conformational change, model proteins with different surface charge properties were monitored in the presence of differently charged CNTs (unpublished data). The selected model proteins were as follows: albumin and α-1-antitrypsin with a negative surface or local negative patch; lysozyme and transferrin that carry a positive surface or local positive patch; and fibrinogen composed of three different subunits (α, β, γ) with different charge properties. The PI values and calculated net charges are listed in Table 2. The surface of the model CNTs was modified into –COOH, –OH, and –NH2. The –OH modification was intended to monitor the effect of the hydrogen bonding donor or acceptor.
 |
Table 2 Structural Changes in Model Proteins on Various Surfaces of Carbon Nanotubes |
The effect on conformational changes would be expected to follow the charge profiles of proteins, with the exception of fibrinogen, as the electrostatic interaction between oppositely charged proteins and NPs may govern this complexation process. However, the order of secondary structure changes in proteins was not consistent with the expectation, while the changes in the Tm values of proteins matched well with the theoretical effect (Table 2). Figures 4 and 5 show the CD spectra for secondary structural changes and Tm value changes. The reference CD spectra of each protein were obtained without CNT (labeled as PBS in the figures) to compare with the spectra of protein-NP complexes. All spectra were measured in phosphate-buffered saline (PBS, pH 7.4). The changes in the secondary structures of the five proteins were analyzed with the corresponding CD analysis software, CDNN,111 and the estimated contents of each secondary structure in the total structure are summarized in Table 3.
 |
Table 3 Secondary Structural Changes Depending on the Surface Characteristics |
Albumin possesses highly negatively charged patches on its surface (Figure 4). Interestingly, the secondary structural changes were most significant in the negatively charged COOH surface, although the difference in changes with other surface-modified CNTs was not large. Generally, the amounts of α-helical content decreased in all NPs, while the amount of β-sheets and random coils increased. The increase in β-sheets or random coils may imply partial unfolding or aggregation of the protein. The surface charge of α-1-antitrypsin was similar to that of albumin (Table 2 and Figure 4), while the most significant changes were unexpectedly found in the OH-modified surface, as shown in Table 3. The decrease in α-helicity and increase in β-sheet and random coil were identified in all proteins. Since the pKa of the terminal α-amino group (–NH2) is known to be around 8.0, the charge strength of the surface –NH2 of CNTs seems to be weak at pH 7.4. This may weaken the interaction between CNT-NH2 and α-1-antitrypsin. Transferrin does not possess a highly charged surface on its structure and shows the similar effect in the CD spectra. All CNTs slightly reduced the α-helicity and slightly elevated the β-sheet content, which may imply that the hydrophobic interactions with the CNTs are important for α-1-antitrypsin. Lysozyme was highly affected by the surface of –COOH due to its positively charged surface. Notably, the change in the Tm value due to the surface of –OH was also significant, as shown in Figure 5, as was the change in the secondary structure. This may suggest that not only the surface charge but also the hydrogen bonding through the surface oxygen of CNTs may affect the conformational change in lysozyme. The largest model protein, fibrinogen, showed a similar pattern of change to those of albumin and lysozyme.
The changes in fibrinogen would be caused by a very complex event since it is composed of three different subunits: α, β, and r. Free fibrinogen without NPs showed two distinct transitions in the Tm curve around 50°C and 60°C (Figure 5). However, the second transition was highly altered with COOH-CNT, and the protein seemed to be less denatured, possibly because each subunit of fibrinogen may be differently affected by NPs.
These results may support the hypothesis that charge profiles of proteins and NPs are highly linked to the conformational changes in proteins. The correlation between charge profiles of proteins and NPs may be reflected by the overall conformation characteristics such as Tm value, while local structural changes (secondary structure) in bound proteins may be difficult to predict based on only charge profiles; i.e., not only surface charge but also the local structural aspect of NPs should be considered to control the secondary structures of corona proteins.
The previous molecular dynamics study showed that the partial denaturation of albumin can be induced by electrostatics and affects the self-assembly of albumin. Through counter-ion association with albumin, hydrophobic aggregation was promoted.112 The partial unfolding of albumin could affect the drug- or toxin-binding properties or amyloid formation. It has been suggested that misfolded albumin contributes to the pathogenesis of aging-related dementia and Alzheimer’s disease.113 α-1 antitrypsin is prevalent in human serum, plays important roles in anti-inflammation, and reduces the damage to the lungs caused by proteases and inflammatory responses.114 The malfunction of α-1antitrypsin can be induced through protein misfolding and could be linked to liver and lung disease. The mutational variant of α-1antitrypsin occasionally partially unfolded with opening of the main β-sheet of the protein, resulting in amyloid-like fiber formation.115 The unfolding of lysozyme is also related to amyloidosis in the body. For the amyloid formation of lysozyme, the structural stability between the folded and the unfolded states was suggested to be an important factor.116 Figure 5 clearly shows that the folding stability was significantly lowered by the various NPs. Forming the transferrin corona with NPs may be beneficial to enhance drug delivery to the brain117 as transferrin-conjugated carbon dots (C-Dots) easily penetrate the BBB through receptor-mediated transport. Interestingly, binding to C-Dots did not induce a large conformational change in transferrin. It was shown that small C-Dots may prevent unfolding of proteins and suppress fibrillation of proteins: the mixture of insulin and C-Dots inhibited the secondary structural changes in insulin and fiber formation.118 In addition, our CD data revealed that the degree of structural changes in transferrin was relatively small compared to other proteins, which may imply that folding stability of transferrin is higher than that of other model proteins. Fibrinogen is essential for the formation of fibrin clots, and the molecular mechanism of clot formation was suggested to begin with the unfolding of the fibrinogen γ-chain.119 The unfolding curves of fibrinogen shown in Figure 5E revealed that a unique unfolding process exists in the presence of COOH-CNT. It is unclear whether this unique unfolding process initially follows the known γ-chain unfolding process or if a completely different unfolding pathway was adopted by the fibrinogen corona state.
