Back to Journals » Infection and Drug Resistance » Volume 13
Plasmid Profiling and Occurrence of β-Lactamase Enzymes in Multidrug-Resistant Uropathogenic Escherichia coli in Kathmandu, Nepal
Authors Thapa Shrestha U , Shrestha S, Adhikari N , Rijal KR , Shrestha B, Adhikari B , Banjara MR , Ghimire P
Received 20 February 2020
Accepted for publication 28 May 2020
Published 23 June 2020 Volume 2020:13 Pages 1905—1917
DOI https://doi.org/10.2147/IDR.S250591
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Upendra Thapa Shrestha,1,2,* Sabnum Shrestha,1,* Nabaraj Adhikari,1,2 Komal Raj Rijal,2 Basudha Shrestha,3 Bipin Adhikari,4 Megha Raj Banjara,2 Prakash Ghimire2
1Kantipur College of Medical Science, Tribhuvan University, Sitapaila, Kathmandu, Nepal; 2Central Department of Microbiology, Tribhuvan University, Kathmandu, Nepal; 3Department of Microbiology, Kathmandu Model Hospital, Kathmandu, Nepal; 4Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
*These authors contributed equally to this work
Correspondence: Komal Raj Rijal
Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu, Nepal
Email [email protected]
Introduction: Extended-spectrum β-lactamases (ESBL) among Gram-negative bacteria, predominantly Escherichia coli (E. coli), in Nepal, have been rising. The main objectives of this study were to determine the prevalence of uropathogenic E. coli, antibiotic resistance, ESBLs, ABLs (AmpC type β-lactamases), MBLs (metallo-β-lactamases) and KPCs (Klebsiella pneumoniae carbapenemases) and their correlation with plasmid profiling patterns among patients with urinary tract infections in a tertiary hospital in Kathmandu, Nepal.
Methods: The mid-stream urine samples collected from patients were inoculated in cystine–lactose–electrolyte-deficient (CLED) agar plates. E. coli producing ESBLs, ABLs, MBLs/KPC were identified phenotypically using standard microbiological methods. Plasmids were extracted by alkaline lysis method from E. coli isolates and profiled using agarose gel electrophoresis.
Results: Out of the total 2661 urine samples, E. coli were isolated in 64.34% (507/788), among which 170 (33.53%) were multidrug-resistant (MDR) isolates. All MDR isolates were resistant to amoxicillin and third-generation cephalosporins but were highly sensitive to imipenem (94.12%, 160/170), amikacin (92.94%, 158/170) and nitrofurantoin (86.47%, 147/170). Among 170 MDR isolates, 78.2% (133/170) were ESBLs, 46.3% (50/170) were AmpC, 11.2% (19/170) were MBL and 0.6% (1/170) were KPC producers. Coproduction of β-lactamases was detected in 24.12% (41/170) of isolates. E. coli isolates showed one plasmid (> 33.5 kb), which was present in all the isolates. Overall, 44 different plasmid profile groups were identified based on molecular weight and number of plasmids. β-Lactamase producers were relatively resistant to the higher number of antibiotics tested (≤ 10) than non-producers (≤ 8), and the number of plasmids were higher in β-lactamase producers (≤ 7) than those in non-producers (≤ 5).
Conclusion: The higher prevalence of the ESBLs, AmpCs, KPCs and MBLs along with their coproduction in E. coli isolates highlights the importance of routine surveillance of ESBLs, AmpCs, KPCs and MBLs in microbiology laboratories using various phenotypic methods.
