Back to Journals » OncoTargets and Therapy » Volume 11
Prognostic and clinicopathological significance of DEPTOR expression in cancer patients: a meta-analysis
Authors Hu B, Shi D, Lv X, Wu F, Chen S, Shao Z
Received 6 March 2018
Accepted for publication 22 June 2018
Published 22 August 2018 Volume 2018:11 Pages 5083—5092
DOI https://doi.org/10.2147/OTT.S167355
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yao Dai
Binwu Hu,1 Deyao Shi,1 Xiao Lv,1 Fashuai Wu,1 Songfeng Chen,2 Zengwu Shao1
1Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; 2Department of Orthopaedic Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Background: DEP domain containing mammalian target of rapamycin (mTOR)-interacting protein (DEPTOR), a recently discovered endogenous inhibitor of mTOR, has been found to be abnormally expressed in various tumors. Recent studies have demonstrated that DEPTOR could serve as a potential prognostic biomarker in several kinds of cancer. However, the prognostic value of DEPTOR is still controversial so far.
Patients and methods: PubMed, Embase and Web of Science were systematically searched to obtain all relevant articles about the prognostic value of DEPTOR in cancer patients. ORs or HRs with corresponding 95% CIs were pooled to estimate the association between DEPTOR expression and the clinicopathological characteristics or survival of cancer patients.
Results: A total of nine eligible studies with 974 cancer patients were included in our meta-analysis. Our results demonstrated that the expression of DEPTOR was not associated with the overall survival (OS) (pooled HR=0.795, 95% CI=0.252–2.509) and event-free survival (EFS) (pooled HR=1.244, 95% CI=0.543–2.848) in cancer patients. Furthermore, subgroup analysis divided by sample size, type of cancer, Newcastle–Ottawa Scale (NOS) score and evaluation of DEPTOR expression showed identical prognostic value. In addition, our analysis also revealed that there was no significant association between expression level of DEPTOR and clinicopathological characteristics, such as tumor stage, lymph node metastasis, differentiation grade and gender.
Conclusion: Our meta-analysis suggested that despite the fact that DEPTOR could be overexpressed or downregulated in cancer patients, it might not be a potential marker to predict the prognosis of cancer patients.
Keywords: DEPTOR, cancer, overall survival, event-free survival, meta-analysis
Introduction
DEP domain containing mammalian target of rapamycin (mTOR)-interacting protein (DEPTOR), a 46 kDa mTOR-binding protein encoded by DEPTOR gene located on the 8q24 region, is primordially found overexpressed in a subset of multiple myeloma (MM) cells.1,2 As the component of mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), DEPTOR exerts its biological functions through inhibiting the activation of them.3 Meanwhile, mTOR can negatively regulate the expression and function of DEPTOR at the transcriptional and posttranslational levels in turn.2 Previous studies have proven that the mTOR signaling pathway is implicated in a wide spectrum of diseases.4 Thus, given the negative feedforward loop regulation between DEPTOR and mTOR, DEPTOR is also considered important in the pathogenesis of many diseases. Correspondingly, many researchers have demonstrated that DEPTOR is involved in cell growth, proliferation, autophagy, apoptosis, transcription regulation and inflammation.5 In addition, accumulating evidence has suggested that DEPTOR plays pivotal roles in tumorigenesis, and the abnormal expression of DEPTOR has been detected in many kinds of tumor, such as MM, breast cancer, prostate cancer and lung cancer.6
Recent studies have revealed that abnormally expressed DEPTOR might be related to the poor prognosis of cancer patients.7–9 However, despite the development of basic and clinical researches about the biological functions of DEPTOR, the prognostic value of abnormally expressed DEPTOR across different tumors is still controversial. The primary problem is that DEPTOR displays variable expression levels in different tumors. Previous studies have revealed that DEPTOR is overexpressed in some kinds of tumor such as MM, cervical cancer, ovarian cancer, thyroid carcinoma, osteosarcoma and T-cell leukemia where its overexpression is essential for cell proliferation and survival.2,10–14 Nevertheless, several other researchers also show that DEPTOR is downregulated in pancreatic ductal adenocarcinoma, esophageal squamous cell carcinoma, colorectal cancer and liver cancer, which indicates quick tumor progression and poor prognosis.7,8,15,16 Thus, DEPTOR may act as a tumor suppressor gene or oncogene depending on the specific tumor type.6 Furthermore, the specific relationship between DEPTOR expression levels and the clinical outcome of cancer patients is also perplexing. Because DEPTOR binds and inhibits the activation of mTOR whose activation is proven to be associated with poor survival of cancer patients, downregulation of DEPTOR is presumed to predict poor prognosis of cancer patients.17 Accordingly, clinical data have illustrated that a lower expression of DEPTOR in MM predicts poor prognosis of patients.18,19 On the other hand, almost opposite results are observed in breast cancer, hepatocellular carcinoma and differentiated thyroid carcinoma, in which the lower expression of DEPTOR is favorable for the outcome of patients.12,20,21 What is more interesting is that even in the same type of tumor, there are almost opposite conclusions about the prognostic value of DEPTOR.8,22,23 Taken together, whether DEPTOR could be regarded as a prognostic biomarker and whether the high or low expression of DEPTOR is more adverse for the prognosis of cancer patients remain unknown. Therefore, in order to obtain a better understanding of the prognostic value of DEPTOR, we performed a quantitative meta-analysis to elucidate the prognostic and clinicopathological significance of DEPTOR expression in patients with cancer.