Perspectives: Application of the Protein Corona
The formation of the protein corona may be disadvantageous for most in vivo applications of NPs. Corona proteins can eliminate the targeting ability of NPs, as the bound proteins may sterically mask the pre-coupled targeting ligands that recognize specific receptors or target molecules on cells. The surface protein ligand of NPs may lose its structure upon interaction with corona proteins. The corona may provoke unintended distribution of NPs in the body and immune responses through conformational changes of bound proteins.
Various trials have focused on the affirmative use of corona formation. NPs pre-incubated and pre-coated with proteins may gain a new biological identity. This may allow alterations in immunogenic responses against NPs by stealth effects, improvement in targeting of NPs, and stabilization of NPs against agglomeration. The PEG NPs coated with clusterin showed reduced uptake by macrophages.99 The agglomeration rate of silver NPs was reduced by the formation of an HSA corona depending on the concentration of HSA as compared to bare NPs.120 Antibody-conjugated NPs used for targeting cancer cells showed improved association with the human ovarian cancer cell line SK-OV-3 through HSA corona formation.33 Protein coronas made from high concentrations of fetal bovine serum could reduce the cytotoxicity of graphene oxide nanomaterials, suggestive of an approach to improve safety.121 Serum corona formation of hyaluronic acid (HA)-based capsules significantly enhanced the targeting specificity of capsules to human mammary gland cancer cell lines by reducing nonspecific capsule-cell interaction.122 A similar effect was also observed with antibody-labeled gold NPs for hard corona.123 Gamma-globulin-coated silica NPs could recruit and accumulate serum immunoglobulins on their corona layer, indicative of a potential tool for the regulation of serum immune proteins.34 PEG-conjugated polymer NPs were proposed as capturing agents for serum Aβ42 in the circulating system, as the corona of these NPs could preferentially bind to Aβ42.124 Casein-coated gold NPs also showed the elimination of Aβ42 in a nonspecific-binding manner.19 These NPs may improve the condition of Alzheimer’s disease. A modified version of liposome assembled with phospholipid and membrane proteins from leukocytes (leukosome) absorbed specific corona proteins on its surface and showed reduced protein corona-mediated uptake by immune cells.125 The acute toxicity of silica NPs was shown to be reduced by the pre-formation of a serum corona complex.126 In addition, metal oxide NPs, Fe3O4-PEG was saturated with protein corona, resulting in the reduction of ROS toxicity and inflammatory response in human macrophages.127
Extensive studies on the protein corona of NPs have improved our knowledge on the behavior of NPs in biological systems. As described above, several proposals have been suggested to control the biological behavior of NPs, and many applications have focused on how to recruit specific proteins on and unload specific proteins from NP surfaces (Figure 6).
Concluding Remarks
To enhance the safety and efficiency of NP application, it is necessary to understand how protein coronas can affect and regulate the functionality of given NP systems. The conformational characteristics of corona proteins recruited by various NPs should be a critical determinant of their functionality and safety. However, the current findings on the structural features of corona proteins at the atomic level are still insufficient to fully evaluate and understand the corona-NP system. In-depth studies on the structural changes of each corona protein on NPs would bridge the gap between “applications” and “basics” of NPs, as the behavior of NPs in vivo is definitely related to the orientation and unfolding of certain proteins within the corona environment.
 |
Figure 1 Size comparison between proteins and NPs. Notes: The sizes of NPs are compared to those of various proteins. The codes in brackets are those from the PDB database. Adapted with permission from Kopp M, Kollenda S, Epple M. Nanoparticle-Protein Interactions: Therapeutic Approaches and Supramolecular Chemistry. Acc Chem Res. 2017; 50(6):1383–1390. Copyright (2019) American Chemical Society.50 Abbreviations: HSA, human serum albumin; IgG, immunoglobulin G. |
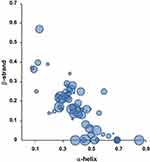 |
Figure 3 The preference of secondary structural elements for MWCNT binding. Notes: About 750 cellular proteins were monitored for binding to MWCNT by mass spectrometry-based proteomics. The secondary structures of the bound proteins were analyzed using the 3D structures deposited in the Protein Data Bank. 269 proteins out of identified 778 proteins were used for the analysis. The proportion of α-helix and β-sheet for each protein was quantified. The protein classes among the CNT-binding proteins are represented with bubbles. The most abundant protein classes contained cytoskeletal proteins, endosomal proteins, and heat shock proteins. The bubble size represents the relative binding affinity to MWCNTs. Adapted from Nanomedicine: Nanotechnology, Biology and Medicine, 9(5), Cai X, Ramalingam R, Wong HS, Cheng J, Ajuh P, Cheng SH, Lam YW. Characterization of carbon nanotube protein corona by using quantitative proteomics. Nanomedicine. 583–593, Copyright (2013), with permission from Elsevier.87 Abbreviation: MWCNT, multi-walled carbon nanotube. |
 |
Figure 5 The Tm curves of protein-NP complexes. Notes: PBS represents the reference Tm curve without NPs. (A) Albumin (PDB code: 2bx8), (B) α-1-antitrypsin (PDB code: 1kct), (C) transferrin (PDB code: 2hau), (D) lysozyme (PDB code: 1lz1), (E) fibrinogen (α, β, r, PDB code: 3ghg). The structure of each protein was obtained from PDB database (http://www.rcsb.org). The Coulumb parameters were ε = 4r and thresholds ± 5.93 kcal/mol⋅e. Negative charge is indicated in red, positive in blue, and neutral in white. Surface charge was calculated using UCSF Chimera. Abbreviations: CD, circular dichroism; NP, nanoparticle; Tm, melting temperature. |
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2018R1D1A1B07050426 and 2018M3A9C8024360). This research was also supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1135).
