Back to Journals » International Journal of Nanomedicine » Volume 12
Preparation, characterization, and transfection efficiency of low molecular weight polyethylenimine-based nanoparticles for delivery of the plasmid encoding CD200 gene
Authors Nouri F , Sadeghpour H, Heidari R , Dehshahri A
Received 29 April 2017
Accepted for publication 6 July 2017
Published 3 August 2017 Volume 2017:12 Pages 5557—5569
DOI https://doi.org/10.2147/IJN.S140734
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster

Fatemeh Nouri,1 Hossein Sadeghpour,2 Reza Heidari,3 Ali Dehshahri,1,4,5
1Department of Pharmaceutical Biotechnology, 2Department of Medicinal Chemistry, 3Department of Pharmacology and Toxicology, School of Pharmacy, 4Center for Nanotechnology in Drug Delivery, School of Pharmacy, 5Pharmaceutical Sciences Research Center, Shiraz University of Medical Sciences, Shiraz, Iran
Abstract: Various strategies have been utilized to improve both gene transfer efficiency and cell-induced toxicity of polyethylenimine (PEI), the most extensively investigated cationic polymeric vector. In this study, we sought to enhance transfection efficiency of low molecular weight PEI (LMW PEI) while maintaining its low toxicity by cross-linking LMW PEI via succinic acid linker. These modifications were designed to improve the hydrophilic–hydrophobic balance of the polymer, by enhancing the buffering capacity and maintaining low cytotoxic effects of the final conjugate. Decreased expression of CD200 in the central nervous system has been considered as one of the proposed mechanisms associated with neuroinflammation in multiple sclerosis; therefore, we selected plasmid-encoding CD200 gene for transfection using the modified PEI derivatives. Dynamic light scattering experiments demonstrated that the modified PEIs were able to condense plasmid DNA and form polyplexes with a size of approximately 130 nm. The highest level of CD200 expression was achieved at a carrier to plasmid ratio of 8, where the expression level was increased by 1.5 fold in the SH-SY5Y cell line, an in vitro model of neurodegenerative disorders. Furthermore, the results of in vivo imaging of the LMW PEI-based nanoparticles in the mouse model of multiple sclerosis revealed that fluorescently labeled plasmid encoding CD200 was distributed from the injection site to various tissues and organs including lymph nodes, liver, brain, and finally, kidneys. The nanoparticles also showed the ability to cross the blood–brain barrier and enter the periventricular area.
Keywords: polyethylenimine, nanoparticle, transfection, CD200, gene delivery
Introduction
The potential of gene therapy in the prevention and treatment of various diseases, including cancer, multiple sclerosis (MS), hemophilia, cystic fibrosis, diabetes, as well as Parkinson and Alzheimer diseases has led researchers to consider gene therapy as a new paradigm in modern medicine.1–3 Transfer of genetic material in the absence of a carrier (naked DNA delivery) is a simple technique. Despite this, the majority of the research is being conducted on vectorization of nucleic acid therapeutics. This is because naked DNA is highly susceptible to the degradation enzymes, which causes its rapid clearance from blood circulation following the systemic administration.4 Despite the remarkable intrinsic capability of viral gene carrier DNAs in the transfection of various genetic materials into different cells and tissues, their immunogenicity, oncogenicity, low-carrying capacity, and expensive production procedures have resulted in attempts to utilize novel non-viral gene vectors.5,6 There are several different reports demonstrating the ability of nanoparticle-forming polymers in the delivery of nucleic acid therapeutics with high efficiency and low toxicity. Among the different polycationic compounds used in transfection, polyethylenimine (PEI) has been considered as the most extensively investigated cationic polymer due to its ability to interact with the negatively charged backbone of the nucleic acids and form nano-sized particles.7–9 The characteristics of PEI are highly related to the high amine content, which causes a significant positive charge density on the surface of the polymer.10,11 Thus, PEI can interact with the negatively charged components of the cell membrane, which results in its translocation into the cell. Furthermore, the substantial amine content plays a critical role in the early escape of the PEI-based formulations from enzymatic degradation inside the endosomes/lysosomes.12 However, a major disadvantage of PEI-based nanoparticles is their toxicity, which in turn, raises concerns about their application in human gene therapy.13 There are several factors that determine cytotoxic effects of PEI, such as high molecular weight, increased branching, and greater density of positive charge on the structure of PEI polymer.14–16 Hence, several chemical modifications as well as conjugation of PEI with various chemical moieties have been examined in order to decrease the toxic effects of PEI.13,17–25 Since high molecular weight PEI induces more toxicity, there are several reports suggesting the application of low molecular weight PEI (LMW PEI) as the core of the transfection vector and modifying it to increase its transfection efficiency.15,26–32 Considering the effect of hydrophobic modification of PEI in increasing the transfection efficiency, we hypothesized that cross-linking LMW PEIs using potentially biodegradable linkages such as amides enhances its gene transfer ability with minimal toxic effects.
CD200 is a membrane glycoprotein which has been shown to cause immune suppression via its receptor CD200R.33,34 The interaction of CD200 and CD200R initiates tyrosine phosphorylation which finally leads to the inactivation of microglia. Microglia are the resident antigen presenting macrophages of the central nervous system (CNS) that play a key role in neuroimmune processes. Protection of CNS against neuronal stress needs an immune-privileged environment; therefore, it is suggested that the interaction between CD200 and CD200R is one of the factors that provide such an environment in the CNS.35 This can be confirmed by the fact that the other immune-suppressed organs such as placenta demonstrates high levels of CD200 expression.34 Furthermore, it has been shown that CD200-deficit mice are more susceptible to different types of inflammatory diseases including MS. Decreased expression of CD200 in MS lesions might promote microglial activation leading to various inflammatory responses; therefore, increased levels of CD200 in patients with MS might be considered as a strategy for neuroprotection.33,36,37
In this study, we hypothesized that cross-linking LMW PEI via amide linkages enhances its poor transfection efficiency while maintaining low cytotoxic effects. The synthesized PEI derivatives were characterized in terms of particle size, zeta potential, buffering capacity, plasmid condensation ability, and protection against nuclease degradation as well as cytotoxicity. Furthermore, we investigated the ability of modified PEI derivatives in transferring the plasmid encoding CD200 gene in SH-SY5Y cell line, an in vitro model of neurodegenerative disorders. We also investigated the ability of PEI conjugates to cross the blood–brain barrier (BBB) by performing in vivo imaging of nanoparticles using a mouse model of MS.
