Back to Journals » OncoTargets and Therapy » Volume 8
Prognostic role of LSD1 in various cancers: evidence from a meta-analysis
Authors Wu J, Hu L, Du Y, Kong F, Pan Y
Received 30 May 2015
Accepted for publication 23 July 2015
Published 15 September 2015 Volume 2015:8 Pages 2565—2570
DOI https://doi.org/10.2147/OTT.S89597
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Jin Wu,1,* Lixia Hu,2,* Yingying Du,3 Fanliang Kong,2 Yueyin Pan3
1The Central Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China; 2Department of Oncology, The Second People’s Hospital of Hefei, Hefei, Anhui, People’s Republic of China; 3Department of Oncology, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work and should be considered as co-first authors
Abstract: The prognostic value of lysine-specific demethylase 1 (LSD1) overexpression in various cancers has been investigated by many studies with inconsistent results. A meta-analysis was performed to assess the association between LSD1 and overall survival (OS) in cancer patients. Eligible studies were identified by searching the online databases PubMed and China National Knowledge Infrastructure up to February 2015. Hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated to clarify the correlation between LSD1 expression and prognosis of different cancers. In total, nine studies with 1,149 cancer patients were included for final analysis. The meta-analysis suggested that LSD1 overexpression was associated with poor OS in cancer patients (HR =1.80, 95% CI: 1.39–2.34, P=0.000). Subgroup analysis by ethnicity, cancer type and HR estimate also showed that high levels of LSD1 were significantly correlated with OS. The meta-analysis showed that LSD1 overexpression may be associated with a worse prognosis in cancer patients.
Keywords: LSD1, cancer, prognosis, meta-analysis, overall survival
Introduction
Lysine-specific demethylase 1 (LSD1) was the first characterized histone demethylase, which could specifically remove the methyl groups from mono- and dimethylated lysine (Lys)4 of histone H3 (H3K4me1/2) and Lys9 of histone H3 (H3K9me1/2).1 LSD1 is essential for mammalian development and is involved in many biological processes, including cell type differentiation, gene activation, and gene repression.2 A recent study indicated that LSD1 might promote cell phase transition (deficiency in LSD1 led to partial cell cycle arrest in G2/M) and cell proliferation, suggesting that its overexpression might promote tumorigenesis.3 The expression of LSD1 has been associated with tumor recurrence during therapy in various cancers, further implicating LSD1 as a tumor promoter.4,5
Many studies investigated the prognostic value of LSD1 in various cancers. Some studies found that the upregulation of LSD1 was associated with worse outcome in cancer patients.6–11 However, some other studies showed insignificant or opposite result.12–14 Therefore, the relation between LSD1 expression and patient survival across different cancers remains controversial. To overcome the limitations of the single study, this meta-analysis was carried out with the aim of evaluating the relationship between LSD1 expression and prognosis of cancer patients.
Materials and methods
Literature search and selection criteria
We searched PubMed and China National Knowledge Infrastructure up to February 2015 to identify relevant studies. We used the search terms: “LSD1”, “lysine specific demethylase 1”, “tumor”, “cancer”, “neoplasm”, “carcinoma”, “malignant”, “survival”, “prognosis”, and “prognostic”. The citation lists associated with the studies were used to identify additional eligible studies. The reviews and bibliographies were also manually inspected to find related articles.
Inclusion and exclusion criteria
The studies were included in our meta-analysis if they met the following inclusion criteria: 1) LSD1 expression evaluated in the human tissues; 2) tumors should be confirmed by pathological or histological examinations; 3) evaluation of the relationship between LSD1 expression and survival; 4) sufficient information provided to estimate the hazard ratios (HRs) with their 95% confidence intervals (CIs) for overall survival (OS). The exclusion criteria were as follows: 1) letters, case reports, reviews, and conference abstracts without original data; 2) articles from which the relevant data could not be extracted. Of the studies which had duplicate data, only the most complete study was included in the analysis.
Data extraction
Data were evaluated and extracted independently from the eligible studies by two investigators (LXH and JW) under the guidelines of a critical review checklist of the Dutch Cochrane Centre proposed by Meta-analysis of Observational Studies in Epidemiology.15 The following items were recorded: first author’s name, year of publication, ethnicity, method, tumor type, total number of patients, and HRs with their 95% CIs for OS. If available, we calculated HRs with their 95% CIs using the data of observed deaths/cancer recurrences, the data of samples in each group, or the data provided by the authors.16 If not, the HRs with their 95% CIs and P-values were collected from the original article. If only Kaplan–Meier curves were available, data were extracted from graphical survival plots to extrapolate HRs with their 95% CIs using previously described methods.17,18 Disagreements were resolved by discussion among all authors.