Patients and methods
Study strategy
The present review was performed in accordance with the standard guidelines for meta-analyses and systematic reviews of tumor marker prognostic studies.24,25 The databases PubMed, Embase and Web of Science were independently searched by two researchers (Binwu Hu and Deyao Shi) to obtain all relevant articles about the prognostic value of abnormally expressed DEPTOR in patients with any type of tumor. The literature search ended on February 1, 2018. The search strategy used both MeSH terminology and free-text words to increase the sensitivity of the search. The following search terms were used: “DEPTOR”, “DEP-domain containing mTOR-interacting protein” and “DEPDC6”. We also screened the references of retrieved relevant articles to identify potentially eligible literatures. Conflicts were solved through group discussion.
Inclusion and exclusion criteria
Studies included in this analysis had to meet the following inclusion criteria: 1) patients were pathologically diagnosed with any type of malignant cancer or neoplasm; 2) DEPTOR expression was determined in human tissues or plasma samples using any technique; 3) patients were divided into high and low expression groups or positive and negative expression groups; the relationship between DEPTOR expression levels and survival outcome was investigated and 4) sufficient published data or the survival curves were provided to calculate HRs for survival rates and their 95% CIs. Exclusion criteria were as follows: studies using nonhuman samples, studies without usable or sufficient data, laboratory articles, reviews, letters, non-English or unpublished articles and conference abstracts. All eligible studies were carefully screened by the same two researchers (Binwu Hu and Deyao Shi), and discrepancies were resolved by discussing with a third researcher (Xiao Lv).
Data extraction
Two investigators (Fashuai Wu and Songfeng Chen) extracted relevant data independently and reached a consensus on all items. For all eligible studies, the following information of each article was collected: author, year of publication, tumor type, characteristics of the study population including country of the population enrolled, sample size, endpoints, assay method, cutoff value, evaluation of DEPTOR expression, Newcastle–Ottawa Scale (NOS) score and source of HR. For endpoints, overall survival (OS), disease-free survival (DFS) and progression-free survival (PFS) were all regarded as end points. In addition, DFS and PFS were redefined as event-free survival (EFS) in our article. We used HR, which was extracted following a methodology suggested previously to evaluate the influence of DEPTOR expression on prognosis of patients.26 If possible, we also asked for original data directly from the authors of the relevant studies.
Quality assessment
Quality of all included studies was assessed independently by three researchers (Binwu Hu, Deyao Shi and Fashuai Wu) using the validated NOS, and disagreements were resolved through discussion with another researcher (Xiao Lv). We considered studies with scores >6 as high-quality studies, and those with scores ≤6 as low-quality studies.
Statistical analyses
Pooled HRs (low/high) and their associated 95% CIs were used to analyze the prognostic value of DEPTOR expression in cancer patients. Pooled ORs (low/high) and their associated 95% CIs were used to analyze the association between DEPTOR expression levels and clinicopathological parameters. The heterogeneity among studies was evaluated using Cochran’s Q and I2 statistics. A P-value of <0.10 or an I2 value of >50% was considered as statistically significant. The fixed effect model was used for analysis without significant heterogeneity among studies (P>0.10, I2<50%). Otherwise, the random effect model was chosen. To explore the source of heterogeneity, subgroup analysis was preformed through classifying the included studies into subgroups according to similar features. We also conducted sensitivity analysis to test the effect of each study on the overall pooled results. In addition, for the studies from which we could obtain clinicopathological characteristics, we calculated the pooled ORs to analyze the relationship between DEPTOR expression levels and clinicopathological characteristics. Owing to the limited number of studies (less than 10) included in this analysis, publication bias was not assessed. Statistical analysis was performed using Stata software 14.0 (StataCorp LP, College Station, TX, USA), and a P-value of <0.05 was considered as significant.