Materials and methods
Materials
Branched PEI (LMW PEI; average MW = 1,800 Da), succinic anhydride, MTT, N-hydroxybenzotriazole, 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride, 1-hydroxybenzotriazole hydrate, and N-[2-hydroxythyl] piperazine-N-[2-ethanesulfonic acid] (HEPES) were purchased from Sigma-Aldrich (Munich, Germany). Human CD200 plasmid (pCMV-XL5-hCD200) was obtained from OriGene (Rockville, MD, USA). Cell culture experiments were performed using FBS and DMEM (Gibco, Gaithersburg, MD, USA). Spectra/Por dialysis membrane was obtained from Spectrum Laboratories (Houston, TX, USA). TransformAid Bacterial Transformation Kit (K-2710) was purchased from Thermo Scientific Company (Hanover, MD, USA). Plasmid purification was performed by Qiagen Endofree Mega Plasmid Kit (Qiagen, Hilden, Germany). cDNA synthesis and Real Time PCR were performed using the PrimeScript™ RT reagent Kit (Perfect Real Time, TaKaRa, Dalian, People’s Republic of China), and RealQ Plus 2x Master Mix Green High ROX™ (AmpliQon, Denmark). All solvents and chemicals were purchased from Sigma-Aldrich (Munich, Germany) and were of the highest purity available.
Synthesis of succinated PEI
To synthesize the succinated PEI derivative, 1,800 Da branched PEI (0.5 g) was dissolved in 8 mL of water and 2 mL of NaCl solution (3 M). The pH of the solution was adjusted to 5 using hydrochloric acid (HCl, 1 M). Then, desired amounts of succinic anhydride were dissolved in DMSO and added to the stirred PEI solution at room temperature. The reaction was allowed to proceed for 3 h. The crude product was dialyzed first against NaCl (0.25 M) to remove unreacted succinate and then twice against double-distilled water followed by lyophilization.
Conjugation of LMW PEI to the succinated PEI
Briefly, succinated PEI (0.1 g) was dissolved in water and stirred with 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride solution for around 1 h. Then, desired amounts of LMW PEI and 1-hydroxybenzotriazole hydrate were dissolved in water and added dropwise to the succinated PEI solution at room temperature. The reaction mixture was allowed to proceed for 24 h with constant shaking. Finally, the reaction mixture was dialyzed against double-distilled water to remove unreacted materials. Following dialysis, the aqueous solutions were lyophilized. The fluffy materials were characterized by 1H-NMR (D2O) spectroscopy using a Bruker Avance DRX-500 MHz NMR spectrometer (Bruker, Ettlingen, Germany) and Fourier transform infrared (FTIR) spectroscopy. Furthermore, the primary amine content of PEI derivatives and the degree of conjugation was determined by 2,4,6-trinitrobenzenesulfonic acid assay.
Preparation and purification of the pDNA
pCMV-XL5-hCD200 plasmid was transfected into Escherichia coli strain DH5α. The transfected bacterial cells were propagated in selective Luria–Bertani medium and then were centrifuged. The cell pellets were used to extract pDNA, which was quantified by using Qiagen Endofree Mega Plasmid Kit according to the manufacturer’s instructions. Purity of the pDNA was measured using a UV spectrophotometer (PerkinElmer, USA). PDNA preparations with A260/A280 ratios around 1.8 were further used in this study.
Buffering capacity
The buffering capacity of PEI and its derivatives were determined by an acid–base titration experiment. Briefly, each PEI derivative (0.4 mg) was dissolved in deionized water and pH of each solution was adjusted to 12 by using 1 M NaOH. Then, polymer solutions were titrated sequentially with aliquots of 3 μL of 1 M HCl and the pH values were recorded using a pH meter (AZ instruments, Taiwan). The addition of HCl was stopped when the pH of the solution reached 2. The slope of plot of pH versus the amount of added HCl demonstrates the buffering capacity of different polymers.
Polyplex preparation
PEI derivatives/pDNA polyplexes were prepared in HBG buffer (HEPES buffered glucose solution; 20 mM HEPES, 5% glucose, pH =7.2) by adding 50 μL of the solution containing PEI conjugates at various concentrations to the same volume of pDNA (40 μg/mL) in the same buffer followed by pipetting up and down. This was incubated at room temperature for 20–30 min to obtain stable complexes. The composition of the prepared formulations was defined by carrier to plasmid (C/P) ratio in which C is the weight of PEI and its derivatives and P represents the weight of pDNA used in the formation of complex.
Ethidium bromide (EtBr) exclusion assay
Quantitative association of pDNA with PEI and its derivatives was measured using a DNA intercalating dye (EtBr). Exclusion of the dye from pDNA upon the addition of polymers leads to a decrease in fluorescence intensity demonstrating the ability of PEI and its derivatives to condense pDNA. A solution of the pDNA (400 ng/mL) and EtBr (0.4 mg/mL) was prepared in HBG buffer and its fluorescence intensity was measured and set to 100% using a spectrofluorometer (LS55, PerkinElmer, MA, USA). Then, PEI solution was added stepwise to the pDNA solution and the decrease in fluorescence intensity was recorded following each addition at an excitation wavelength 510 nm and emission wavelength 590 nm with a 5 nm slit width. The obtained graph represents the quantitative condensation ability of PEI derivatives at various C/P ratios. All measurements were performed in triplicates.
Gel retardation assay
Agarose gel (1%, w/v) containing 0.5 μg/mL GelRed (Biotium, Fremont, CA, USA) was prepared in buffer (Tris, acetate, and ethylenediaminetetraacetic acid at pH =8). Each PEI/pDNA complex was prepared by mixing 10 μL of pDNA solution (40 μg/mL) with an equal volume of the polymer solution at different C/P ratios. The samples (10 μL) were mixed with 2 μL of 6X loading dye (CinnaGen, Iran) and the mixtures were loaded onto an agarose gel. The gel was run for 30 min at 100 V and location of DNA bands was visualized using a UV illuminator.
