Back to Journals » International Journal of Nanomedicine » Volume 12
Polyethyleneimine-capped silver nanoclusters for microRNA oligonucleotide delivery and bacterial inhibition
Authors Du C, Yan H, Liang J, Luo A, Wang L, Zhu J , Xiong H, Chen Y
Received 21 July 2017
Accepted for publication 17 October 2017
Published 1 December 2017 Volume 2017:12 Pages 8599—8613
DOI https://doi.org/10.2147/IJN.S146968
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Chunyuan Du,1,* Haibo Yan,2,* Jichao Liang,1 Ailing Luo,1 Lingqian Wang,1 Jing Zhu,1 Huayu Xiong,3 Yong Chen1
1Hubei Province Key Laboratory of Biotechnology of Chinese Traditional Medicine, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei University, Wuhan, 2Department of Internal Medicine, Shandong Medical College, Linyi, 3Ministry-of-Education Key Laboratory for the Synthesis and Application of Organic Functional Molecules, Hubei University, Wuhan, China
*These authors contributed equally to this work
Abstract: Efficient and safe nonviral gene delivery systems are a prerequisite for the clinical application of therapeutic genes. In this paper, polyethyleneimine-capped silver nanoclusters (PEI-AgNCs) were prepared for the purpose of microRNA (miRNA) delivery. The resultant PEI-AgNCs were characterized by a photoluminescence assay and transmission electron microscopy. A cytotoxicity assay showed that PEI-AgNCs exhibit relatively low cytotoxicity. Interestingly, PEI-AgNCs were confirmed to transfect miRNA mimics more effectively than PEI in HepG2 and 293A cells. In this regard, hsa-miR-21 or hsa-miR-221 mimics (miR-21/221m) were transported into HepG2 cells by using PEI-AgNCs. The miR-21/221 expression was determined post-transfection by quantitative real-time polymerase chain reaction. Compared with the negative control, PEI-AgNCs/miR-21/221m groups exhibited higher miR-21/221 levels. In addition, AgNCs endow PEI with stronger antibacterial activity, and this advantage provided PEI-AgNCs the potential to prevent bacterial contamination during the transfection process. Furthermore, we showed that PEI-AgNCs are viable nanomaterials for plain imaging of the cells by laser scanning confocal microscopy, indicating great potential as an ideal fluorescent probe to track the transfection behavior. These results demonstrated that PEI-AgNCs are promising and novel nonviral vectors for gene delivery.
Keywords: gene delivery, silver nanoclusters, polyethylenimine, cell imaging, antibacterial activity
Introduction
Metal nanoclusters have been recognized as a novel class of nanomaterials in recent decades because of their promising applications in biological labeling, optical sensing, imaging, catalysis, and other fields.1–3 Among the various metal clusters that have been reported, silver nanoclusters (AgNCs) are of intense interest because they are a novel class of fluorophores widely recognized for their photophysical properties, application potential for detection of a variety of targets, and remarkable biocompatibility.4–6 Additionally, recent studies have demonstrated the usefulness of fluorescent AgNCs in biodetection and biological imaging.7,8 Meanwhile, AgNCs have been shown to effectively inactivate bacteria and inhibit microbial growth, but the mechanism has yet to be sufficiently elucidated.9 Therefore, fluorescent AgNCs hold great potential as novel optical probes and may find wide application in biological research.