Keywords: uropathogenic Escherichia coli, antibiotic resistance, ESBL, extended-spectrum β-lactamases, AmpC type β-lactamases, MBL, metallo-β-lactamases, KPC, Klebsiella pneumoniae carbapenemases
Introduction
Antimicrobial resistance is one of the greatest challenges in modern empirical treatment of infectious diseases.1 For the treatment of infections, especially caused by Gram-negative bacteria, fluoroquinolones, cephalosporins, β-lactams and β-lactamases inhibitors alone or in combination are frequently used.2,3 However, resistance to such drugs is the major burden in developing countries.1 Among them, prevalence of beta-lactamases-producing pathogens is on the rise causing a threat to patients4–6 with spread of CTX-M genes; encoding genes have been seized from the chromosome of Kluyvera spp. The ctx gene is retained on a bacterial plasmid that is spread among the members of family Enterobacteriaceae, particularly in E. coli.7
The selective pressure induced by continuous and uncontrolled use of β-lactam antibiotics has triggered the emergence of diverse form of mutated novel beta-lactamase enzymes.8,9 Increasingly, E. coli have emerged as prominent bacteria for producing such newer β-lactamases. They comprise of plasmid-mediated AmpC β-lactamases (eg, CMY types), extended-spectrum β-lactamases (ESBLs, eg, TEM, SHV, CTX-M types), and carbapenemases (Klebsiella pneumoniae carbapenemase (KPC) types, metallo-β-lactamases (MBLs) and OXA types).10,11 CMY, CTX-M, and NDM types of β-lactamases in E. coli are predominantly responsible for the growing resistance to β-lactam antibiotics.12
Evidence of integron mediated resistant is sufficient in bacterium. These genes are usually found on plasmids encoding resistance to aminoglycosides, sulfonamides, tetracyclines and other antibiotics.13 Those carrying R-plasmids cause serious problems among antibiotic-resistant bacteria because of their propensity to spread rapidly.14 A laboratory evidence of horizontal transfer of multi-antibiotic-resistant plasmids among clinical isolates indicates further spread.15
Antibiotic-resistant genes in Gram-negative bacteria are located on the plasmids, which are responsible for resistance to a number of antimicrobial agents.16 Assigned plasmids are self-ruling DNA molecules equipped for self-transmission between cells, including their capacity to activate a part of the chromosome through high-recurrence recombination.17 The ability to acquire novel genes by plasmids through mobile genetic elements such as transposons or insertion sequences, and their propensity to replicate in a wide range of hosts, makes them perfect vectors for the spread of AMR.18 Identifying the characteristics of plasmids and their behavior in different bacterial hosts provides significant knowledge regarding the transmission of AMR. Molecular characterization of plasmid and genotypes can help to understand whether the spread of AMR genes is caused by epidemic plasmids in different hosts or by clonal spread of bacterial organisms. Plasmids are important for horizontal gene transfer in a close family of bacteria. These are responsible for the spread of antibiotic resistance genes in the environment. Thus, plasmid profiling helps to identify the potential of the spread of resistant genes. Plasmid profiles will be useful in the surveillance of outbreaks and in tracing antibiotics resistance.19
UTI is very common and can affect patients well beyond reproductive age.20 Although recognized as a most prevalent illness in infectious diseases wards in the hospitals, routine laboratory diagnoses with antibiotic profiles and susceptibility tests are rarely done.4,20–22 In most cases, only basic routine urine microbiological tests are conducted before prescribing the empirical treatment, and this may be even more prominent in non-tertiary hospitals and peripheral health facilities.4
Prescribing antibiotics without routinely identifying the organisms and their antimicrobial susceptibility can contribute to antibiotic resistance including ESBLs-producing organisms.23 Additionally, in Nepal, antibiotics are available over the counter (without a prescription) from drug shops and dispensers.24 High empirical treatment and unnecessary use of antibiotics in viral infections can further facilitate the development of AMR.4 This is a major problem and can increase the ESBLs-, AmpCs-, KPCs-, MBLs-mediated resistance.
Several studies in the past have investigated the prevalence of pathogens producing ESBLs among inpatients. Studies have shown the varying prevalence of ESBLs producers, for instance, 27.7% in Pokhara,23 55% in Kathmandu,25 64% in Chitwan,26 and 43% in Children Hospital in Kathmandu in 2018.27 Another study reported 35.9% of ESBLs-producing E. coil isolates among outpatients at a tertiary care hospital in Kathmandu.28 However, a study from Lalitpur district reported 6.8% of ESBLs-producing isolates.29 Studies have shown the wide range in prevalence of ESBLs producers (10% to 64%) in different hospitals/settings from various samples. However, there are very few studies that have explored the organisms producing ESBLs amongst the uropathogens from the clinical samples of patients suspected or diagnosed with urinary tract infections. In addition, studies so far have used only phenotypical methods to detect and determine the prevalence of ESBL organisms. It is established that ESBLs and AmpC are the main mechanisms of resistance to β-lactam antibiotics, but the genotypic mechanism of resistance varies globally. Therefore, providing the prevalence of circulating β-lactamase genes in each geographic region can be important. Furthermore, multidrug resistance in bacteria may result from the accumulation of multiple genes by resistance plasmids (R-Plasmids) and/or by the increased expression of genes that code for multidrug efflux pumps.30
The main objectives of this study were to determine the prevalence of E. coli producing ESBLs, AmpC (AmpC type β-lactamases), MBL (metallo-β-lactamase) and KPC (Klebsiella pneumoniae carbapenemases), and their correlation with plasmid profiling (genotyping) patterns among the patients suspected of or with urinary tract infection in a tertiary hospital in Kathmandu, Nepal.