Measurements of the size and zeta potential of the polyplexes
Particle size and surface charge of PEI/DNA complexes were evaluated using dynamic light scattering (DLS) and laser Doppler velocimetry, respectively, using Malvern Nano ZS (Malvern Instruments, Malvern, UK). Polymer/pDNA complexes were prepared in HBG buffer at C/P ratio of 8 by mixing equal volumes of buffer containing PEI and plasmid. Data were collected for 30 cycles in automatic mode, and the results are reported as mean ± standard error of mean (SEM) (n=3).
Resistance of pDNA against DNase I degradation
To demonstrate the ability of PEI and its derivatives in the protection of pDNA against enzymatic degradation, protection and release assay was performed as described elsewhere.38 Briefly, polyplexes were prepared at different C/P ratios (ranging from 0.25:1 to 8:1) and mixed with 1 μL of DNase I enzyme in PBS or DNase/Mg2+ digestion buffer (50 mM Tris–Cl, pH 7.6 and 10 mM MgCl2) and incubated for 30 min at 37°C. Instantly following the incubation, 4 mL of EDTA (250 mM) was added to inactivate the enzyme and then mixed with 1% sodium dodecyl sulfate (SDS), dissolved in 1 M NaOH (pH 7.2). To allow complete dissociation of the complexes, all the samples were incubated for approximately 2 h at room temperature. Finally, agarose gel electrophoresis was performed to visualize the location of plasmid bands.
Cell culture and cytotoxicity assay
SH-SY5Y human neuroblastoma cells (C611, NCBI, Tehran, Iran) were incubated at 37°C in a humidified (100% humidity) atmosphere of 5% CO2 in 100 μL DMEM (1 g/L glucose, 2 mM glutamine) supplemented with 10% FBS, 100 μg/mL streptomycin, and 100 IU/mL penicillin. The toxicity of PEI and its derivatives complexed with pDNA was evaluated using MTT assay. Cells were cultured at a density of 1×104 cells per well in a 96-well plate for 1 day before the evaluation of toxicity. To prepare the polyplexes at various C/P ratios of 0.25:1–8:1, desired concentrations of PEI and its derivatives in HBG buffer were prepared. Next, pDNA solutions (40 μg/mL) were prepared in the same buffer in separate tubes. Then, 50 μL of the PEI solution was added to the plasmid solution to prepare the polyplex formulations at the C/P ratios of 0.25, 4, and 8 at the final volume of 100 μL. MTT assay was performed by the addition of 10 μL of polyplex formulation to the 96-well plates followed by the replacement of medium with 100 μL fresh DMEM growth medium containing FBS after 4 h. Following 24 h of incubation, the medium was aspirated and MTT solution (5 mg/mL) was added to each well and incubated for another 1.5 h. Finally, formazan crystals formed were dissolved in 100 μL/well DMSO, and the absorbance was measured by an ELISA reader (ELx800, BioTek, Germany) at 590 nm and background corrected at 630 nm. Data are presented as mean ± standard deviation (SD); n=3.
Transfection procedure and evaluation of the transgene expression
To assess the transfection efficiency of PEI and its derivatives in transfecting plasmid encoding CD200 gene into the SH-SY5Y cells, in vitro transfection experiment was performed using the plasmid pCMV-XL5-hCD200 at a final concentration of 200 ng/well. Briefly, polyplexes were prepared at various C/P ratios as described for the MTT assay. Then, 10 μL of polyplex formulation was added to each well and incubated for 4 h at 37°C followed by the replacement of medium with fresh DMEM containing 10% FBS and incubation for an additional 48 h. At the end of the transfection procedure, cells were harvested and suspended in FACS buffer (PBS, 2% FBS and 0.1% Na3N) at 5×106 cells/mL. Then, 5 μL of APC anti-human CD200 antibody (Biolegend, Germany) was added followed by incubation on ice for 5–10 min in dark. Finally, cells were centrifuged and resuspended in the appropriate buffer and analyzed by flow cytometry (FACS Calibur, Becton Dickinson, Mountain View, CA) using FlowJo software (TreeStar Inc., San Carlos, CA). The cells treated with no plasmid or polyplex (with medium only) were considered as negative control.
To evaluate transgene expression at mRNA level, real-time quantitative PCR was performed. The RNA extraction was performed using RNA isolation kit (Jena Bioscience, Germany) according to the manufacturer’s instructions. Following the isolation of RNA and treatment with DNase (Thermo Fisher Scientific, Waltham, MA, USA), levels of CD200 transcripts expression were quantified using PrimeScript™ RT reagent Kit (Perfect Real Time, TaKaRa, Dalian, People’s Republic of China), and RealQ Plus 2x Master Mix Green High ROX™ (AmpliQon, Denmark) kit. Following CD200 primers were used; forward: 5′-AATACCTTTGTTTTTGGGAAGATCT-3′ and reverse: 5′-GGTGGTCTTCAGAGAATTTGTAGTGA-3′. Transcript levels were normalized against GAPDH primers; forward: 5′-ACTTCAACAGCGACACCCACT-3′ and reverse: 5′-GCCAAATTCGTTGTCATACCAG-3′. All the reactions were performed using MJ mini thermal cycler (BIO-RAD, Germany). Relative gene expression of CD200 was calculated by the 2−(ΔΔCT) method using GAPDH as the reference gene.