Results
Characteristics of studies
According to our search strategy, a total of 552 studies were retrieved. Among these researches, the following studies were excluded: duplicates (n=310), reviews (n=19), letters (n=1), patents (n=6), meeting abstracts (n=66), studies describing noncancer topics (n=88), studies belonging to basic research (n=45) and studies lacking relevant data (n=8). Eventually, nine studies meeting the inclusion criteria were included in this meta-analysis. The screening process and results are shown in Figure 1, and the main characteristics of the included studies are shown in Table 1. Among these studies, a total of 974 patients were included, with a maximum sample size of 220 and a minimum sample size of 24 patients (mean=108.0). The accrual period of these studies ranged from 2011 to 2017. The regions represented in the studies included the Asian origin (six) and Caucasian descent (three). Six different types of cancer were evaluated with the greatest number being digestive system malignancies (three esophageal squamous cell carcinoma, one hepatocellular carcinoma and one colorectal cancer). Other types of cancer were also included (two MM, one breast cancer and one differentiated thyroid carcinoma). Among these studies, OS, DFS and PFS were estimated as survival outcome in 78% (7/9), 22% (2/9) and 22% (2/9) of the studies, respectively. DFS and PFS were combined together into EFS, which was regarded as a prognostic parameter in our study. To evaluate the expression of DEPTOR, six studies used immunohistochemistry (IHC), while GeneChip, microarray and capillary electrophoresis immunoassays were also applied. Because the cutoff definitions were various, the cutoff values were different in these studies.
  | Figure 1 The flow diagram indicated the process of study selection. |
Association between DEPTOR expression levels and OS of cancer patients
Among the nine included articles, seven studies involving 789 patients reported the relationship between abnormal expression levels of DEPTOR and OS of cancer patients. Owing to the heterogeneity, we used the random effect model to calculate the pooled HR. The pooled HR for OS was 0.795 (95% CI=0.252–2.509, P=0.696), which indicated that there was no significant association between expression levels of DEPTOR and the OS of cancer patients (Figure 2). Given that significant heterogeneity existed among studies (χ2=51.97, P<0.0001, I2=88.5%), we further conducted subgroup analyses by factors of sample size (>100 or <100), type of cancer (digestive system or nondigestive system malignancies), paper quality (NOS scores ≥7 or <7) and evaluation of DEPTOR expression (high/low or positive/negative) to explore the sources of heterogeneity (Figure 3). The subgroup analyses illustrated almost the same results that there was no significant difference in the influences of high or low expression of DEPTOR on the prognosis of cancer patients in the subgroup of all above factors (Table 2), for sample size, >100 (HR=0.332, 95% CI=0.092–1.195, P=0.092) and <100 (HR=1.406, 95% CI=0.351–5.629, P=0.631; Figure 3A); for type of cancer, digestive system (HR=0.755, 95% CI=0.195–2.921, P=0.684) and nondigestive system malignancies (HR=0.902, 95% CI=0.056–14.41, P=0.942; Figure 3B); for paper quality, NOS scores ≥7 (HR=0.619, 95% CI=0.131–2.925, P=0.545) and NOS scores <7 (HR=1.153, 95% CI=0.167–7.984, P=0.885; Figure 3C) and for evaluation of DEPTOR expression, high/low (HR=0.470, 95% CI=0.085–2.587, P=0.385) and positive/negative (HR=1.225, 95% CI=0.343–4.368, P=0.755; Figure 3D). To further explore the sources of heterogeneity, we performed meta-regression by the covariates including above factors. Meta-regression revealed that all above factors were not the sources of heterogeneity (Table 2).