Methods
A hospital-based cross-sectional study was conducted from October 2013 to March 2014 at Kathmandu Model Hospital, Kathmandu, Nepal. A total of 2661 mid-stream urine samples were collected from out-patients department (OPD) and intensive care unit (ICU) during the study period. The samples were collected in a clean, leak-proof container with no visible signs of contamination and were labeled properly with demographic information of patients. The samples from patients with a recent history of antibiotics therapy were excluded from this study. A written informed consent was obtained from the patients for participation and collection of samples. The cultured urine samples were considered significant when the number of colonies were more than 105 cfu/mL.
Identification of E. coli
Urine samples were cultured on cystine–lactose–electrolyte-deficient (CLED) agar plates by semi-quantitative culture technique.31 The isolation and identification of E. coli were performed following standard microbiological techniques as described by the American Society of Microbiology (ASM).32
“Antimicrobial Susceptibility Testing”
“Antimicrobial susceptibility testing” (AST) was performed using modified Kirby–Bauer disk diffusion method following the guidelines of Clinical and Laboratory Standards Institute guidelines (CLSI).33 Sixteen common antibiotic discs from HiMedia: meropenem (10µg), amoxycillin (10µg), cefotaxime (30µg), cefixime (30µg), ceftriaxone (30µg), ceftazidime (30µg), cefoxitin (30µg), cotrimoxazole (25µg), nitrofurantoin (300µg), levofloxacin (5µg), amikacin (30µg), gentamicin (10µg), cefoperazone/sulbactam (75/30µg), piperacillin/tazobactam (100/10µg), imipenem (10µg), doxycycline (30µg) were used. E. coli that were resistant to at least one agent in three or more antibiotic categories were characterized as multidrug-resistant E. coli.34
Screening and Phenotypic Confirmation of E. coli Producing ESBLs, AmpCs, MBLs and KPCs
Detection of ESBLs Producers
E. coli producing ESBLs was screened by using cefotaxime (30μg), ceftazidime (30μg) or ceftriaxone (30μg) following CLSI guidelines.33 ESBLs producers were confirmed by combined disc assay using SD240 Hi-Media ESBL detection kit consisting of ceftazidime (30μg) and ceftazidime (30μg) plus clavulanic acid (10μg).
Detection of AmpCs Production
MDR isolates were screened for AmpCs production using cefoxitin (30μg) with screening cutoffs of ≤18 (ie, the CLSI susceptible breakpoints)35 and were further subjected to phenotypic confirmation (Combined Disc Assay) using cefoxitin (30μg) alone and in combination with cloxacillin (200μg). Increment of the zone of inhibition by ≥4 mm was considered as positive result.36
Detection of MBLs and KPCs
E. coli isolates were considered as MBLs producers when the inhibitory zone diameters around the meropenem disc with ethylene diamine tetra-acetic acid (EDTA) and the meropenem disc with phenylboronic acid, PBA+EDTA had increased to ≥5mm as compared to the inhibitory zone diameter of meropenem disc alone. Similarly, KPCs producers among E. coli isolates were considered when the inhibitory zone diameter around the meropenem disc with PBA and the meropenem disc with PBA+EDTA had increased to ≥5mm as compared to the inhibitory zone diameter of meropenem disc alone.37 The reference strains, ESBLs producing E. coli NCTC 13351, non-ESBLs producing E. coli ATCC 25922, MBLs producing E. coli NCTC 13476 and non-MBLs producing E. coli ATCC 25922 from Institue of Medicine, Tribhuvan University Teaching Hospital, were included as controls in the study.
Extraction and Profiling of Plasmid DNA
Plasmid DNA was extracted by alkaline lysis method.38 The extracted DNA was separated using 0.8% agarose gel electrophoresis at 120V for 1 hour, stained with SYBR safe stain solution and a photograph of the stained gel was taken after exposure on UV trans-illuminator. Reference as super mix plasmid DNA marker (GeNei TM, Genei Laboratories Private Limited, India) was used for the estimation of plasmid sizes. Plasmid profiles were created by grouping strains possessing the same number of plasmid bands and molecular size.39
Results
Out of 2661 urine samples, 29.61% (788/2661) were positive for a urinary pathogen. E. coli was the predominant bacterial species accounting for 64.34% (507/788) of total isolates in which 170 (33.53%) were multidrug-resistant. All MDR isolates were resistant to amoxicillin and third-generation cephalosporins (ceftriaxone, cefotaxime, cefixime and ceftazidime) but were highly sensitive to imipenem (94.12%, 160/170), amikacin (92.94%, 158/170) and nitrofurantoin (86.47%, 147/170) (Table 1).