Fluorescence microscopy and in vivo imaging of the nanoparticles in mouse model of MS
All animal experiments were conducted with the approval of the Institutional Ethical Committee and Research Advisory Committee of Shiraz University of Medical Sciences (SUMS; Shiraz, Iran) and were based on the Ethical Guidelines for the Care and Use of Animals in Medical Research (SUMS protocol#7409). C57BL/6 male mice were obtained from the animal laboratory of SUMS. The induction of demyelination was performed by feeding 8-week-old C57BL-6 mice with a diet containing 0.2% (w:w) cuprizone (bis-cyclohexanone oxaldihydrazone; Sigma-Aldrich, Munich, Germany) for 6 weeks followed by a normal chow for additional 6 weeks. The demyelination was confirmed by Luxol Fast Blue staining according to Kluver–Barrera procedure as described elsewhere.39 To track nanoparticles in the body of the mice, fluorescently labeled plasmid encoding CD200 by GelRed was used, and the polyplexes at the highest C/P ratio of 8 were prepared as described earlier (vide supra). Mice were injected via the tail vein at a final volume of 100 μL of the polyplex formulation. Control mice received the same volume of HBG buffer. At the end of each time point (0, 15, 30, 60, 120, 180 min, and 24 h post injection) mice were deeply anesthetized with a ketamine-xylazine cocktail via intramuscular injection and then immediately imaged by the Kodak In-Vivo Imaging System F Pro (Kodak, USA) at the excitation and emission wavelengths of 510 and 600 nm, respectively. The captured images were processed and analyzed with the Kodak In-Vivo F Pro Imaging System equipped with Carestream MI software. Furthermore, the presence of nanoparticles containing the fluorescently labeled plasmids inside the brain tissue was shown using a fluorescence microscope (Olympus, Japan).
Statistical analysis
Data are presented as the mean ± SD. The statistical significance was determined using Student’s t-test and P-values <0.05 were considered as significant.
Results and discussion
Synthesis of PEI derivatives
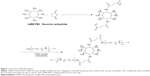
Figure 1 shows the strategy for the synthesis of PEI derivatives. LMW branched PEI (1,800 Da) was initially modified by the conjugation of predominantly primary amines with succinic anhydride to improve its hydrophilicity–hydrophobicity balance. This modification also provided the terminal carboxylate groups to which another LMW PEI could be grafted in the next step. Next, the PEI derivatives were labeled as PEI-SUC and PEI-SUC-PEI conjugates. The structure of modified PEI was confirmed by 1H-NMR spectra in which the methylene of succinate appeared at 2.5 ppm, whereas PEI protons were found between 2.6 and 3.6 ppm. FTIR spectra showed two new peaks at 1,658 and 1,715 cm−1 for PEI-SUC conjugate corresponding to amide and carboxylic acid groups, respectively. Following the conjugation of LMW PEI to the PEI-SUC structure, carboxylate peak weakened to 1,660 cm−1 confirming the formation of an amide bond. Then, 2,4,6-trinitrobenzenesulfonic acid assay was performed to determine the extent of LMW PEI primary amine substitution by succinic acid. According to the results, the degree of conjugation was around 10 mol%. Furthermore, the same assay was performed to determine the grafting degree of LMW PEI on the previous structure. The results of this assay demonstrated that the yields for the coupling of LMW PEI to the succinated PEI derivative was less than 100% so that the final conjugates contained an average of 75.6%±0.7% of the primary amine content of the original unmodified LMW PEI. The reproducibility of the synthesis procedure was determined by performing several independent preparations.
Biophysical properties of PEI conjugates and the polyplexes
Evaluation of the buffering capacity of PEI and its derivatives
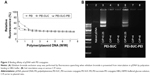
Proton sponge effect has been considered as the major mechanism by which the polycationic-based nanocarriers induce early escape from endosomal/lysosomal compartments. This process results in the protection of nucleic acid materials from acid hydrolases inside the lysosomes.8,12,13,19,25 To measure the buffering capacity of PEI and its derivatives following modification, an acid–base titration was performed and the buffering capacity was illustrated by plotting the pH versus the added acid. The polymers with higher buffering capacity needed larger amounts of HCl for the alteration of their pH. Figure 2 shows that the unmodified LMW PEI had substantial buffering capacity over almost the whole pH range. As shown in Figure 2, the conjugation of succinic acid on the structure of LMW PEI decreased the buffering capacity, whereas the subsequent conjugation of LMW PEI to PEI-SUC conjugate drastically increased the buffering capacity to considerably more than that of parent unmodified LMW PEI. This could be the effect of high amine content of the PEI-SUC-PEI conjugate.12 Reduction of buffering capacity following the hydrophobic modification of various PEI molecules has been reported in several previous investigations.13,19–23,25 This decrease is highly associated with the conjugation strategies used for the grafting of various moieties on PEI structure. Since majority of conjugation methods lead to the conversion of the surface primary amines to the secondary ones, the buffering capacity of the conjugates decreases. The buffering capacity of PEI at the pH range of 9–11 is highly attributed to the primary and secondary amines with the reported pKa values of 8–9, whereas the buffing capacity in the pH range of 5.5–7 is attributed to the tertiary amines with the pKa values of 6–7. It is supposed that the critical pH range of the endosomal compartment is around 5.5–7 which acts as a driving force for osmotic burst mechanism by which swelling and disruption of the endosomal/lysosomal membranes lead to the early escape of polyplexes.40 It is obvious that PEI-SUC-PEI derivative exhibited the highest buffering capacity in the critical pH range.