Nucleic acid-based therapeutics is a promising method for the treatment of diseases that are currently considered incurable, particularly acquired and inherited diseases.10 Among therapeutic genes, the role of microRNA (miRNA or miR) has been emphasized in biological and fundamental cellular processes. For example, miR-21 and miR-221 are valid links between obesity and obesity-associated diseases such as metabolic syndrome, type 2 diabetes, and cancer.11–13 The success of gene therapy primarily depends on delivering foreign genes into targeted cells. However, it is a challenge to develop a novel vector that can efficiently deliver DNA/RNA molecules into the target cells with minimal toxicity and high bioavailability during the delivery process. Currently, commonly used gene vectors are divided into two major categories: viral vectors and nonviral vectors. Viral vectors are used for gene delivery with high transfection efficiency, but the potential immunogenicity and infectivity of those viral vectors has been a primary disadvantage for their effective utilization.14 Because of the limitation of viral vectors, a variety of nonviral gene carriers with clinical potential and ease of production have been designed for miRNA delivery, such as micelles, liposomes and nanoparticles formed with cationic polymers, polysaccharides, and peptides.15–17 In particular, in the cationic polymer-based gene carriers, branched polyethylenimine with a weight-average molecular weight of 25 kDa (PEI25K) has been considered the “gold standard” due to its superior transfection efficiency both in vitro and in vivo.18,19 However, the high cationic density of PEI25K would simultaneously cause severe problems, especially high cytotoxicity and hemolysis due to the membrane-disruptive properties.20 Thus, how to decrease the cytotoxicity and/or improve the transfection efficiency is a key issue to be resolved in the design of gene delivery systems. To improve these disadvantages of PEI, the polyethyleneimine-capped silver nanoclusters (PEI-AgNCs) had been established and utilized as an efficient gene carrier to transfect therapeutic miRNAs into human hepatoma cells.
In this work, a PEI-AgNCs copolymer was synthesized based on the previous work.21 It was then employed as a carrier to realize the delivery of miR-21 or miR-221 mimics (miR-21/221m), using the human hepatoma cell line HepG2 and human embryonic kidney cell line 293A as models. Furthermore, AgNCs enhanced the antibacterial activity of PEI, which gave a chance to prevent bacterial contamination in the transfection process. In addition, with the excellent photoluminescent properties, PEI-AgNCs might be applied in cell imaging. These results made it possible to apply the copolymer in biological systems.
Materials and methods
Reagents
Silver nitrate (AgNO3), ascorbic acid (AA), acetic acid (HAc), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), PEI25K, PTEN, and GAPDH antibodies were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Dulbecco’s Minimal Eagle’s Medium (DMEM), fetal bovine serum (FBS), Trizol reagent, and the DNA-staining reagent (0.5% DAPI) were obtained from Thermo Fisher Scientific (Waltham, MA, USA). MiR-21/221 mimics, miR-21/221 inhibitors, and the negative miRNA controls were purchased from RiboBio Co., Ltd. (Guangzhou, China). The fluorescein amidite (FAM)-labeled miR-21/221m (F-miR-21/221m) were purchased from GenePharma (Shanghai, China). LipoFiter™ Liposomal Transfection Reagent (LipoFiter) was purchased from Hanbio Biotechnology Co., Ltd (Shanghai, China).
Synthesis of PEI-AgNCs
The highly fluorescent and water-soluble PEI-AgNCs was synthesized according to the previously reported method with minor modifications.21 Briefly, 2.5 μmol of PEI25K and 2.5 μmol of AgNO3 were dissolved in 10 mL of ultrapure water and stirred for 2 hours to facilitate complexation between Ag+ and the amine ligands. Subsequently, the pH was adjusted to 4 with HAc, and 6 μmol of AA was added to the solution and stirred continuously for 5 minutes. Then, the solution was subjected to an ultraviolet light source (8 W, wavelength 365 nm) for various time intervals. All reactions were carried out at room temperature.
Characterization of PEI-AgNCs
The fluorescence of water-soluble PEI-AgNCs was analyzed using RF-5301 fluorometer (Shimadzu, Kyoto, Japan). The color of water-soluble PEI-AgNCs was measured under visible and ultraviolet (UV) light. The shape and size of the PEI-AgNCs were measured by transmission electron microscopy (TEM, JEM-100SX microscope; JEOL, Tokyo, Japan). The aqueous solution of PEI-AgNCs was dropped into the copper net. TEM observation was carried out after the samples were air-dried at room temperature.