  | Figure 2 Meta-analysis of the pooled HRs of OS for cancer patients. |
  | Table 2 Subgroup analyses of pooled HRs for OS in cancer patients with abnormal expression level of DEPTOR |
Association between DEPTOR expression levels and EFS of cancer patients
A total of four studies including 370 patients reported the impact of abnormally expressed DEPTOR on DFS or PFS of cancer patients. In the current study, we defined DFS and PFS as EFS. The consequence displayed that high or low expression of DEPTOR made no difference in predicting the EFS of cancer patients (HR=1.244, 95% CI=0.543–2.848, P=0.606; Figure 4). There was significant heterogeneity among studies (χ2=27.87, P<0.0001, I2=89.2%). However, due to the limited number of included studies, we did not perform the subgroup analyses.
  | Figure 4 Meta-analysis of the pooled HRs of EFS for cancer patients. |
Association between DEPTOR expression levels and clinicopathological characteristics of cancer patients
As shown in Table 3, we analyzed the association between DEPTOR expression levels and clinicopathological characteristics of cancers patients. The meta-analytic results showed that there was no significant association between expression levels of DEPTOR and differentiation grade (OR=1.210, 95% CI=0.776–1.886, P=0.401), lymph node metastasis (OR=1.021, 95% CI=0.339–3.072, P=0.971), tumor stage (OR=1.995, 95% CI=0.594–6.695, P=0.264) and gender (OR=1.002, 95% CI=0.739–1.358, P=0.990), which was consistent with the results of prognostic analyses.
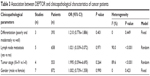  | Table 3 Association between DEPTOR and clinicopathological characteristics of cancer patients |
Sensitivity analysis and publication bias
Sensitivity analysis was performed to examine the effects of individual study on the overall results. For OS, sensitivity analysis showed that HRs and their 95% CIs did not change significantly after the exclusion of any of the studies (Figure 5A), which indicated that individual study had little influence on our eventual outcome, and proved that our analysis was relatively stable and credible. For EFS, the sensitivity analysis identified that results from Parvani et al and Pei et al affected results greatly, indicating that these studies were possible to be the main source of heterogeneity. However, after excluding either of them, we still observed that overexpression or low expression of DEPTOR made no difference in predicting the EFS of cancer patients (Figure 5B). As for publication bias analysis, because of the limited number of studies included in each analysis (<10), publication bias was not assessed.
  | Figure 5 Sensitivity analysis plot of pooled HRs of OS (A) and EFS (B) for cancer patients with abnormally expressed DEPTOR. |
Discussion
DEPTOR is a recently discovered endogenous inhibitor of mTOR, which is initially identified as being overexpressed in a subset of MM cells.2 As a natural inhibitor of mTOR, DEPTOR could suppress the activity of both mTORC1 and mTORC2. In addition, the aberrant expression of DEPTOR could induce cell growth, apoptosis, autophagy and endoplasmic reticulum stress response.27,28 Accumulating studies have revealed that DEPTOR could be abnormally expressed in numerous kinds of tumor and plays pivotal roles in the pathogenesis and progression of tumor.5,28–30 Nevertheless, the relationship between abnormally expressed DEPTOR with the prognosis of cancer patients is still controversial, especially when it comes to whether the high or low expression of DEPTOR is more adverse.
Here, we performed current meta-analysis to explore the prognostic value of abnormally expressed DEPTOR and the relation between DEPTOR expression levels and clinicopathological characteristics of cancer patients. Through systematic analysis, our results demonstrated that there was no significant difference in the influences of high or low expression of DEPTOR on the OS of cancer patients. In addition, the subgroup analyses and meta-regression analysis displayed that factors including sample size, type of cancer, paper quality and evaluation of DEPTOR expression did not alter above results. DFS and PFS are important parameters reflecting the progression of tumor. In this article, we defined DFS and PFS as EFS. By combining the HRs, we found a similar result that there was no difference in predicting the EFS of cancer patients for high or low expression of DEPTOR.
Our results were consistent with some previous discoveries from Lai et al7 and Dong et al22 that in colorectal cancer and esophageal squamous cell carcinoma, high or low expression of DEPTOR made no difference in predicting the prognosis of cancer patients. However, on the other hand, former studies have also demonstrated that overexpression of DEPTOR was worse for outcome of patients in triple-negative breast cancer, hepatocellular carcinoma and differentiated thyroid carcinoma.12,20,21 In addition, results from Quwaider et al19 and Boyd et al18 showed that a lower expression of DEPTOR was more adverse for MM. Thus, we speculated that the different prognostic roles of DEPTOR in different tumors might be because of the limitation of the sample size and the different clinicopathological characteristics of the patients recruited. Dong et al22 have shown that OS did not differ significantly between DEPTOR expression levels of all patients, while high expression of DEPTOR benefited patients in the early stage but not advanced stage of esophageal squamous cell carcinoma. Therefore, large sample sizes studies are desperately needed to elucidate the impact of abnormally expressed DEPTOR on the prognosis of cancer patients.