Binding affinity of PEI derivatives to pDNA
Nucleic acid compaction by polycationic compounds is a common process that occurs inside the cell’s nucleus which in turn causes DNA packaging. A similar process occurs following the electrostatic interaction between positively charged polymers and negatively charged backbone of pDNA. The outcome of this interaction is the formation of nano-sized complexes, namely, polyplexes. The formation of such nanostructures is a prerequisite step in polymer-based gene delivery.41 To measure the binding strength of PEI and its derivatives to pDNA, EtBr exclusion assay was performed. This technique is based on the addition of PEI to the pDNA solution which in turn leads to the exclusion of EtBr from the base pairs of the plasmid and consequently the decrease in fluorescence intensity. In other words, polymers with higher affinity to pDNA decrease the fluorescence intensity at lower concentrations than that of higher concentrations. Figure 3A shows the results of fluorescence quenching analysis. According to the results, all the PEI derivatives were able to efficiently interact with pDNA. As shown in Figure 3A, at a C/P ratio of ≥2, all the polycationic compounds decreased the fluorescence intensity by at least 70%. The pattern in the reduction of fluorescence intensity of PEI and its derivatives was similar. However, at the lowest C/P ratio tested (C/P=0.25), unmodified PEI decreased the fluorescence intensity by around 60%, whereas PEI-SUC conjugate decreased the fluorescence intensity by around 35%. However, PEI-SUC-PEI polymers were able to decrease the fluorescence intensity to 54%. By the addition of more polymers, sufficient positive charge was provided to condense pDNA. The full condensation of pDNA was not observed at C/P ratio of 0.25. It seems that the complete condensation of nucleic acid material occurs while the polycations with high amine content result in the formation of complexes by “wraparound” of pDNA. However, polycations with lower amine content induce simple electrostatic interaction between the positively charged amines and the negatively charged backbone of the nucleic acid materials. In this study, the complexes formed by the interaction between PEI-SUC-PEI conjugate and pDNA at the C/P ratio of 0.25 could be considered as looser complex with lower protection effect, whereas the complexes formed by the PEI-SUC-PEI derivative at higher C/P ratios induced full condensation. At the highest C/P ratio of 8, all the polymers were able to decrease the fluorescence intensity to around 25%. The attachment of LMW PEI onto the PEI-SUC structure resulted in the formation of PEI derivatives showing higher affinity for pDNA than that of unmodified parent polymer. This observation is consistent with the previous studies in which increase in the content of amine in the polymer and its charge density led to the formation of tighter complexes than that of unmodified parent polymer.11,14,19,24,25 Based on our results, the polymers with the highest affinity for pDNA binding were also the most effective at gene transfer experiments (“vide infra”). Although there are some reports suggesting that facilitation of dissociation of pDNA from its carrier (vector unpackaging) is an effective strategy to improve gene transfer efficiency of the polymers.17 Our data demonstrated that the most powerful polymers in pDNA condensation were the most effective ones in gene delivery. Hence, the dissociation of polyplexes will not necessarily lead to high transfection efficiency as this phase is not the rate-limiting step in the whole process of polycation-based gene delivery.
Gel retardation assay
Electrostatic interaction between polycationic polymers and pDNA resulted in the retardation of plasmid migration during gel agarose electrophoresis. As illustrated in Figure 3B, all the polyplexes prepared either by PEI-SUC or by PEI-SUC-PEI derivatives fully condensed the pDNA at C/P ratios ≥4, whereas the lowest C/P ratio tested (C/P=0.25) resulted in the weakest interaction between the polymer and pDNA than that of higher C/P ratios. This observation is consistent with the results obtained by EtBr quenching assay in which the weakest condensation was associated with the lowest C/P ratio tested in this study (C/P=0.25).
Particle size and zeta potential measurements
The electrostatic interaction between positively charged polycations and negatively charged plasmid leads to the formation of complexes with a desirable surface charge which plays a key role in polymer-induced toxicity as well as gene transfer ability. Having demonstrated that PEI-SUC and PEI-SUC-PEI derivatives can interact with the pDNA and form complexes, the polyplexes were formed at their optimum C/P ratio in which the highest level of transgene expression was achieved (C/P=8). Their particle size and zeta potential were measured using dynamic light scattering and laser Doppler velocimetry, respectively (Figure 4). According to the results, the unmodified LMW PEI formed nanoparticles with a size of 137 nm, whereas PEI-SUC conjugate resulted in the formation of larger polyplexes with a size of 192 nm. However, coupling PEI-SUC conjugate to LMW PEI led to the formation of smaller nanoparticles with a size of 128 nm. These findings were in good agreement with previous studies in which the conjugation of alkyl moieties on PEI structure led to the formation of larger nanoparticles.13,17,19,22,24,25,42 These substituted groups alter the surface interaction between the polymer and plasmid. As LMW PEI was coupled to the PEI-SUC structure, the primary amine density and consequently the net positive charge increased, which led to the formation of complexes with smaller size (128 nm). To assess the stability of particle size during the experiment, the particle size was measured 2 h after the preparation of polyplex. According to the results, there was no significant increase in the size of complexes showing the relative stability of polyplexes during experiments.
The results of laser Doppler velocimetry revealed that the conjugation of succinic acid onto the structure of PEI led to the formation of polyplexes with lower surface charge density than that of unmodified parent polymer (P<0.05). As expected, by the attachment of LMW PEI to the PEI-SUC structure the net positive charge of the complexes increased to values even higher than that of the unmodified PEI polymer (Figure 4). As demonstrated in Figure 4, in unmodified LMW PEI and PEI-SUC, the zeta potential decreased from 14 mV to 6 mV, respectively, which further increased to 23 mV for PEI-SUC-PEI (P<0.05). The increased value of zeta potential of PEI-SUC-PEI derivatives is associated with the elevated primary amine content of the polymer. Furthermore, high amine content of the polymer provides enough positive charge on the polyplexes, thereby preventing aggregation by electrostatic repulsion between the positively charged complexes. These polyplexes will be able to interact electrostatically with the negatively charged components on the cell surface facilitating their cell entry.24
Resistance of pDNA against DNase I degradation
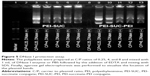