Cell culture and transfection
HepG2, Huh7, and 293A cells (obtained from National Infrastructure of Cell Line Resource, Peking Union Medical College) were grown in DMEM supplemented with 10% FBS and 100 μg/mL penicillin-streptomycin antibiotic solution. The cells were kept at 37°C in a humidified atmosphere containing 5% CO2. For transfection, cells were seeded into 24-well plates 24 hours before transfection in DMEM containing 10% FBS. The enhanced green fluorescent protein (EGFP)-C1 plasmid (800 ng) or miRNAs (100 nM) were added to the PEI or PEI-AgNCs (2.3 μg/mL) solutions. The complex particle solutions were mixed and incubated for 15 minutes at room temperature. After replacing with fresh serum-free DMEM, the complex particles were added to the cells. The medium was replaced by fresh complete medium 24 hours later and maintained for another 24 hours at 37°C. Then, the total cellular protein or RNA was extracted for Western blot and real-time PCR (RT-PCR) analysis.
Agarose gel retardation assay
The agarose gel retardation assay was conducted to determine the ability of PEI-AgNCs to condense DNA and RNA molecules. Briefly, DNA or RNA complexes containing different polymers at various nitrogen:phosphate (N/P) ratios were prepared. After 30 minutes of incubation at room temperature, 20 μL of complexes was loaded onto a 1.5% agarose gel and electrophoresed at 100 V for 15 minutes in Tris-acetate-EDTA buffer. The bands were visualized with a UV lamp and recorded with the gel imaging system.
Cytotoxicity assays
The cell viability was evaluated by an MTT reduction assay. HepG2 or 293A cells were seeded into a 96-well plate with ~104 cells per well and incubated for 24 hours to allow for cell attachment. Then, the as-prepared PEI, PEI-AgNCs, PEI/DNA, and PEI-AgNCs/DNA complexes at the designed concentrations were added into basic medium with free serum and incubated for 24 hours. After being reincubated in fresh medium containing FBS for 24 hours, 20 μL of 5 mg/mL MTT was added at 4 hours before measurement. Subsequently, 150 μL of dimethyl sulfoxide was added to dissolve the formazan crystals formed by live cells. The absorbance of formazan was measured at 570 nm using a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Fluorescence imaging
To assess the transfection efficiency of PEI-AgNCs for miRNA, we carried out fluorescence microscopic imaging with F-miR-21/221m. HepG2 cells were plated in 24-well plates at ~5×104 cells and allowed to adhere for 24 hours. Then, the medium was exchanged with 500 μL of fresh basic medium with free serum. F-miR-21/221m (100 nM) was added to the PEI or PEI-AgNCs (2.3 μg/mL) solutions. The transfection was performed as described in the “Cell culture and transfection” section. After 12 hours, the cells were fixed with 4% paraformaldehyde for 30 minutes and subsequently stained with the DNA-staining reagent (0.5% DAPI) for another 10 minutes. The cells were washed three times with PBS before every reagent change. 239A cells were used to judge the transfection efficiency of PEI-AgNCs in the same way as described earlier. Fluorescent FAM labeled-miRNA mimics (F-miRm) within cells were imaged using an Olympus IX-73 optical camera (Olympus Corporation, Tokyo, Japan). LipoFiter was used as positive control and applied according to the manufacturer’s procedures.
Quantification of miR-21 or miR-221 levels
HepG2 cells were transfected with 100 nM of miR-221 mimics (miR-221m) or miR-221 inhibitors (miR-221i) using PEI or PEI-AgNCs (2.3 μg/mL) or the LipoFiter. The same test condition was applied to miR-21. The total RNA was obtained using Trizol reagent according to the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed using TaqMan microRNA Assays (Thermo Fisher Scientific) on a CFX Connect Bio-Rad system. Reactions were typically run in triplicate.
Western blot analysis
HepG2 cells were transfected with 100 nM of miR-221m or miR-221i using PEI-AgNCs in serum-free DMEM. After 24 hours of transfection, cells were washed and reincubated in 2 mL of fresh medium containing 10% FBS for 24 hours. Whole cell lysates were prepared using RIPA lysis buffer according to the manufacturer’s instructions (Beyotime Biotechnology, Shanghai, China). Protein concentrations were determined using a Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Aliquots containing 70 μg of protein were subjected to gel electrophoresis on 7.5% or 10% sodium dodecyl sulfate (SDS) polyacrylamide gel-electrophoresis gels and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% nonfat milk in PBS for 2 hours and then incubated overnight with primary antibody at 4°C. After the membrane had been washed with 0.1% Tween 20 in PBS five times, it was further incubated with a horseradish peroxidase-conjugated secondary antibody for 80 minutes. The membrane was washed and developed with enhanced chemiluminescence using ECL Plus (Beyotime Biotechnology).