 |
Table 1 Antibiotic Susceptibility Pattern of MDR E. coli Isolates (n=170) |
Antibiotic-resistant pattern of MDR E. coli showed 18 different resistant phenotypes (Table 2). The isolates seemed to be resistant to the number of antimicrobials that ranged from 6 to 16 of the commonly used antibiotics. The resistance pattern to a different class of antibiotics among β-lactamase producer and non-producer MDR isolates of E. coli is shown in Table 3.
 |
Table 2 Antibiotic Resistance Pattern of Clinical Isolates of MDR E. coli Isolates |
 |
Table 3 Resistance to Different Classes of Antibiotics Among β-Lactamase Producer and Non-Producer MDR Isolates |
β-Lactamase production was observed in 162 bacterial isolates, out of which 133 (82.1%) were ESBLs producers, followed by 50 (30.84%) AmpCs producers, 19 (11.72%) MBLs producers and 1 (0.62%) KPCs producers. The co-production of ESBLs and AmpCs was observed in 33 (20.37) isolates, followed by 5 (3.09%) ESBLs and MBLs, 1 (0.62%) MBLs and KPCs and 2 (1.23%) MBLs and AmpCs co-producers (Figure 1).
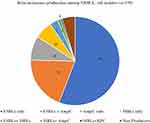 |
Figure 1 Distribution of different β-lactamases among MDR E. coli isolates. |
The β-lactamase producers were relatively resistant to a higher number of antibiotics used (highest: 16 antibiotics and nine different categories) while only a few isolates [1.76% (3/170)] were resistant to non-ESBLs producers to a maximum of 12 antibiotics (only seven different antibiotic categories) in this study. Seven plasmids were found in β-lactamase producers and a maximum of 5 plasmids were found in non-ESBLs producers (Table 3). E. coli isolates on plasmid profiling showed that one plasmid of greater than 33.5 kb size was present in all the isolates. The number of plasmids in the isolates varied from 1 to 7 along with the size ranging from 0.8 kb to >33.5 kb. Forty-four different plasmid profile patterns were obtained based on molecular weight and number of plasmids content. The number of isolates per plasmid profile group varied from 1 to 20. The maximum number of isolates had two plasmids [67 (39.41%)], followed by three plasmids [29 (17.06%)], and one plasmid [20 (11.76%)]. The most frequent molecular weight pattern obtained among 20 (11.76%) isolates was >33.5 kb, followed by 19 (11.18%) isolates possessing two plasmids of 33.5kb and >33.5kb sizes (Table 4).
 |
Table 4 Plasmid Profile of MDR E. coli Isolates |
Non-β-lactamase-producing isolates were found to possess one, two and five plasmids, whereas β-lactamase producers had multiple plasmids. Most of the uropathogenic E. coli producing ESBLs, AmpC and ESBLs/AmpCs had a variable number of plasmids with one plasmid size (>33.5 kb) in common (Table 5). MBLs producers and co-producers of MBLs with AmpCs, and ESBLs and KPCs had few number of plasmids; however, all of them contained plasmid size greater or equal to 33.5 kb in common. The lower molecular size plasmids showed no significant role in β-lactamases production. The lowest molecular weight plasmid, ie, 0.8 kb, was found in non β-lactamase-producing isolates. Plasmid DNA of clinical isolates of MDR E. coli was separated by molecular weight on agarose gel stained with ethidium bromide (Figure 2).
 |
Table 5 Plasmid Profiling in Relation to β-Lactamase Production in MDR E. coli Isolates |
Discussion
E. coli is a predominant cause of both community and nosocomial urinary tract infections.40 The trend of emerging multiple antibiotic-resistant E. coli among clinical samples raises a major concern for the clinician. The findings of this study show a high level of resistance among E. coli against antibiotics with sensitivity to imipenem (94.12%), amikacin (92.94%) and nitrofurantoin (86.47%). Consistent results of resistance have been reported from Saudi Arabia,41 Egypt42 and India.43 E. coli isolates from clinical settings showed resistance to nitrofurantoin that ranged from 2.4% to 6.5% and resistance to amikacin (2%) while resistance to imipenem was only 0.3%.41 Our findings are consistent with the study from India which showed least resistance to amikacin (13.9%) and imipenem (2.3%).43 Many studies have shown that meropenem has greater potency than imipenem by 4–16 folds even in E. coli and other members of Enterobacteriaceae.44,45 However, contrastingly, this study found a higher sensitivity of imipenem (94.12%) than meropenem (12.35%). One study from India has shown consistent findings with our study.46 The differences in resistance profile with the other studies can result from differences in infection epidemiology, prescription patterns, and socio-demographic features of the population. In this study, we included only patients who presented with suspected UTI in contrast to other studies that explored all patients or all the infectious diseases. In addition, the commonly used antibiotics in Nepal both at the hospital and over the counter include mostly oral penicillins and cephalosporins, which might explain the high resistance among these groups of antimicrobials.47,48.