Disclosure
The author reports no conflicts of interest in this work.
References
1. Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotechnol. 2012;7(12):779–786. doi:10.1038/nnano.2012.207
2. Sun T, Zhang YS, Pang B, Hyun DC, Yang M, Xia Y. Engineered nanoparticles for drug delivery in cancer therapy. Angew Chem Int Ed. 2014;53:12320–12364.
3. Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioeng Transl Med. 2016;1:10–29.
4. Bobo D, Robinson KJ, Islam J, et al. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm Res. 2016;33(10):2373–2387. doi:10.1007/s11095-016-1958-5
5. Papi M, Caputo D, Palmieri V, et al. Clinically approved PEGylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cells. Nanoscale. 2017;9:10327–10334. doi:10.1039/C7NR03042H
6. Guttmann A, Krasnokutsky S, Pillinger MH, Berhanu A. Pegloticase in gout treatment—safety issues, latest evidence and clinical considerations. Ther Adv Drug Saf. 2017;8:379–388. doi:10.1177/2042098617727714
7. English C, Aloi JJ. New FDA-approved disease-modifying therapies for multiple sclerosis. Clin Ther. 2015;37:691–715. doi:10.1016/j.clinthera.2015.03.001
8. Edgar JYC, Wang H. Introduction for design of nanoparticle based drug delivery systems. Curr Pharm Des. 2017;23(14):2108–2112. doi:10.2174/1381612822666161025154003
9. Matoba T, Koga JI, Nakano K, Egashira K, Tsutsui H. Nanoparticle-mediated drug delivery system for atherosclerotic cardiovascular disease. J Cardiol. 2017;70(3):206–211. doi:10.1016/j.jjcc.2017.03.005
10. Mishra D, Hubenak JR, Mathur AB. Nanoparticle systems as tools to improve drug delivery and therapeutic efficacy. J Biomed Mater Res A. 2013;101(12):3646–3660. doi:10.1002/jbm.a.34642
11. Conte R, Marturano V, Peluso G, Calarco A, Cerruti P. Recent advances in nanoparticle-mediated delivery of anti-inflammatory phytocompounds. Int J Mol Sci. 2017;18(4):709. doi:10.3390/ijms18040709
12. Lin YS, Lee MY, Yang CH, Huang KS. Active targeted drug delivery for microbes using nano-carriers. Curr Top Med Chem. 2015;15(15):1525–1531. doi:10.2174/1568026615666150414123157
13. Yeon Kyung L, Eun-Ju C, Thomas JW, Sang-Hyun K, Dongwoo K. Effect of the protein corona on nanoparticles for modulating cytotoxicity and immunotoxicity. Int J Nanomedicine. 2015;10:97–113. doi:10.2147/IJN.S72998
14. Szebeni J, Simberg D, González-Fernández Á, Barenholz Y, Dobrovolskaia MA. Roadmap and strategy for overcoming infusion reactions to nanomedicines. Nat Nanotechnol. 2018;13(12):1100–1108. doi:10.1038/s41565-018-0273-1
15. Ernsting MJ, Murakami M, Roy A, Li SD. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles. J Control Release. 2013;172(3):782–794. doi:10.1016/j.jconrel.2013.09.013
16. Shalini K, Puja K, Pritam KP, Nandini P, Suresh KV, Mallick MA. Gold nanoparticle from andrographis peniculata photosystem II and their in vivo biological effect on embryonic zebrafish (Danio rerio). Environ Nanotechnol Monit Manag. 2019;11:100201. doi:10.1016/j.enmm.2018.100201
17. Xu C, Zhao X, Wang L, et al. Protein Conjugation with gold nanoparticles: spectroscopic and thermodynamic analysis on the conformational and activity of serum albumin. J Nanosci Nanotechnol. 2018;18(11):7818–7823. doi:10.1166/jnn.2018.15215
18. Verma SK, Jha E, Panda PK, et al. Molecular investigation to RNA and protein based interaction induced in vivo biocompatibility of phytofabricated AuNP with embryonic zebrafish. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S671–S684. doi:10.1080/21691401.2018.1505746
19. Javed I, Peng G, Xing Y, et al. Inhibition of amyloid beta toxicity in zebrafish with a chaperone-gold nanoparticle dual strategy. Nat Commun. 2019;10(1):3780. doi:10.1038/s41467-019-11762-0
20. Behzadi E, Sarsharzadeh R, Nouri M, et al. Albumin binding and anticancer effect of magnesium oxide nanoparticles. Int J Nanomedicine. 2018;14:257–270. doi:10.2147/IJN.S186428
21. Verma SK, Nisha K, Panda PK, et al. Green synthesized MgO nanoparticles infer biocompatibility by reducing in vivo molecular nanotoxicity in embryonic zebrafish through arginine interaction elicited apoptosis. Sci Total Environ. 2020;713:136521. doi:10.1016/j.scitotenv.2020.136521
22. Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi:10.1038/nature02261
23. Chetty PS, Mayne L, Lund-Katz S, Englander SW, Phillips MC. Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci U S A. 2017;114(5):968–973. doi:10.1073/pnas.1617523114