The free pDNA is highly susceptible to degradation by various enzymes including serum nucleases. Therefore, the formation of compact nanoparticles not only condenses pDNA but also protects it against enzymatic digestion. The protective ability of PEI derivatives was evaluated using DNase I as a model enzyme and the integrity of the loaded plasmids following the enzyme treatment was demonstrated by agarose gel electrophoresis. As presented in Figure 5, all the polyplexes at the lowest C/P ratio of 0.25 could not optimally protect the pDNA from digestion by the enzyme, whereas higher C/P ratios of 4 and 8 resulted in a remarkable protective effect. This observation is consistent with the results obtained in EtBr exclusion assay and gel retardation experiment. It could be concluded that full condensation of pDNA by polycationic carriers is a necessity for their substantial protection. However, it has been demonstrated in previous studies that the full condensation of nucleic acid materials by polycations may not necessarily lead to the complete protection effect against enzyme digestion.9 This behavior might be associated with the other polyplex properties, such as particle size, zeta potential, or the particle shape. Loose complexes are formed by the electrostatic interactions between the polycations and pDNA. It would presumably result in the exposure of more pDNA segments on the exterior parts of the nanoparticles which causes increased susceptibility of pDNA to enzyme degradation. However, tight complexes are formed by wraparound of pDNA leading to the more protective effects.8–13 As the concentration of nuclease in animal or human body is lower than that of DNase I used in this study (0.25 units), the administration of the polyplexes even at a C/P ratio of 0.25 might lead to protection at the physiological condition.38,43,44
Biological studies
Cytotoxicity
The cytotoxicity of PEI/pDNA polyplexes at different C/P ratios was evaluated in SH-SY5Y cell line in 96-well plates using MTT colorimetric assay. Cytotoxicity of different PEI conjugates was compared with that of nanoparticles prepared from unmodified 1,800 Da branched PEI at the same C/P ratio (Figure 6A). At the lowest C/P ratio tested, all the unmodified and modified PEI derivatives were almost non-toxic. By increasing C/P ratios from 0.25 to 8, the cell survival of the cells treated with PEI-SUC-PEI derivatives decreased from 98% to 85%. Similarly, no significant decrease in the cell viability was observed for the cell treated either with PEI-SUC or unmodified PEI polyplexes (P>0.05). In other words, PEI-SUC-PEI polyplexes induced more cytotoxicity, particularly at higher C/P ratios. This observation is consistent with the fact that PEI-SUC-PEI conjugates contain more amine and positive charge density on their surfaces. According to some previous studies, the main factor determining the toxicity of polycationic polymers is the positive charge density of the polymer which causes the electrostatic interaction with the negatively charged components of the cell membrane. These interactions lead to membrane perturbation and finally cell death.45,46 A two-stage cytotoxicity mechanism has been defined for PEI-based polyplexes. According to this, the interaction of positively charged polyplexes with the negatively charged components on the plasma membrane results in the perturbation of cell membrane. These necrotic-like changes occur within 30 min followed by the apoptotic program induced by the formation of channels in the outer membrane of mitochondria and causes the release of cytochrome c and finally the activation of caspase 3 which occur 24 h later.45 The positive charge of polycationic polymers is crucial for their role in gene delivery;47 however, this charge density must be modulated to achieve a polyplex with improved characteristics. Based on the results obtained in our study, cross-linking LMW PEI molecules through a succinic acid linker increased the positive charge of the final conjugate; however, cytotoxicity of this new derivative was less than 15%. According to our previous studies, the cell viability for the polyplexes prepared by unmodified 25 kDa PEI at a C/P ratio of 8 was around 40%,48 whereas cell survival following the treatment of cells with PEI-SUC-PEI polyplexes was around 85%. This result demonstrates that the modulation of positive charge density is still the most effective strategy to achieve PEI-based gene transfer with low toxicity.
In vitro gene transfer experiments
The immune privilege status of CNS results from a highly well-balanced anti-inflammatory microenvironment within the system. The wide, but not a ubiquitous expression of CD200 on the surface of various cells suggests that this membrane glycoprotein regulates myeloid cell activity in different cells and organs.33,34 As it has been proved that CD200 suppresses immune activity through its receptor (CD200R), the role of the interaction between CD200 and CD200R has been highlighted not only in Alzheimer and Parkinson diseases but also in MS.34,36,37 The susceptibility of CD200 deficient mice to several inflammatory diseases including MS has been found to increase, which is highly associated with the activation of microglia.36,37 However, great attention has been directed to gene therapy as a new paradigm in the treatment of various complicated diseases including MS. Furthermore, the selection of immunotherapy as the breakthrough of the year for 2013 has encouraged many types of research to utilize this method in addition to various other conventional treatments.49 CD200 creates an immune-privileged environment in the CNS and can downregulate the immune responses by binding to CD200R, which causes the downregulation of CD200–CD200R in neuroinflammatory diseases such as MS. In addition, CD200 has the potential in gene therapy, thus we hypothesized that the delivery of the plasmid encoding CD200 gene using a non-toxic polymer-based nanoparticle could be an effective route to increase the expression of CD200 in target cells. In this study, transfection efficiency of the polyplexes based on cross-linked LMW PEI was evaluated on SH-SY5Y cells using the plasmid pCMV-XL5-hCD200. The transfection experiments were performed using the polyplexes at C/P ratios (w/w) of 0.25, 4, and 8 and compared to the unmodified LMW PEI at the same C/P ratios. The result of CD200 expression (Figure 6B) demonstrated that the unmodified LMW PEI even at highest C/P ratio of 8 could not increase the expression level of CD200, whereas the level of CD200 in the cells treated with PEI-SUC-PEI nanoparticles increased to 92%. In other words, treatment of cells with PEI-SUC-PEI polyplexes enhanced the expression of CD200 by up to around 1.5 fold relative to unmodified LMW PEI (P<0.05). A similar trend was observed in the mRNA level where the highest level of mRNA was achieved by PEI-SUC-PEI nanoparticles at a C/P ratio of 8 (data not shown). One of the most probable reasons for the elevated gene transfer ability of the PEI derivative is the improved hydrophobic–hydrophilic balance of the new conjugate. There are several studies that have reported the effect of hydrophobic modification of various PEI derivatives in increasing their transfection efficiency.13,17,19,24,25,32,44,48 These modifications result in more efficient interaction of the polymer with cell membrane which in turn led to cell entry. Furthermore, greater buffering capacity of the modified PEI derivative could be an explanation for improved transfection efficiency.11 As these PEI derivatives exhibited higher buffering capacity in the endosomal pH range than that of unmodified polymer, they may be responsible for early escape from endosomes resulting in higher transfection efficiency. However, high transfection efficiency can be achieved not only by higher buffering capacity but also by an optimal size, zeta potential, plasmid’s condensation ability, stability against enzymatic degradation, and with improved low toxicity.