Statistical analysis
HRs with their 95% CIs were calculated on the basis of the association between LSD1 expression and the OS of cancer patients. The χ2 test and the I2 statistic were used to evaluate the heterogeneity among studies.19 If the heterogeneity was significant between studies (I2>50% or P<0.10), the random effects model was used; otherwise, the fixed effects model was used.20 Sensitivity analysis was also conducted by sequential omission of individual studies to evaluate stability of the results. Publication bias was estimated by Egger’s linear regression test with a funnel plot.21 The statistical analyses were performed using STATA version 12.0 software (Stata Corporation, Collage Station, TX, USA). All P-values were two-sided, and P<0.05 was considered statistically significant.
Results
Study characteristics
The results of the search strategy are described in Figure 1. With our retrieval strategy, a total of 73 references were found. After review of abstracts, we identified 29 potential studies eligible for inclusion in the evaluation. Upon full-text review, nine studies6–14 were selected for our meta-analysis, and the study characteristics are summarized in Table 1. The total number of patients included was 1,149, ranging from 63 to 261 patients per study. Eight studies6–11,13,14 evaluated Asians and one12 evaluated Caucasian. The types of cancers in these studies included esophageal cancer, non-small-cell lung cancer, colon cancer, hepatocellular carcinoma, breast cancer, human melanomas, and tongue cancer. The method of LSD1 detection was based on immunohistochemistry. HRs with 95% CIs were reported directly in five studies,8,10–13 calculated from available data in one study,14 and extrapolated from Kaplan–Meier curves in three studies.6,7,9
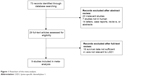  | Figure 1 Flowchart of the meta-analysis. |
  | Table 1 Main characteristics and results of the eligible studies |
Meta-analysis results
The main results of this meta-analysis are listed in Table 2. Our analysis suggested that LSD1 overexpression was associated with poor OS in cancer patients (HR =1.80, 95% CI: 1.39–2.34, P=0.000) with heterogeneity (I2=53.6%, P=0.028) (Figure 2).
  | Table 2 Main meta-analysis results of LSD1 expression in cancer patients |
  | Figure 2 Forest plots for the relationship between LSD1 expression and overall survival. |
To explain the heterogeneity in OS, subgroup analysis was performed by ethnicity, cancer type, and HR estimate. Subgroup analysis by ethnicity suggested a significant association in Asian patients (HR =1.97, 95% CI: 1.61–2.41, P=0.000). When grouped according to cancer type, a significant relationship between LSD1 expression and OS was observed in esophageal cancer patients (HR =1.77, 95% CI: 1.34–2.33, P=0.000). When stratifying by HR estimate, significant relevance was observed both in “reported directly from articles” subgroup (HR =1.63, 95% CI: 1.17–2.29, P=0.004) and “survival curves” subgroup (HR =2.20, 95% CI: 1.63–2.96, P=0.000).
Sensitivity analysis and publication bias
Sensitivity analysis indicated that the pooled HRs were not significantly influenced by omitting any single study (Figure 3). The shape of the funnel plot did not reveal any evidence of obvious asymmetry (Figure 4). The P-value of Egger’s regression intercept was 0.134, indicating that there was no significant publication bias in the meta-analysis.
  | Figure 3 Sensitivity analysis for meta-analysis of LSD1. |
  | Figure 4 Funnel plot of publication bias on the relationship between LSD1 expression and overall survival. |
Discussion
LSD1 consists of several domains, including an N-terminal SWIRM domain, a conserved motif shared by many chromatin regulatory complexes, an amine oxidase domain, and a C-terminal tower domain.22–24 It cooperates with the CoREST and CtBP24 corepressor complex and demethylates histone H3K4 and H3K9 through this interaction.25,26 Epigenetic changes in LSD1 have been shown to play a key role in carcinogenesis.27 LSD1 can prevent the accumulation of the dimethyl groups of p53, repressing p53-mediated transcriptional upregulation, preventing apoptosis, and contributing to human carcinogenesis via a chromatin modification mechanism. Recently, many studies have been carried out to identify the prognostic role of LSD1 in various cancers. Zhao et al8 demonstrated that high-level LSD1 predicts unfavorable overall survival in hepatocellular carcinoma patients (HR =2.456, 95% CI: 1.234–3.932, P<0.001). Similar results were obtained in reports by Lin et al10 and Yuan et al11 with pooled HR for OS 1.645 (95% CI: 1.182–2.500, P=0.020) and 3.908 (95% CI: 1.238–12.339, P=0.020), respectively. However, insignificant or opposite results were also observed in some studies. Since the prognostic value of LSD1 for tumor patients remains controversial, a meta-analysis was needed to explore the issue clearly.