24. Poirier Y, Grimm A, Schmitt K, Eckert A. Link between the unfolded protein response and dysregulation of mitochondrial bioenergetics in alzheimer’s disease. Cell Mol Life Sci. 2019;76(7):1419–1431. doi:10.1007/s00018-019-03009-4
25. von Bergen M, Barghorn S, Biernat J, Mandelkow EM, Mandelkow E. Tau aggregation is driven by a transition from random coil to beta sheet structure. Biochim Biophys Acta. 2005;1739(2–3):158–166. doi:10.1016/j.bbadis.2004.09.010
26. Grassi D, Howard S, Zhou M, et al. Identification of a highly neurotoxic α-synuclein species inducing mitochondrial damage and mitophagy in parkinson’s disease. Proc Natl Acad Sci U S A. 2018;115(11):E2634–E2643. doi:10.1073/pnas.1713849115
27. Dettmer U
28. Ge X, Yang Y, Sun Y, Cao W, Ding F. Islet amyloid polypeptide promotes amyloid-beta aggregation by binding-induced helix-unfolding of the amyloidogenic core. ACS Chem Neurosci. 2018;9(5):967–975. doi:10.1021/acschemneuro.7b00396
29. Schmidt M, Wiese S, Adak V, et al. Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat Commun. 2019;10(1):5008. doi:10.1038/s41467-019-13038-z
30. Liu HL, Wu YC, Zhao JH, et al. Insights into the conformational changes of several human lysozyme variants associated with hereditary systemic amyloidosis. Biotechnol Prog. 2007;23(1):246–254. doi:10.1021/bp060264a
31. Rennella E, Morgan GJ, Yan N, Kelly JW, Kay LE. The role of protein thermodynamics and primary structure in fibrillogenesis of variable domains from immunoglobulin light chains. J Am Chem Soc. 2019;141(34):13562–13571. doi:10.1021/jacs.9b05499
32. Sukhanova A, Poly S, Bozrova S, et al. Nanoparticles with a specific size and surface charge promote disruption of the secondary structure and amyloid-like fibrillation of human insulin under physiological conditions. Front Chem. 2019;7:480. doi:10.3389/fchem.2019.00480
33. Qiong D, Yan Y, Junling G, et al. Targeting ability of antibody-functionalized particles is enhanced by albumin but inhibited by serum coronas. ACS Macro Lett. 2015;4:1259–1263. doi:10.1021/acsmacrolett.5b00627
34. Mirshafiee V, Kim R, Park S, Mahmoudi M, Kraft ML. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials. 2016;75:295–304. doi:10.1016/j.biomaterials.2015.10.019
35. Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein-nanoparticle interactions: opportunities and challenges. Chem Rev. 2011;111(9):5610–5637. doi:10.1021/cr100440g
36. Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano. 2010;4:3623–3632. doi:10.1021/nn901372t
37. Deng ZJ, Mortimer G, Schiller T, Musumeci A, Martin D, Minchin RF. Differential plasma protein binding to metal oxide nanoparticles. Nanotechnology. 2009;20:455101. doi:10.1088/0957-4484/20/45/455101
38. Verma SK, Jha E, Panda PK, et al. Mechanistic insight into size-dependent enhanced cytotoxicity of industrial antibacterial titanium oxide nanoparticles on colon cells because of reactive oxygen species quenching and neutral lipid alteration. ACS Omega. 2018;3(1):1244–1262. doi:10.1021/acsomega.7b01522
39. Lundqvist M, Stigler J, Elia G, et al. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci USA. 2009;105:14265–14270. doi:10.1073/pnas.0805135105
40. Monopoli MP, Walczyk D, Campbell A, et al. Physical−chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc. 2011;133(8):2525–2534. doi:10.1021/ja107583h
41. Cedervall T, Lynch I, Lindman S, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA. 2007;104(7):2050–2055. doi:10.1073/pnas.0608582104
42. Linse S, Cabaleiro-Lago C, Xue WF, et al. Nucleation of protein fibrillation by nanoparticles. Proc Natl Acad Sci USA. 2007;104:8691–8696. doi:10.1073/pnas.0701250104
43. Li F, Sarah P. Effect of nanoparticles on protein folding and fibrillogenesis. Int J Mol Sci. 2009;10:646–655.
44. Zia F, Kendall M, Watson SP, Mendes PM. Platelet aggregation induced by polystyrene and platinum nanoparticles is dependent on surface area. RSC Adv. 2018;8(66):37789–37794. doi:10.1039/C8RA07315E
45. Ge C, Du J, Zhao L, et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc Natl Acad Sci U S A. 2011;108(41):16968–16973. doi:10.1073/pnas.1105270108
46. Khan S, Gupta A, Chaudhary A, Nandi CK. Orientational switching of protein conformation as a function of nanoparticle curvature and their geometrical fitting. J Chem Phys. 2014;141(8):084707. doi:10.1063/1.4893441
47. Vertegel AA, Siegel RW, Dordick JS. Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir. 2004;20:6800–6807. doi:10.1021/la0497200
48. Shang W, Nuffer JH, Dordick JS, Siegel RW. Unfolding of ribonuclease A on silica nanoparticle surfaces. Nano Lett. 2007;7(7):1991–1995.