Cell imaging
HepG2, Huh7 or 293A cells were seeded onto 18 mm sterile coverslips in a six-well plate. After 24 hours, cells were incubated with the PEI-AgNCs for 6 hours at the final concentration of 12 μg/mL. Then, cells were washed five times with PBS to remove the unbound PEI-AgNCs. Later, cells binding with PEI-AgNCs were fixed for 20 minutes in 1 mL of 4% paraformaldehyde and then washed with PBS for five times. The images were captured using a confocal laser scanning microscope (Carl Zeiss AG, Oberkochen, Germany). PEI-AgNCs were excited at 405 nm. For atomic force microscope imaging and fluorescence microscopic images of EGFP-C1 gene expression, please see supplementary materials.
Evaluation of antibacterial activity
Escherichia coli (E. coli) were obtained from China Center of Industrial Culture Collection (CICC, Beijing, China). As E. coli grow well in lysogeny broth (LB) medium and are commonly used in antibacterial experiments, they were cultivated in LB medium, consisting of 10 g/L tryptone, 5 g/L bacto-yeast extract, and 10 g/L NaCl.22 After inoculation, the growth medium was placed into a thermostatic incubator at 37°C with shaking at 200 rpm. After 9–10 hours, the bacterial cells were brought into log phase until an optical density at 600 nm (OD600) of 0.5–0.6 was reached. One and a half microliters of the log-phase bacterial cells (~1.0×109 colony forming units/mL [CFU/mL]) was inoculated into 150 μL of fresh LB medium containing 14.25 μg/mL of PEI and PEI-AgNCs in the 96-well plate and cultured under shaking for 68 hours at 37°C. During the culture process, the optical density of each well at 600 nm was monitored at different time intervals.
The number of the viable bacterial cells in the wells was analyzed using CFU counting. In brief, each culture in the 96-well plate with a dilution factor of 50,000 was plated in triplicate on LB agar plates and incubated at 37°C for 16 hours, and the bacterial colonies formed were counted and recorded.
Morphological characterization of bacteria
The morphological changes in the bacteria were characterized by scanning electron microscopy (SEM) as previously described.23 Briefly, log-phase bacteria incubated with 14.25 μg/mL of PEI and PEI-AgNCs for 48 hours were collected by centrifugation at 5,000 rpm for 5 minutes and fixed with 2.5% glutaraldehyde overnight at 4°C. After washing with PBS three times, the bacterial cells were dehydrated through a sequential treatment of 30%, 50%, 70%, 80%, 90%, 95%, and 100% ethanol for 15 minutes and imaged using an SEM (ULTRA Plus; Carl Zeiss AG).
Statistical analysis
All studies were repeated at least three times. Data are presented as the mean ± standard deviation of the three independent experiments. The statistical analysis was performed with one-way analysis of variance, followed by the Student’s t-test. A p-value <0.05 was considered statistically significant.
Results and discussion
Characterizations of PEI-AgNCs
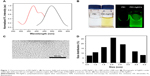
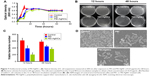
The synthesizing conditions for the highly fluorescent and water-soluble PEI-AgNCs were optimized (Figure S1). Under the optimal conditions, PEI-AgNCs were directly synthesized under UV light for 6 hours with stirring, making the synthetic process more facile, as compared to previous studies which stirred for 2 days.21,24 The formation of PEI-AgNCs was characterized based on the fluorescence emission spectra and TEM images. Upon excitation at 380 nm, PEI-AgNCs displayed an intense emission band with a maximum at ~500 nm (Figure 1A). Under a UV lamp, the solution exhibited bright green emission (Figure 1B). TEM images showed the cluster with an average diameter of ~2 nm. No larger Ag particles or aggregations were observed (Figure 1C). The corresponding size distribution histogram revealed that the as-prepared AgNCs were well dispersed with an average diameter of ~2 nm (Figure 1D). As it is known, the most important parameter of a gene carrier is the particle size, which has been demonstrated to play key roles in determining cellular uptake efficiency and toxic effects on cells.25,26 The small size of PEI-AgNCs has the potential for utilization in experimental gene delivery applications. Furthermore, these synthesized clusters were stable for longer than 6 months (Figure S2), which endowed these PEI-AgNCs with a promising application in bioimaging.