Fluorescent microscopy and in vivo imaging in mouse model of MS
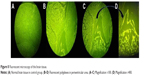
To evaluate the systemic circulation of the nanoparticles and their potential to cross the BBB, a mouse model (cuprizone model) of MS was prepared as described elsewhere.39 Following the confirmation of the efficacy of this model of toxic demyelination in the CNS, the polyplex formulations were injected followed by imaging the animal at 0, 15, 30, 60, 120, 180 min, and 24 h post injection. Figure 7 shows the distribution of the fluorescently labeled plasmid encoding CD200 from the injection site to the various tissues and organs including the lymph nodes, liver, brain, and finally kidneys. Mouse imaging after 24 h post injection demonstrated that all the injected nanoparticles have been cleared over this period of time. According to the previous investigation, the polyplexes formed from LMW PEI accumulated in the liver, kidney, and spleen 2 h post injection.26 Although the experiment to detect organ distribution of the nanoparticles was performed just for 2 h, their results demonstrated some differences between the behavior of LMW PEI and high molecular weight PEI in organ accumulation. The accumulation of LMW PEI in the kidneys was found to be significantly higher than that of high molecular weight PEI.26 This is consistent with our results showing that the polyplexes were removed from the body 24 h post injection. Furthermore, we performed fluorescent microscopy to visualize the presence of labeled nanoparticles in the CNS. As illustrated in Figure 8, no fluorescence was detected in the control group (Figure 8A), whereas the polyplexes were able to cross the BBB and enter the periventricular area (Figure 8B–D). Our results showed that the fluorescent nanoparticles crossed the BBB in the mouse model of MS. Although it is unlikely for unmodified PEI to transport across the BBB following the intravenous administration, the more probable reason for the presence of PEI derivatives inside the brain tissue could be the improved membrane biophysical properties of the PEI conjugates created by the conjugation of alkyl chains on the parent PEI molecules.50 Therefore, these polyplexes are expected to be used in our future studies aimed at developing CNS-targeted gene delivery systems.
Conclusion
Cross-linking LMW PEI using succinic anhydride yielded nanocarriers with optimal transfection efficiency to transfer the plasmid encoding CD200 gene, which is more powerful than those of unmodified LMW PEI. Furthermore, these nanocarriers showed the ability to cross BBB making them as a potential candidate for further investigations to create CNS-targeted delivery systems.
Acknowledgments
This work was funded by Shiraz University of Medical Sciences, Iran (grant number: 7409). Financial support from Iran National Science Foundation (INSF) (#89000844) and Iranian Nanotechnology Initiative Council (INIC) is acknowledged. This work was a part of Ph.D. thesis of Fatemeh Nouri. Furthermore, we would like to thank Dr Samira Hossaini Alhashemi, Dr Negar Azarpira, Dr Bahman Khalvati, and Dr Mohammad Taheri for their helpful assistance.
Author contributions
Fatemeh Nouri performed the experiments and prepared the first draft of the manuscript. Reza Heidari prepared the MS mouse model. Ali Dehshahri and Hossein Sadeghpour conceived the idea and supervised all the experiments. All the authors contributed toward data analysis, drafting and critically revising the paper, gave the final approval of the revision to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Wirth T, Parker N, Ylä-Herttuala S. History of gene therapy. Gene. 2013;525(2):162–169. | ||
Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012–an update. J Gene Med. 2013;15(2):65–77. | ||
Libutti SK. New horizons for cancer gene therapy. Cancer Gene Ther. 2014;21(1):1. | ||
Ibraheem D, Elaissari A, Fessi H. Gene therapy and DNA delivery systems. Int J Pharm. 2014;459(1–2):70–83. | ||
McErlean EM, McCrudden CM, McCarthy HO. Delivery of nucleic acids for cancer gene therapy: overcoming extra-and intra-cellular barriers. Ther Deliv. 2016;7(9):619–637. | ||
Kozielski KL, Rui Y, Green JJ. Non-viral nucleic acid containing nanoparticles as cancer therapeutics. Expert Opin Drug Deliv. 2016;13(10):1475–1487. | ||
Pandey AP, Sawant KK. Polyethylenimine: a versatile, multifunctional non-viral vector for nucleic acid delivery. Mater Sci Eng C Mater Biol Appl. 2016;68:904–918. | ||
Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. | ||
Dehshahri A, Alhashemi SH, Jamshidzadeh A, et al. Comparison of the effectiveness of polyethylenimine, polyamidoamine and chitosan in transferring plasmid encoding interleukin-12 gene into hepatocytes. Macromol Res. 2013;21(12):1322–1330. | ||
Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Deliv Rev. 2002;54(5):715–758. | ||
Dehshahri A, Oskuee RK, Shier WT, Hatefi A, Ramezani M. Gene transfer efficiency of high primary amine content, hydrophobic, alkyl-oligoamine derivatives of polyethylenimine. Biomaterials. 2009;30(25):4187–4194. | ||
Lungwitz U, Breunig M, Blunk T, Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60(2):247–266. | ||
Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem. 2008;19(7):1448–1455. | ||
Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. | ||
Fischer D, Bieber T, Li Y, Elsässer HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16(8):1273–1279. | ||
Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly (ethyleneimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45(3):268–275. | ||
Forrest ML, Meister GE, Koerber JT, Pack DW. Partial acetylation of polyethylenimine enhances in vitro gene delivery. Pharm Res. 2004;21(2):365–371. | ||
Thomas M, Klibanov AM. Enhancing polyethylenimine’s delivery of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2002;99(23):14640–14645. | ||