49. Lacerda SH, Park JJ, Meuse C, et al. Interaction of gold nanoparticles with common human blood proteins. ACS Nano. 2010;4(1):365–379. doi:10.1021/nn9011187
50. Kopp M, Kollenda S, Epple M. Nanoparticle-protein interactions: therapeutic approaches and supramolecular chemistry. Acc Chem Res. 2017;50(6):1383–1390. doi:10.1021/acs.accounts.7b00051
51. Boselli L, Polo E, Castagnola V, Dawson KA. Regimes of biomolecular ultrasmall nanoparticle interactions. Angew Chem Int Ed Engl. 2017;56(15):4215–4218. doi:10.1002/anie.201700343
52. Jia G, Wang H, Yan L, et al. Cytotoxicity of carbon nanomaterials: single-wall nanotube, multi-wall nanotube, and fullerene. Environ Sci Technol. 2005;39:1378–1383. doi:10.1021/es048729l
53. Zeinabad HA, Zarrabian A, Saboury AA, Alizadeh AM, Falahati M. Interaction of single and multi wall carbon nanotubes with the biological systems: tau protein and PC12 cells as targets. Sci Rep. 2016;6:26508. doi:10.1038/srep26508
54. Dahia I, Hanane M, Aicha M, et al. Scattering correlation spectroscopy and raman spectroscopy of thiophenol on gold nanoparticles: comparative study between nanospheres and nanourchins. J Phys Chem C. 2017;121:18254–18262. doi:10.1021/acs.jpcc.7b05355
55. Hanane M, Saber J, Djeddi I, et al. Protein corona study by scattering correlation spectroscopy: a comparative study between spherical and urchin-shaped gold nanoparticles. Nanoscale. 2019;11(8):3665–3673. doi:10.1039/C8NR09891C
56. Cha SH, Hong J, McGuffie M, Yeom B, VanEpps JS, Kotov NA. Shape-dependent biomimetic inhibition of enzyme by nanoparticles and their antibacterial Activity. ACS Nano. 2015;9(9):9097–9105. doi:10.1021/acsnano.5b03247
57. Stefan T, Docter D, Kuharev J, et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nanotechnol. 2013;8(10):772–781. doi:10.1038/nnano.2013.181
58. Moyano DF, Liu Y, Peer D, Rotello VM. Rotello modulation of immune response using engineered nanoparticle surfaces. Small. 2016;12(1):76–82. doi:10.1002/smll.201502273
59. Maiorano G, Sabella S, Sorce B, et al. Effects of cell culture media on the dynamic formation of protein–nanoparticle complexes and influence on the cellular response. ACS Nano. 2010;4(12):7481–7491. doi:10.1021/nn101557e
60. Lundqvist M, Stigler J, Cedervall T, et al. The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 2011;5(9):7503–7509. doi:10.1021/nn202458g
61. Ke PC, Lin S, Parak WJ, Davis TP, Caruso FA. Decade of the protein corona. ACS Nano. 2017;11(12):11773–11776. doi:10.1021/acsnano.7b08008
62. Vroman L, Adams AL, Fischer GC, Munoz PC. Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces. Blood. 1980;55(1):156–159. doi:10.1182/blood.V55.1.156.156
63. Vroman L. Effect of Adsorbed proteins on the wettability of hydrophilic and hydrophobic solids. Nature. 1962;196(4853):476–477. doi:10.1038/196476a0
64. Baimanov D, Cai R, Chen C. Understanding the chemical nature of nanoparticle-protein interactions. Bioconjug Chem. 2019;30(7):1923–1937. doi:10.1021/acs.bioconjchem.9b00348
65. Kihara S, van der Heijden NJ, Seal CK, et al. Soft and hard interactions between polystyrene nanoplastics and human serum albumin protein corona. Bioconjug Chem. 2019;30(4):1067–1076. doi:10.1021/acs.bioconjchem.9b00015
66. Winzen S, Schoettler S, Baier G, et al. Complementary analysis of the hard and soft protein corona: sample preparation critically effects corona composition. Nanoscale. 2015;7(7):2992–3001. doi:10.1039/C4NR05982D
67. Weber C, Simon J, Mailänder V, et al. Preservation of the soft protein corona in distinct flow allows identification of weakly bound proteins. Acta Biomater. 2018;76:217–224. doi:10.1016/j.actbio.2018.05.057
68. Ali MS, Altaf M, Al-Lohedan HA. Green synthesis of biogenic silver nanoparticles using Solanum tuberosum extract and their interaction with human serum albumin: evidence of “corona” formation through a multi-spectroscopic and molecular docking analysis. J Photochem Photobiol B. 2017;173:108–119. doi:10.1016/j.jphotobiol.2017.05.015
69. de Barros CHN, Cruz GCF, Mayrink W, Tasic L. Bio-based synthesis of silver nanoparticles from orange waste: effects of distinct biomolecule coatings on size, morphology, and antimicrobial activity. Nanotechnol Sci Appl. 2018;11:1–14. doi:10.2147/NSA.S156115
70. Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3(1):40–47. doi:10.1016/S1748-0132(08)70014-8
71. Auer S, Trovato A, Vendruscolo M, Hummer G. A condensation-ordering mechanism in nanoparticle-catalyzed peptide aggregation. PLoS Comput Biol. 2009;5(8):e1000458. doi:10.1371/journal.pcbi.1000458
72. Colvin VL, Kulinowski KM. Nanoparticles as catalysts for protein fibrillation. Proc Natl Acad Sci U S A. 2007;104(21):8679–8680. doi:10.1073/pnas.0703194104
73. Rocha S, Thünemann AF, Pereira Mdo C, Coelho M, Möhwald H, Brezesinski G. Influence of fluorinated and hydrogenated nanoparticles on the structure and fibrillogenesis of amyloid beta-peptide. Biophys Chem. 2008;137(1):35–42. doi:10.1016/j.bpc.2008.06.010
74. Konar M, Mathew A, Dasgupta S. Effect of silica nanoparticles on the amyloid fibrillation of lysozyme. ACS Omega. 2019;4(1):1015–1026. doi:10.1021/acsomega.8b03169
75. Sung Jean P, Dongwoo K. Conformational changes of fibrinogen in dispersed carbon nanotubes. Int J Nanomed. 2012;7:4325–4333.
76. Zhang T, Tang M, Yao Y, Ma Y. Pu Y MWCNT interactions with protein: surface-induced changes in protein adsorption and the impact of protein corona on cellular uptake and cytotoxicity. Int J Nanomed. 2019;14:993–1009.
77. Spaeth P, Adhikari S, Le L, et al. Circular dichroism measurement of single metal nanoparticles using photothermal imaging. Nano Lett. 2019;19(12):8934–8940. doi:10.1021/acs.nanolett.9b03853
78. Fardanesh A, Zibaie S, Shariati B, et al. Amorphous aggregation of tau in the presence of titanium dioxide nanoparticles: biophysical, computational, and cellular studies. Int J Nanomedicine. 2019;14:901–911.