Gel retardation assay and characterization of PEI-AgNCs/DNA nanoparticles
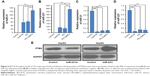
The nucleic acid loading efficiency was a key criterion affecting the efficacy of gene transfection.27 An agarose gel electrophoresis assay was used to investigate the ability of PEI-AgNCs to condense DNA or oligonucleotide at different complex ratios. As shown in Figure 2, PEI-AgNCs could totally complex with miR-21 mimics (miR-21m) and miR-21 inhibitors (miR-21i) to form particles when the N/P ratios were greater than 11.2 and 20, respectively. In addition, PEI-AgNCs could completely condense plasmid at the N/P ratio of 2.4, which is lower than that of PEI (3.2), suggesting that PEI-AgNCs showed greater ability to absorb DNA. These results indicated that PEI-AgNCs have great capacity to absorb DNA or oligonucleotide.
The particle size of complexes is also an important property of gene delivery systems.28 The morphology and average size of PEI-AgNCs/DNA complexes were further explored by atomic force microscopy (AFM) (Figure S3). AFM showed that the average size of PEI-AgNCs/DNA complexes at the optimum transfection N/P ratio was smaller than 150 nm. These results confirmed that PEI-AgNCs could be qualified as a transfection carrier.
Cytotoxicity of PEI-AgNCs
Cytotoxicity is also a vital issue that needed to be solved before the gene delivery vector was utilized for transfection study. To estimate the cytotoxicity of PEI-AgNCs, the relative viabilities of HepG2 and 293A cells exposed to PEI-AgNCs or PEI-AgNCs/DNA complexes were measured using the MTT assay; PEI25K was used as a control. All the samples exhibited a dose-dependent cytotoxicity. Compared to the PEI25K and PEI25K/DNA controls, PEI-AgNCs and PEI-AgNCs/DNA complexes exhibited less toxicity in HepG2 and 293A cells at the same concentrations or complexing ratios, respectively. As shown in Figure 3, almost 70% of HepG2 cells and 90% of 293A cells were viable for PEI-AgNCs/DNA even at an N/P ratio of 15, implying that PEI-AgNCs were compatible for safe gene therapy applications. The reduced cytotoxicity was presumably due to the biocompatibility of AgNC.29
MiR-21/221 expression in vitro
Motivated by the capacity to absorb nucleic acid, the appropriate size, as well as the low cytotoxicity, PEI-AgNCs was then assessed for the ability to act as an efficient carrier in gene delivery. EGFP-C1 gene expression assay demonstrated that PEI-AgNCs is an efficient carrier in gene delivery (Figure S4). However, DNA transfection data cannot readily be implemented for other genes, such as small RNAs.30 Therefore, we further investigated whether PEI-AgNCs was also capable of driving oligonucleotides into targeted cells efficiently. MiR-21 and miR-221 have been widely studied as the oncogenes in multiple tumors.31,32 In this study, miR-21m or miR-21i was transfected into HepG2 cells using PEI-AgNCs carriers. In miR-21 gain-of-function experiments, the qRT-PCR results (Figure 4A) showed a significant increase in the miR-21 expression level when cells were transfected with miR-21m using different transfection carriers compared to the control group. In miR-21 loss-of-function experiments, the qRT-PCR results (Figure 4C) showed a significant decrease in the miR-21 expression level when cells were transfected with miR-21i using different transfection carriers compared to the control group. Interestingly, compared to PEI, PEI-AgNCs exhibited higher transfection efficiency to miR-21m, which enhanced miR-21 expression in HepG2 cells. The parallel result was also found in miR-221 expression (Figure 4B and D). This fascinating result provided an attractive chance for tumor treatment that depends on miRNA replacement.