Nimesh S, Aggarwal A, Kumar P, Singh Y, Gupta KC, Chandra R. Influence of acyl chain length on transfection mediated by acylated PEI nanoparticles. Int J Pharm. 2007;337(1–2):265–274. | ||
Khalvati B, Sheikhsaran F, Sharifzadeh S, et al. Delivery of plasmid encoding interleukin-12 gene into hepatocytes by conjugated polyethylenimine-based nanoparticles. Artif Cells Nanomed Biotechnol. 2017;45(5):1036–1044. | ||
Dehshahri A, Sadeghpour H, Keykhaee M, Khalvati B, Sheikhsaran F. Enhanced delivery of plasmid encoding interleukin-12 gene by diethylene triamine penta-acetic acid (DTPA)-conjugated PEI nanoparticles. Appl Biochem Biotechnol. 2016;179(2):251–269. | ||
Dehshahri A, Oskuee RK, Ramezani M. Plasmid DNA delivery into hepatocytes using a multifunctional nanocarrier based on sugar-conjugated polyethylenimine. Gene Ther Mol Biol. 2012;14:62–71. | ||
Oskuee RK, Dehshahri A, Shier WT, Ramezani M. Modified polyethylenimine: self assemble nanoparticle forming polymer for pDNA delivery. Iran J Basic Med Sci. 2008;11(1):33–40. | ||
Gabrielson NP, Pack DW. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules. 2006;7(8):2427–2435. | ||
Putnam D, Gentry CA, Pack DW, Langer R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc Natl Acad Sci U S A. 2001;98(3):1200–1205. | ||
Kunath K, von Harpe A, Fischer D, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89(1):113–125. | ||
Johnson ME, Shon J, Guan BM, et al. Fluorocarbon modified low-molecular-weight polyethylenimine for siRNA delivery. Bioconjug Chem. 2016;27(8):1784–1788. | ||
Lee YH, Park HI, Choi JS. Novel glycol chitosan-based polymeric gene carrier synthesized by a Michael addition reaction with low molecular weight polyethylenimine. Carbohydr Polym. 2016;137:669–677. | ||
Zhao J, Yang L, Huang P, et al. Synthesis and characterization of low molecular weight polyethyleneimine-terminated Poly (β-amino ester) for highly efficient gene delivery of minicircle DNA. J Colloid Interface Sci. 2016;463:93–98. | ||
Wang H, Chen J, Ying J, Xu Y, Sheng R. Hydrophobic chain modified low molecular weight polyethylenimine for efficient antigen delivery. RSC Adv. 2016;6(17):13636–13643. | ||
Liu S, Huang W, Jin MJ, et al. High gene delivery efficiency of alkylated low-molecular-weight polyethylenimine through gemini surfactant-like effect. Int J Nanomedicine. 2014;9:3567–3581. | ||
Thomas M, Ge Q, Lu JJ, Chen J, Klibanov AM. Cross-linked small polyethylenimines: while still nontoxic, deliver DNA efficiently to mammalian cells in vitro and in vivo. Pharm Res. 2005;22(3):373–380. | ||
Chitnis T, Imitola J, Wang Y, et al. Elevated neuronal expression of CD200 protects Wlds mice from inflammation-mediated neurodegeneration. Am J Pathol. 2007;170(5):1695–1712. | ||
Wright GJ, Jones M, Puklavec MJ, Brown MH, Barclay AN. The unusual distribution of the neuronal/lymphoid cell surface CD200 (OX2) glycoprotein is conserved in humans. Immunology. 2001;102(2):173–179. | ||
Koning N, Swaab DF, Hoek RM, Huitinga I. Distribution of the immune inhibitory molecules CD200 and CD200R in the normal central nervous system and multiple sclerosis lesions suggests neuron-glia and glia-glia interactions. J Neuropathol Exp Neurol. 2009;68(2):159–167. | ||
Hoek RM, Ruuls SR, Murphy CA, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science. 2000;290(5497):1768–1771. | ||
Broderick C, Hoek RM, Forrester JV, Liversidge J, Sedgwick JD, Dick AD. Constitutive retinal CD200 expression regulates resident microglia and activation state of inflammatory cells during experimental autoimmune uveoretinitis. Am J Pathol. 2002;161(5):1669–1677. | ||
Jiang HL, Kim YK, Arote R, et al. Chitosan-graft-polyethylenimine as a gene carrier. J Control Release. 2007;117(2):273–280. | ||
Skripuletz T, Lindner M, Kotsiari A, et al. Cortical demyelination is prominent in the murine cuprizone model and is strain-dependent. Am J Pathol. 2008;172(4):1053–1061. | ||
Wang DA, Narang AS, Kotb M, et al. Novel branched poly (ethylenimine)-cholesterol water-soluble lipopolymers for gene delivery. Biomacromolecules. 2002;3(6):1197–1207. | ||
Lin Z, Wang C, Feng X, Liu M, Li J, Bai C. The observation of the local ordering characteristics of spermidine-condensed DNA: atomic force microscopy and polarizing microscopy studies. Nucleic Acids Res. 1998;26(13):3228–3234. | ||
Sheikhsaran F, Sadeghpour H, Khalvati B, Entezar-Almahdi E, Dehshahri A. Tetraiodothyroacetic acid-conjugated polyethylenimine for integrin receptor mediated delivery of the plasmid encoding IL-12 gene. Colloids Surf B Biointerfaces. 2017;150:426–436. | ||
Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5(10):1425–1433. | ||
Sabahi Z, Samani SM, Dehshahri A. Conjugation of poly (amidoamine) dendrimers with various acrylates for improved delivery of plasmid encoding interleukin-12 gene. J Biomater Appl. 2015;29(7):941–953. | ||
Parhamifar L, Larsen AK, Hunter AC, Andresen TL, Moghimi SM. Polycation cytotoxicity: a delicate matter for nucleic acid therapy-focus on polyethylenimine. Soft Matter. 2010;6(17):4001–4009. | ||
Hall A, Lächelt U, Bartek J, Wagner E, Moghimi SM. Polyplex evolution: understanding biology, optimizing performance. Mol Ther. 2017;25(7):1476–1490. | ||
Amin ZR, Rahimizadeh M, Eshghi H, Dehshahri A, Ramezani M. The effect of cationic charge density change on transfection efficiency of polyethylenimine. Iran J Basic Med Sci. 2013;16(2):150–156. | ||
Oskuee RK, Dehshahri A, Shier WT, Ramezani M. Alkylcarboxylate grafting to polyethylenimine: a simple approach to producing a DNA nanocarrier with low toxicity. J Gene Med. 2009;11(10):921–932. | ||
Couzin-Frankel J. Cancer immunotherapy. Science. 2013;342(6165):1432–1433. | ||
Meng Q, Yu M, Gu B, et al. Myristic acid-conjugated polyethylenimine for brain-targeting delivery: in vivo and ex vivo imaging evaluation. J Drug Target. 2010;18(6):438–446. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.