79. Yadav I, Kumar S, Aswal VK, Kohlbrecher J. Structure and interaction in the ph-dependent phase behavior of nanoparticle-protein systems. Langmuir. 2017;33(5):1227–1238. doi:10.1021/acs.langmuir.6b04127
80. Riek R, Pervushin K, Wüthrich K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem Sci. 2000;25(10):462–468. doi:10.1016/S0968-0004(00)01665-0
81. Woods KE, Perera YR, Davidson MB, Wilks CA, Yadav DK, Fitzkee NC. Understanding protein structure deformation on the surface of gold nanoparticles of varying size. J Phys Chem C Nanomater Interfaces. 2016;120(49):27944–27953. doi:10.1021/acs.jpcc.6b08089
82. Shang L, Wang Y, Jiang J, Dong S. pH-Dependent protein conformational changes in albumin: gold nanoparticle bioconjugates: a spectroscopic study. Langmuir. 2007;23:2714–2721. doi:10.1021/la062064e
83. Capomaccio R, Jimenez IO, Colpo P, et al. Determination of the structure and morphology of gold nanoparticle-HSA protein complexes. Nanoscale. 2015;7(42):17653–17657. doi:10.1039/C5NR05147A
84. Ramezani F, Rafii-Tabar H. An in-depth view of human serum albumin corona on gold nanoparticles. Mol Biosyst. 2015;11(2):454–462. doi:10.1039/C4MB00591K
85. Zhang W, Zhang Q, Wang F, et al. Comparison of interactions between human serum albumin and silver nanoparticles of different sizes using spectroscopic methods. Luminescence. 2015;30(4):397–404. doi:10.1002/bio.2748
86. Imaninezhad M, Schober J, Griggs D, Ruminski P, Kuljanishvili I, Zustiak SP. Cell attachment and spreading on carbon nanotubes is facilitated by integrin binding. Front Bioeng Biotechnol. 2018;6:12. doi:10.3389/fbioe.2018.00129
87. Cai X, Ramalingam R, Wong HS, et al. Characterization of carbon nanotube protein corona by using quantitative proteomics. Nanomedicine. 2013;9(5):583–593. doi:10.1016/j.nano.2012.09.004
88. Holt BD, Short PA, Rape AD, Wang YL, Islam MF, Dahl KN. Carbon nanotubes reorganize actin structures in cells and ex vivo. ACS Nano. 2010;4(8):4872–4878. doi:10.1021/nn101151x
89. Sarkar B, Verma SK, Akhtar J, et al. Molecular aspect of silver nanoparticles regulated embryonic development in Zebrafish (Danio rerio) by Oct- 4expression. Chemosphere. 2018;206:560–567. doi:10.1016/j.chemosphere.2018.05.018
90. Verma SK, Jha E, Panda PK, et al. Molecular insights to alkaline based bio-fabrication of silver nanoparticles for inverse cytotoxicity and enhanced antibacterial activity. Mater Sci Eng C Mater Biol Appl. 2018;92:807–818. doi:10.1016/j.msec.2018.07.037
91. Chen M, von Mikecz A. Formation of nucleoplasmic protein aggregates impairs nuclear function in response to SiO2 nanoparticles. Exp Cell Res. 2005;305(1):51–62. doi:10.1016/j.yexcr.2004.12.021
92. Márquez A, Berger T, Feinle A, et al. Bovine serum albumin adsorption on TiO2 colloids: the effect of particle agglomeration and surface composition. Langmuir. 2017;33(10):2551–2558. doi:10.1021/acs.langmuir.6b03785
93. Maiolo D, Bergese P, Mahon E, Dawson KA, Monopoli M. Surfactant titration of nanoparticle-protein corona. Anal Chem. 2014;86(24):12055–12063. doi:10.1021/ac5027176
94. Schulze C, Schulze C, Kroll A, et al. Not ready to use overcoming pitfalls when dispersing nanoparticles in physiological media. Nanotoxicology. 2008;2(2):51–61. doi:10.1080/17435390802018378
95. My Kieu H, Yoo Jin S, Tae Hyun Y. Effects of agglomeration on in vitro dosimetry and cellular association of silver nanoparticles. Environ Sci Nano. 2018;5:446–455.
96. Settanni G, Zhou J, Suo T, et al. Protein corona composition of poly(ethylene glycol)- and poly(phosphoester)-coated nanoparticles correlates strongly with the amino acid composition of the protein surface. Nanoscale. 2017;9(6):2138–2144. doi:10.1039/C6NR07022A
97. Wang J, Bai R, Yang R, et al. Size- and surface chemistry-dependent pharmacokinetics and tumor accumulation of engineered gold nanoparticles after intravenous administration. Metallomics. 2015;7(3):516–524. doi:10.1039/C4MT00340C
98. Dai Q, Walkey C, Chan WC. Polyethylene glycol backfilling mitigates the negative impact of the protein corona on nanoparticle cell targeting. Angew Chem Int Ed Engl. 2014;53(20):5093–5096. doi:10.1002/anie.201309464
99. Schöttler S, Becker G, Winzen S, et al. Protein adsorption is required for stealth effect of poly (ethylene glycol)-and poly (phosphoester)-coated nanocarriers. Nat Nanotechnol. 2016;11(4):372–377. doi:10.1038/nnano.2015.330
100. Cai C, Wang M, Wang L, Wang B, Feng W, Chen C. Thermal unfolding process of lysozyme on PEGylated gold nanoparticles reveals length-dependent effects of PEG layer. J Nanosci Nanotechnol. 2018;18(8):5542–5550. doi:10.1166/jnn.2018.15416
101. Chakraborti S, Sarwar S, Chakrabarti P. The effect of the binding of ZnO nanoparticle on the structure and stability of α-lactalbumin: a comparative study. J Phys Chem B. 2013;117(43):13397–13408. doi:10.1021/jp404411b
102. Deng ZJ, Liang M, Toth I, Monteiro M, Minchin RF. Plasma protein binding of positively and negatively charged polymer-coated gold nanoparticles elicits different biological responses. Nanotoxicology. 2013;7:314–322. doi:10.3109/17435390.2012.655342
103. Fertsch-Gapp S, Semmler-Behnke M, Wenk A, Kreyling WG. Binding of polystyrene and carbon black nanoparticles to blood serum proteins. Inhal Toxicol. 2011;23:468–475. doi:10.3109/08958378.2011.583944
104. Mukhopadhyay A, Basu S, Singha S, Patra HK. Inner-view of nanomaterial incited protein conformational changes: insights into designable interaction. Research. 2018;2018:9712832.