Although the results indicated that the transfection efficiency of PEI-AgNCs on miR-221i modestly increased compared to the PEI, the difference was not statistically significant (Figure 4D). The transfection efficiency of PEI-AgNCs was much higher than that of PEI when using miR-221 mimics (Figure 4). One possible explanation for this result is that the complex formed by miR-221 mimics (double-stranded RNA) and PEI-AgNCs is more easily endocytosed by cells than that formed by miR-221 inhibitors (single-stranded RNA).
The biological function induced by PEI-AgNCs/miR-221 complexes in HepG2 cells was confirmed by evaluating the protein expression levels of the downstream targets of miR-221, such as PTEN.33 As expected, an increase in PTEN protein expression was observed with the treated PEI-AgNCs/miR-221i (Figure 4E, left panel). In contrast, an obvious decrease in PTEN protein expression was observed with treated PEI-AgNCs/miR-221m (Figure 4E, right panel). These results suggested that PEI-AgNCs carried miR-221i or miR-221m into the cytoplasm efficiently and specifically silenced or enhanced the miR-221 expression in HepG2 cells. These results further demonstrated that PEI-AgNCs can transfect miRNA into HepG2 cells to function with a practical application.
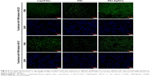
To further confirm the ability of PEI-AgNCs to carry miRNA mimics into cells, the fluorescent F-miRm were assayed by inverted fluorescence microscopy. PEI-AgNCs-mediated transport in HepG2 and 293A cells are shown in Figures 5 and 6, respectively. In the inverted fluorescence microscopy images, green fluorescence for F-miRm marker and blue fluorescence for DAPI staining of the nuclei were visualized. The visible fluorescence intensity of PEI-AgNCs/F-miR-21m was significantly higher than that of PEI/F-miR-21m. Similar results were observed when transfected with miR-221. These data were consistent with the results that were obtained by qRT-PCR and further demonstrated that PEI-AgNCs have great potential to transfect miRNAs.
Antibacterial activity
AgNC and its compounds have been known for their antimicrobial activities, and PEI also has been explored as an antiseptic.34,35 However, there have been few reports about the antimicrobial activities of PEI-AgNCs. To investigate the bactericidal effect, we compared the antibacterial ability of PEI-AgNCs with PEI against E. coli. Approximately, 1.0×109 CFU of log-phase E. coli were inoculated into 150 μL of fresh LB medium containing 14.25 μg/mL of PEI and PEI-AgNCs in a 96-well plate. As shown in Figure 7A, no significant differences in the growth of E. coli were observed for the first 4 hours among the different groups. However, after 6 hours, the OD600 of PEI- and PEI-AgNCs-treated cells markedly decreased compared with the control group. Meanwhile, the number of viable bacterial cells was measured using the CFU counting method (Figure 7B and C). The number of viable cells in the PEI-AgNCs-treated group was significantly lower than that in the PEI-treated group at the same concentration (14.25 μg/mL). To further explore the antibacterial mechanism of PEI-AgNCs, morphological changes of E. coli were studied by SEM (Figure 7D). The cell walls treated with PEI-AgNCs were wrinkled and damaged, which is in contrast to that observed in the untreated cells, which are smooth and intact. These data indicated that after the conjugation of positively charged PEI to AgNC, PEI-AgNCs exhibited higher antibacterial activity than PEI alone, which made it possible to prevent bacterial contamination in the process of transfection.