As for the clinicopathological characteristics, our analysis also revealed that there was no significant relation between expression levels of DEPTOR and clinicopathological characteristics, including tumor stage, lymph node metastasis, differentiation grade and gender, which was consistent with the prognostic value of DEPTOR.
Mechanisms underlying the regulatory roles of DEPTOR in tumorigenesis and tumor progression have been extensively investigated. DEPTOR is an endogenous inhibitor of mTOR. Despite the fact that DEPTOR could be overexpressed or downregulated in tumor tissues, previous studies have proven that both overexpression and low expression of DEPTOR could active the PI3K/AKT pathway. Downregulation of DEPTOR could active the PI3K/AKT pathway directly via promoting the mTOR activity.8 On the other hand, overexpression of DEPTOR could inhibit mTORC1, which relieves the inhibitory feedback signal normally transmitted from mTORC1 to PI3K, leading to hyperactive PI3K signaling.2 A meta-analysis conducted by Ocana et al17 have demonstrated that activation of the PI3K/mTOR/AKT pathway was associated with significantly worse 5-year survival of solid tumor. Considering that both overexpression and low expression of DEPTOR could active the PI3K/AKT pathway, it was understandable that high and low expression levels of DEPTOR might make no difference in predicting the prognosis and clinicopathological characteristics of cancer patients.
In our study, a few limitations should be underlined. First, only nine studies were included in our meta-analysis and even fewer articles, seven and four articles, respectively, were included for the OS and EFS analyses; this restricted our ability to evaluate the prognostic value of DEPTOR in subgroup analyses and might have led to the bias of the results. Second, due to the limited number of included studies, we could not perform the publication bias analysis, which was possible to exist in our meta-analysis. Third, the cutoff values of overexpression or low expression of DEPTOR were different among studies, although most of them were set to median or a result compared with adjacent normal tissues. Fourth, differences in paper quality and sample size across the studies might cause bias in the meta-analysis, although subgroup analyses and meta-regression did not show the paper quality or sample size as the resource of heterogeneity. Fifth, some HRs could not be directly obtained from the publications. Thus, calculating them through survival curves might not be precise enough. Sixth, significant heterogeneity existed among our analysis, which might cause bias of results. Therefore, larger scale, multicenter and high-quality studies are desperately necessary to confirm our findings.
Conclusion
Our study revealed that despite the fact that DEPTOR could be overexpressed or downregulated in cancer patients, it might not be an appropriate biomarker to predict the prognosis of cancer patients. Moreover, the expression level of DEPTOR was not associated with clinicopathological features including TNM stage, lymph node metastasis, differentiation grade and gender. This is the first meta-analysis to evaluate the relationship between expression levels of DEPTOR and prognosis of cancer patients. In the future, more relevant studies are warranted to investigate the role of DEPTOR in human cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
Panic N, Larghi A, Amore R, et al. Single nucleotide polymorphisms within the 8Q24 region are not associated with the risk of intraductal papillary mucinous neoplasms of the pancreas. J Gastrointestin Liver Dis. 2016;25(3):311–315. | ||
Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. | ||
Duan S, Skaar JR, Kuchay S, et al. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell. 2011;44(2):317–324. | ||
Alayev A, Holz MK. mTOR signaling for biological control and cancer. J Cell Physiol. 2013;228(8):1658–1664. | ||
Catena V, Fanciulli M. Deptor: not only a mTOR inhibitor. J Exp Clin Cancer Res. 2017;36(1):12. | ||
Wang Z, Zhong J, Inuzuka H, et al. An evolving role for DEPTOR in tumor development and progression. Neoplasia. 2012;14(5):368–375. | ||