To the best of our knowledge, this is the first meta-analysis focused on the association between LSD1 expression and patient survival. The present study pooled the survival data of 1,149 cancer patients from nine studies and found that LSD1 overexpression was associated with poor OS in cancer patients (HR =1.80, 95% CI: 1.39–2.34, P=0.000). The subgroup analyses grouped by ethnicity, cancer type, and HR estimate were consistent with the overall analysis. It may suggest that detected LSD1 expression could be a prognostic factor in cancers.
Our meta-analysis also has several limitations that should be acknowledged. First, only one study focused on Caucasian patients, which made it difficult to draw a firm conclusion on the prognostic value of LSD1 for Caucasian patients. Second, the number of prognostic studies dealing with each type of cancer was relatively small, which might weaken the reliability of our results. Moreover, well-designed clinical studies with a large number of cases for each specific cancer should be performed in the future to validate the relationship between LSD1 expression level and prognosis of patients with cancer. Third, although the method for detecting LSD1 level in all included studies was immunohistochemistry, it was difficult to follow entirely consistent monitoring standards for the dyeing process, antibody concentration, and cutoff value of different tissues. Fourth, we extracted data from survival curves because not all survival data of the enrolled studies were presented directly. These calculated HRs with their 95% CIs might be less reliable than the directly given data.
Conclusion
The present meta-analysis indicated that increased LSD1 level was significantly associated with poor OS. More multicenter clinical investigations with larger sample sizes should be conducted to confirm these findings.
Disclosure
The authors report no conflicts of interest in this work.
References
Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. | ||
Wang J, Scully K, Zhu X, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446(7138):882–887. | ||
Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J Biol Chem. 2007;282(21):15471–15475. | ||
Hayami S, Kelly JD, Cho HS, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128(3):574–586. | ||
Lim S, Janzer A, Becker A, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31(3):512–520. | ||
Lv T, Yuan D, Miao X, et al. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One. 2012;7(4):e35065. | ||
Ding J, Zhang Z, Liao G, et al. Positive expression of LSD1 and negative expression of E-cadherin correlate with metastasis and poor prognosis of colon cancer. Dig Dis Sci. 2013;58(6):1581–1589. | ||
Zhao ZK, Yu HF, Wang DR, et al. Overexpression of lysine specific demethylase 1 predicts worse prognosis in primary hepatocellular carcinoma patients. World J Gastroenterol. 2012;18(45): 6651–6656. | ||
Yu Y, Wang B, Zhang K, et al. High expression of lysine-specific demethylase 1 correlates with poor prognosis of patients with esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2013;437(2):192–198. | ||
Lin S, Chen Y, Zhu W, et al. Correlation between LSD1 expression and prognosis in patients with esophageal squamous cell carcinoma. Chin J Cancer Prev Treat. 2014;21(4):280–283. | ||
Yuan C, Li Z, Qi B, et al. High expression of the histone demethylase LSD1 associates with cancer cell proliferation and unfavorable prognosis in tongue cancer. J Oral Pathol Med. 2015;44(2):159–165. | ||
Derr RS, van Hoesel AQ, Benard A, et al. High nuclear expression levels of histone-modifying enzymes LSD1, HDAC2 and SIRT1 in tumor cells correlate with decreased survival and increased relapse in breast cancer patients. BMC Cancer. 2014;14:604. | ||
Chen L, Xu Y, Xu B, et al. Over-expression of lysine-specific demethylase 1 predicts tumor progression and poor prognosis in human esophageal cancer. Int J Clin Exp Pathol. 2014;7(12): 8929–8934. | ||
Miura S, Maesawa C, Shibazaki M, et al. Immunohistochemistry for histone h3 lysine 9 methyltransferase and demethylase proteins in human melanomas. Am J Dermatopathol. 2014;36(3):211–216. | ||
Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012. | ||
Parmar MK, Torri V Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. | ||
Williamson PR, Smith CT, Hutton JL, et al. Aggregate data meta-analysis with time-to-event outcomes. Stat Med. 2002;21(22):3337–3351. | ||
Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. | ||
Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25(1):1–14. | ||
Mimasu S, Sengoku T, Fukuzawa S, et al. Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25 A. Biochem Biophys Res Commun. 2008;366(1):15–22. | ||
Yang M, Culhane JC, Szewczuk LM, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14(6):535–539. | ||
Lee MG, Wynder C, Cooch N, et al. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. | ||
Gatta R, Mantovani R. NF-Y substitutes H2A-H2B on active cell-cycle promoters: recruitment of CoREST-KDM1 and fine-tuning of H3 methylations. Nucleic Acids Res. 2008;36(20):6592–6607. | ||
Lim S, Metzger E, Schule R, et al. Epigenetic regulation of cancer growth by histone demethylases. Int J Cancer. 2010;127(9):1991–1998. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