In vitro bioimaging
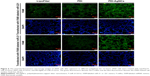
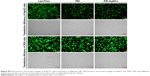
Because of the fascinating optical properties and the superiority of easy synthesis by designing different ligands as stabilizing agents, AgNCs can be employed for biological imaging. Figure 8 shows the photoluminescence images of different cells after incubation with PEI-AgNCs for 6 hours. Notably, the cells revealed an extremely intense green fluorescence, potently suggesting that PEI-AgNCs could act as a promising optical label for cell imaging, which endows PEI with great potential to be an ideal fluorescent probe to track transfection behavior.36
Conclusion
In this study, a cationic transfection polymer, PEI-AgNCs, was synthesized and used for the purpose of miRNA delivery. The obtained PEI-AgNCs polymer showed the capacity to condense the plasmid and oligonucleotide and form stable nanoparticles that had a size on the nanoscale. Compared to its parental PEI, PEI-AgNCs exhibited relatively low cytotoxicity and was able to mediate higher transfection efficiency to miR-21/221m in HepG2 and 293A cells in vitro. In addition, PEI-AgNCs was demonstrated to transfect miR-221i or miR-221m into HepG2 cells to regulate the expression of PTEN, which provided a chance to tumor treatment. Moreover, AgNCs endowed PEI with stronger antibacterial activity, which conferred PEI-AgNCs with the potential to prevent bacterial contamination in the process of transfection. Furthermore, without the need for fluorophore labeling, PEI-AgNCs possess superior photoluminescent properties, allowing bioimaging and tracking the transfection behavior inside cells. Therefore, PEI-AgNCs can be utilized as efficient gene delivery agents.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 81400791, 21505036), the Young Talent project of the Education Department of Hubei Province (grant number Q20141002), the major projects of technological innovation of Hubei Province (grant number 2016ACA140), and the Industry University Cooperation project of the Ministry of Education of China (grant number 201601031006).
Disclosure
The authors report no conflicts of interest in this work.
References
Tanaka S, Miyazaki J, Tiwari DK, et al. Fluorescent platinum nanoclusters: synthesis, purification, characterization, and application to bioimaging. Angew Chem Int Ed Engl. 2011;50(2):431–435. | ||
Yu J, Choi S, Dickson R. Shuttle-based fluorogenic silver-cluster biolabels. Angew Chem Int Ed Engl. 2009;48(2):318–320. | ||
Vosch T, Antoku Y, Hsiang JC, et al. Strongly emissive individual DNA-encapsulated Ag nanoclusters as single-molecule fluorophores. Proc Natl Acad Sci U S A. 2007;104(31):12616–12621. | ||
Peyser LA, Vinson AE, Bartko AP, et al. Photoactivated fluorescence from individual silver nanoclusters. Science. 2001;291(5501):103–106. | ||
Yang SW, Vosch T. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal Chem. 2011;83(18):6935–6939. | ||
Song J, Wu F, Wan Y, et al. Visual test for melamine using silver nanoparticles modified with chromotropic acid. Microchimica Acta. 2014;181(11–12):1267–1274. | ||
Shang L, Dörlich RM, Trouillet V, et al. Ultrasmall fluorescent silver nanoclusters: protein adsorption and its effects on cellular responses. Nano Res. 2012;5(8):531–542. | ||
Shang L, Dong S, Nienhaus GU. Ultra-small fluorescent metal nanoclusters: synthesis and biological applications. Nano Today. 2011;6(4):401–418. | ||
Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346–2353. | ||
Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genetics. 2007;8(3):173–184. | ||
Luo A, Yan H, Liang J, et al. MicroRNA-21 regulates hepatic glucose metabolism by targeting FOXO1. Gene. 2017;627:194–201. | ||
Meerson A, Traurig M, Ossowski V, et al. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-α. Diabetologia. 2013;56(9):1971–1979. | ||
Lustig Y, Barhod E, Ashwal-Fluss R, et al. RNA-binding protein PTB and microRNA-221 coregulate AdipoR1 translation and adiponectin signaling. Diabetes. 2014;63(2):433–445. | ||
Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. | ||
Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–1122. | ||
Hu QL, Jiang QY, Jin X, et al. Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials. 2013;34(9):2265–2276. | ||
Gao S, Tian H, Guo Y, et al. miRNA oligonucleotide and sponge for miRNA-21 inhibition mediated by PEI-PLL in breast cancer therapy. Acta Biomater. 2015;25:184–193. | ||