105. Harush-Frenkel O, Debotton N, Benita S, Altschuler Y. Targeting of nanoparticles to the clathrin-mediated endocytic pathway. Biochem Biophys Res Commun. 2007;353(1):26–32. doi:10.1016/j.bbrc.2006.11.135
106. Zhang LW, Monteiro-Riviere NA. Mechanisms of quantum dot nanoparticle cellular uptake. Toxicol Sci. 2009;110(1):138–155. doi:10.1093/toxsci/kfp087
107. Lunov O, Syrovets T, Loos C, et al. Differential uptake of functionalized polystyrene nanoparticles by human macrophages and a monocytic cell line. ACS Nano. 2011;5(3):1657–1669. doi:10.1021/nn2000756
108. Kharazian B, Hadipour NL, Ejtehadi MR. Understanding the nanoparticle-protein corona complexes using computational and experimental methods. Int J Biochem Cell Biol. 2016;75:162–174. doi:10.1016/j.biocel.2016.02.008
109. Gessner A, Lieske A, Paulke BR, Muller RH. Functional groups on polystyrene model nanoparticles: influence on protein adsorption. J Biomed Mater Res A. 2003;65:319–326. doi:10.1002/jbm.a.10371
110. Gessner A, Lieske A, Paulke B, Muller R. Influence of surface charge density on protein adsorption on polymeric nanoparticles: analysis by two-dimensional electrophoresis. Eur J Pharm Biopharm. 2002;54:165–170. doi:10.1016/S0939-6411(02)00081-4
111. Böhm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi:10.1093/protein/5.3.191
112. Baler K, Martin OA, Carignano MA, Ameer GA, Vila JA, Szleifer I. Electrostatic unfolding and interactions of albumin driven by pH changes: a molecular dynamics study. J Phys Chem B. 2014;118(4):921–930. doi:10.1021/jp409936v
113. Tsao FHC, Barnes JN, Amessoudji A, Li Z, Meyer KC. Aging-related and gender specific albumin misfolding in alzheimer’s disease. J Alzheimers Dis Rep. 2020;4(1):67–77. doi:10.3233/ADR-200168
114. Strnad P, McElvaney NG, Lomas DA. Alpha1-antitrypsin deficiency. N Engl J Med. 2020;382(15):1443–1455. doi:10.1056/NEJMra1910234
115. Elliott PR, Lomas DA, Carrell RW, Abrahams JP. Inhibitory conformation of the reactive loop of α-1antitrypsin. Nat Struct Biol. 1996;3(8):676–681. doi:10.1038/nsb0896-676
116. Sziegat F, Wirmer-Bartoschek J, Schwalbe H. Characteristics of human lysozyme and its disease-related mutants in their unfolded states. Angew Chem Int Ed Engl. 2011;50(24):5514–5518. doi:10.1002/anie.201008040
117. Li S, Peng Z, Dallman J, et al. Crossing the blood-brain-barrier with transferrin conjugated carbon dots: a zebrafish model study. Colloids Surf B Biointerfaces. 2016;145:251–256. doi:10.1016/j.colsurfb.2016.05.007
118. Shanghao L, Lingyu W, Charles CC, et al. Nontoxic carbon dots potently inhibit human insulin fibrillation. Chem Mater. 2015;27(5):1764–1771. doi:10.1021/cm504572b
119. Zhmurov A, Brown AE, Litvinov RI, Dima RI, Weisel JW, Barsegov V. Mechanism of fibrin(ogen) forced unfolding. Structure. 2011;19(11):1615–1624. doi:10.1016/j.str.2011.08.013
120. Gebauer JS, Malissek M, Simon S, et al. Impact of the nanoparticle-protein corona on colloidal stability and protein structure. Langmuir. 2012;28(25):9673–9679. doi:10.1021/la301104a
121. Hu W, Peng C, Lv M, et al. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano. 2011;5(5):3693–3700. doi:10.1021/nn200021j
122. Ju Y, Dai Q, Cui J, et al. Improving targeting of metal-phenolic capsules by the presence of protein coronas. ACS Appl Mater Interfaces. 2016;8(35):22914–22922. doi:10.1021/acsami.6b07613
123. Kelly PM, Åberg C, Polo E, et al. Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat Nanotechnol. 2015;10(5):472–479. doi:10.1038/nnano.2015.47
124. Brambilla D
125. Corbo C, Molinaro R, Taraballi F, et al. Unveiling the in vivo protein corona of circulating leukocyte-like carriers. ACS Nano. 2017;11(3):3262–3273. doi:10.1021/acsnano.7b00376
126. Leibe R, Hsiao IL, Fritsch-Decker S, et al. The protein corona suppresses the cytotoxic and pro-inflammatory response in lung epithelial cells and macrophages upon exposure to nanosilica. Arch Toxicol. 2019;93(4):871–885. doi:10.1007/s00204-019-02422-9
127. Escamilla-Rivera V, Uribe-Ramírez M, González-Pozos S, Lozano O, Lucas S, De Vizcaya-ruiz A. Protein corona acts as a protective shield against Fe3O4-PEG inflammation and ROS-induced toxicity in human macrophages. Toxicol Lett. 2016;240(1):172–184. doi:10.1016/j.toxlet.2015.10.018
128. Ebrahim-Habibi MB, Ghobeh M, Aghakhani Mahyari F, Rafii-Tabar H, Sasanpour P. Protein G selects two binding sites for carbon nanotube with dissimilar behavior; a molecular dynamics study. J Mol Graph Model. 2019;87:257–267. doi:10.1016/j.jmgm.2018.12.007