Lai EY, Chen ZG, Zhou X, et al. DEPTOR expression negatively correlates with mTORC1 activity and tumor progression in colorectal cancer. Asian Pac J Cancer Prev. 2014;15(11):4589–4594. | ||
Ji YM, Zhou XF, Zhang J, et al. DEPTOR suppresses the progression of esophageal squamous cell carcinoma and predicts poor prognosis. Oncotarget. 2016;7(12):14188–14198. | ||
Quwaider D, Corchete LA, Misiewicz-Krzeminska I, et al. DEPTOR maintains plasma cell differentiation and favorably affects prognosis in multiple myeloma. J Hematol Oncol. 2017;10(1):92. | ||
Srinivas KP, Viji R, Dan VM, et al. DEPTOR promotes survival of cervical squamous cell carcinoma cells and its silencing induces apoptosis through downregulating PI3K/AKT and by up-regulating p38 MAP kinase. Oncotarget. 2016;7(17):24154–24171. | ||
Rogers-Broadway KR, Chudasama D, Pados G, et al. Differential effects of rapalogues, dual kinase inhibitors on human ovarian carcinoma cells in vitro. Int J Oncol. 2016;49(1):133–143. | ||
Pei L, Xie P, Zhou E, Yang Q, Luo Y, Tang Z. Overexpression of DEP domain containing mTOR-interacting protein correlates with poor prognosis in differentiated thyroid carcinoma. Mol Med Rep. 2011;4(5):817–823. | ||
Hu Y, Su H, Ye Q, et al. DEPTOR is a direct NOTCH1 target that regulates proliferation, metabolism and glucocorticoid resistance in T cell leukemia. Cancer Research Conference: 106th Annual Meeting of the American Association for Cancer Research, AACR. Vol. 75. 2015. | ||
Hu B, Lv X, Gao F, et al. Downregulation of DEPTOR inhibits the proliferation, migration, and survival of osteosarcoma through PI3K/Akt/mTOR pathway. Onco Targets Ther. 2017;10:4379–4391. | ||
Li H, Sun GY, Zhao Y, et al. DEPTOR has growth suppression activity against pancreatic cancer cells. Oncotarget. 2014;5(24):12811–12819. | ||
Obara A, Fujita Y, Abudukadier A, et al. DEPTOR-related mTOR suppression is involved in metformin’s anti-cancer action in human liver cancer cells. Biochem Biophys Res Commun. 2015;460(4):1047–1052. | ||
Ocana A, vera-Badillo F, Al-Mubarak M, et al. Activation of the PI3K/mTOR/AKT pathway and survival in solid tumors: systematic review and meta-analysis. PLoS One. 2014;9(4):e95219. | ||
Boyd KD, Walker BA, Wardell CP, et al. High expression levels of the mammalian target of rapamycin inhibitor DEPTOR are predictive of response to thalidomide in myeloma. Leuk Lymphoma. 2010;51(11):2126–2129. | ||
Quwaider D, Herrero AB, Krzeminski P, et al. A novel function of DEPTOR in multiple myeloma: commitment to plasma cell maturation. Clin Lymphoma Myeloma Leuk. 2015;15:e215–e216. | ||
Parvani JG, Davuluri G, Wendt MK, et al. Deptor enhances triple-negative breast cancer metastasis and chemoresistance through coupling to survivin expression. Neoplasia. 2015;17(3):317–328. | ||
Yen CH, Lu YC, Li CH, et al. Functional characterization of glycine N-methyltransferase and its interactive protein DEPDC6/DEPTOR in hepatocellular carcinoma. Mol Med. 2012;18:1–296. | ||
Dong X, Wang L, Han Z, et al. Different functions of DEPTOR in modulating sensitivity to chemotherapy for esophageal squamous cell carcinoma. Exp Cell Res. 2017;353(1):35–45. | ||
Liu NB, Zhang JH, Liu YF, et al. High DEPTOR expression correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Onco Targets Ther. 2015;8:3449–3455. | ||
Altman DG, Mcshane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med. 2012;9(5):e1001216. | ||
Mcshane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97(16):1180–1184. | ||
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. | ||
Zhao Y, Xiong X, Sun Y, Deptor SY. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44(2):304–316. | ||
Catena V, Bruno T, de Nicola F, et al. Deptor transcriptionally regulates endoplasmic reticulum homeostasis in multiple myeloma cells. Oncotarget. 2016;7(43):70546–70558. | ||
Ohshima K, Nojima S, Tahara S, et al. Argininosuccinate synthase 1-deficiency enhances the cell sensitivity to arginine through decreased DEPTOR expression in endometrial cancer. Sci Rep. 2017;7:45504. | ||
Foster H, Coley HM, Goumenou A, Pados G, Harvey A, Karteris E. Differential expression of mTOR signalling components in drug resistance in ovarian cancer. Anticancer Res. 2010;30(9):3529–3534. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