Jia H, Luo X, Cheng H, et al. Extraordinarily enhanced gene transfection and cellular uptake by aromatic hydrophobicization to PEI25K. J Mater Chem. 2012;22(45):24092–24101. | ||
Nel AE, Mädler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. | ||
Fischer D, Li Y, Ahlemeyer B, et al. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. | ||
Yuan Z, Cai N, Du Y, et al. Sensitive and selective detection of copper ions with highly stable polyethyleneimine-protected silver nanoclusters. Anal Chem. 2014;86(1):419–426. | ||
Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189(23):8746–8749. | ||
Wang HY, Hua XW, Wu FG, et al. Synthesis of ultrastable copper sulfide nanoclusters via trapping the reaction intermediate: potential anticancer and antibacterial applications. ACS Appl Mater Interfaces. 2015;7(13):7082–7092. | ||
Wen T, Qu F, Li NB, et al. Polyethyleneimine-capped silver nanoclusters as a fluorescence probe for sensitive detection of hydrogen peroxide and glucose. Anal Chim Acta. 2012;749(20):56–62. | ||
Lewin M, Carlesso N, Tung CH, et al. Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat Biotechnol. 2000;18(4):410–414. | ||
Zhu Y, Liang G, Sun B, et al. A novel type of self-assembled nanoparticles as targeted gene carriers: an application for plasmid DNA and antimicroRNA oligonucleotide delivery. Int J Nanomedicine. 2016;11(1):399–411. | ||
Yin H, Kanasty RL, Eltoukhy AA, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. | ||
Chen M, Cooper HM, Zhou JZ, et al. Reduction in the size of layered double hydroxide nanoparticles enhances the efficiency of siRNA delivery. J Colloid Interface Sci. 2013;390(1):275–281. | ||
Lee KJ, Nallathamby PD, Browning LM, et al. In vivo imaging of transport and biocompatibility of single silver nanoparticles in early development of zebrafish embryos. Acs Nano. 2011;1(2):133–143. | ||
Höbel S, Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5(5):484–501. | ||
Liu C, Yu J, Yu S, et al. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. 2010;53(1) :98–107. | ||
Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. | ||
Garofalo M, Leva GD, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. | ||
Shahverdi AR, Fakhimi A, Shahverdi HR, et al. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine. 2007;3(2):168–171. | ||
Lin J, Qiu S, Lewis K, et al. Mechanism of bactericidal and fungicidal activities of textiles covalently modified with alkylated polyethylenimine. Biotechnol Bioeng. 2003;83(2):168–172. | ||
Díez I, Ras RH. Fluorescent silver nanoclusters. Nanoscale. 2011;3(5):1963–1970. |
Supplementary materials
Methods
Atomic force microscope imaging
Polyethyleneimine-capped silver nanoclusters (PEI-AgNCs)/DNA samples with different nitrogen:phosphate (N/P) ratios were deposited onto freshly cleaved mica substrates. After 15 minutes of incubation, pour off remaining PEI-AgNCs/DNA solution, then rinse off any excess buffer using a syringe or micropipettor to squirt clean Millipore water on the mica substrate, and then the sample was dried under nitrogen. Atomic force imaging (AFM) topography images of immobilized complexes were acquired in air using the tapping mode on an AFM (Dimension EDGE, Bruker Corporation, Billerica, MA, USA).
Fluorescence microscopic images of EGFP-C1 gene expression
In vitro transfection efficiency of PEI-AgNCs was compared with PEI and the commercial reagents. The study was carried out under optimized conditions of N/P ratio in different cells. HepG2 cells were plated in 24-well plates at ~5×104 cells and allowed to adhere for 24 hours. Then the medium was exchanged with 500 μL of fresh basic medium with free serum. The PEI/DNA and PEI-AgNCs/DNA complexes containing 800 ng of DNA at the N/P ratio of 10.8 were incubated with cells for 24 hours. Cells were then washed and reincubated in 2 mL of medium containing 10% fetal bovine serum for 24 hours. 239A cells were used to judge the transfection efficiency of PEI-AgNCs in the same way as described earlier (N/P =4.8). Expression of EGFP-C1 within cells was imaged using an Olympus IX-73 optical camera (Olympus Corporation, Tokyo, Japan). LipoFiter™ Liposomal Transfection Reagent (Hanbio Biotechnology Co., Ltd., Shanghai, China) was used as positive control and applied according to the manufacturer’s procedures. The results of the experiments are presented in Figures S1–S4.
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.