129. Chen X, Wang Y, Wang P. Peptide-induced affinity binding of carbonic anhydrase to carbon nanotubes. Langmuir. 2015;31(1):397–403. doi:10.1021/la504321q
130. Vaitheeswaran S, Garcia AE. Protein stability at a carbon nanotube interface. J Chem Phys. 2011;134(12):125101. doi:10.1063/1.3558776
131. Mahendra WIP, Gandhi S, Ju Nie T, et al. Protein/carbon nanotubes interaction: the effect of carboxylic groups on conformational and conductance changes. Appl Phys Lett. 2009;95(7):073704. doi:10.1063/1.3211328
132. Liu X, Liu T, Song J, et al. Understanding the interaction of single-walled carbon nanotube (SWCNT) on estrogen receptor: a combined molecular dynamics and experimental study. Ecotoxicol Environ Saf. 2019;172:373–379. doi:10.1016/j.ecoenv.2019.01.101
133. Asuri P, Bale SS, Pangule RC, Shah DA, Kane RS, Dordick JS. Structure, function, and stability of enzymes covalently attached to single-walled carbon nanotubes. Langmuir. 2007;23(24):12318–12321. doi:10.1021/la702091c
134. Lu N, Sui Y, Tian R, Peng YY. Adsorption of plasma proteins on single-walled carbon nanotubes reduced cytotoxicity and modulated neutrophil activation. Chem Res Toxicol. 2018;31(10):1061–1068. doi:10.1021/acs.chemrestox.8b00141
135. Noordadi M, Mehrnejad F, Sajedi RH, Jafari M, Ranjbar B, Lebedev N. The potential impact of carboxylic-functionalized multi-walled carbon nanotubes on trypsin: a comprehensive spectroscopic and molecular dynamics simulation study. PLoS One. 2018;13(6):e0198519. doi:10.1371/journal.pone.0198519
136. Sekar G, Sivakumar A, Mukherjee A, Chandrasekaran N. Existence of hydroxylated MWCNTs demotes the catalysis effect of amylases against starch degradation. Int J Biol Macromol. 2016;86:250–261. doi:10.1016/j.ijbiomac.2016.01.071
137. Yang M, Meng J, Mao X, et al. Carbon nanotubes induce secondary structure changes of bovine albumin in aqueous phase. J Nanosci Nanotechnol. 2010;10(11):7550–7553. doi:10.1166/jnn.2010.2825
138. De Paoli SH, Diduch LL, Tegegn TZ, et al. The effect of protein corona composition on the interaction of carbon nanotubes with human blood platelets. Biomaterials. 2014;35(24):6182–6194. doi:10.1016/j.biomaterials.2014.04.067
139. Worrall JW, Verma A, Yan H, Rotello VM. “Cleaning” of nanoparticle inhibitors via proteolysis of adsorbed proteins. Chem Commun (Camb). 2006;22:2338–2340. doi:10.1039/B517421J
140. Srivastava S, Verma A, Frankamp BL, Rotello VM. Controlled assembly of protein-nanoparticle composites through protein surface recognition. Adv Mater. 2005;17(5):617–621. doi:10.1002/adma.200400776
141. Zarabi MF, Farhangi A, Mazdeh SK, et al. Synthesis of gold nanoparticles coated with aspartic acid and their conjugation with FVIII protein and FVIII antibody. Indian J Clin Biochem. 2014;29(2):154–160. doi:10.1007/s12291-013-0323-2
142. Wangoo N, Suri CR, Shekhawat G. Interaction of gold nanoparticles with protein: a spectroscopic study to monitor protein conformational changes. Appl Phys Lett. 2008;92:133104.
143. Wangoo N, Bhasin KK, Mehta SK, Suri CR. Synthesis and capping of water-dispersed gold nanoparticles by an amino acid: bioconjugation and binding studies. J Colloid Interface Sci. 2008;323(2):247–254. doi:10.1016/j.jcis.2008.04.043
144. Tsai DH, DelRio FW, Keene AM, et al. Adsorption and conformation of serum albumin protein on gold nanoparticles investigated using dimensional measurements and in situ spectroscopic methods. Langmuir. 2011;27(6):2464–2477. doi:10.1021/la104124d
145. Chaudhary A, Gupta A, Khan S, Nandi CK. Morphological effect of gold nanoparticles on the adsorption of bovine serum albumin. Phys Chem Chem Phys. 2014;16(38):20471–20482. doi:10.1039/C4CP01515K
146. Chakraborty S, Joshi P, Shanker V, Ansari ZA, Singh SP, Chakrabarti P. Contrasting effect of gold nanoparticles and nanorods with different surface modifications on the structure and activity of bovine serum albumin. Langmuir. 2011;27(12):7722–7731. doi:10.1021/la200787t
147. Gagner JE, Lopez MD, Dordick JS, Siegel RW. Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials. 2011;32(29):7241–7252. doi:10.1016/j.biomaterials.2011.05.091
148. Banerjee V, Das KP. Structure and functional properties of a multimeric protein α-a-crystallin adsorbed on silver nanoparticle surface. Langmuir. 2014;30(16):4775–4783. doi:10.1021/la5007007
149. Wang G, Lu Y, Hou H, Liu Y. Probing the binding behavior and kinetics of silver nanoparticles with bovine serum albumin. RSC Adv. 2017;7(15):9393–9401. doi:10.1039/C6RA26089F
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.