More than 70% of the isolates were ESBLs producers, nearly half of the isolates were AmpCs producers and fewer percent of MBLs and KPCs producers were found in this study. The continuous exposure of bacteria to a variety of β-lactams has been well established for the production of β-lactamases, the most common mechanism of conferring β-lactams resistance among Enterobacteriaceae.8 The β-lactamase enzyme production mediated by both chromosomal and plasmid genes (subject to inducible expression) is a pivotal means of antibiotic resistance. These characteristic features are usually coded on transferable genes.8 β-Lactamase genes are usually found on plasmids encoding resistance to aminoglycosides, fluoroquinolones, sulfonamides, tetracyclines and other antibiotics,49 thus leading to cross resistance. Pathogen producing ESBLs, AmpCs and carbapenemases like MBLs and KPCs leaves us with limited treatment options. Further, MBLs just like ESBLs and AmpCs producers can be transferred between species by plasmids.50
In Nepal, there were limited studies demonstrating a high level of ESBLs producers among which Enterobacteriaceae were 28% to 67%.51 Most of the studies conducted showed that the ESBLs producers range as low as 22.3% to as high as 86.9%. Studies conducted in hospital settings reported 55% of ESBLs producers in Kathmandu,51 27.7% in Pokhara23 and 63.27% in Kathmandu Model Hospital.52 Previous studies from India have reported ESBLs productions in uropathogenic E. coli ranging from 18.5% to 60.7%.53
AmpC β-lactamases are plasmid-mediated that hydrolyze all cephalosporins except cefepime and the carbapenems. In AmpCs, the inducible chromosomal genes are mobilized as plasmids.54 In this study, the rate of AmpCs detection was 46.3%. The production of AmpCs in our hospital setting is higher compared to other studies that showed 12.6% at the tertiary care transplant center, in Kathmandu, Nepal,55 19.8% over a period of 6 months at various tertiary care hospitals in India,56 11.9% at a teaching hospital in Nigeria,57 and 5% in three major hospitals of Iran.58 The higher rate of AmpCs in this study might be due to the use of penicillin, cephalosporins and oxymino-β-lactams drugs as the primary choice of empiric therapy for the infections caused by Gram-negative bacteria.
Carbapenems are the ultimate resort for the treatment of isolates resistant to penicillin and cephalosporins.59 Carbapenem resistance due to carbapenemase production was firstly discovered in 1988 in New York, America.60 In our study, the rate of MBLs detection was 11.2%. Government medical college in Uttarakhand, India, reported 15.3% MBLs-producing isolates61 and a medical college in South India reported 13.4% MBLs-producing E. coli,62 which are almost consistent with this study. MBLs-producing E. coli were similar across other studies from India (2.87%), Dubai (0.3%) and Nigeria (41.2%).63–65 In this study, KPCs production was 0.6% in contrast to a study conducted in Iran which reported the prevalence of KPCs to be 1.4%.64 This particular difference could have been due to the variation in the settings.
A total of 41 (24.12%) isolates co-produced β-lactamase enzymes in this study, whereas other studies from our similar settings, with different clinical samples, have reported to be 6.2%.65 However, a study from Punjab, India, showed a similar prevalence of 19.04%.66 The co-existence of different classes of β-lactamases in a single bacterial isolate imposes diagnostic and treatment challenges with limited available drugs that are potentially toxic that include polymyxin and colistin.62 Furthermore, the outcomes for infected patients treated with such drugs remain unknown.67
E. coli isolates on plasmid profiling showed one plasmid >33.5 kb and was present in all the isolates. Many of the isolates possessed common plasmids of similar molecular size and these isolates also show the same β-lactamase production. Existence of common plasmid among the isolates implies the spread of resistant plasmid in the community. Plasmids of molecular size ranging from 0.8 kb to >33.5 kb were isolated from all the MDR isolates. Contrastingly, plasmid sizes ranging from 0.12 kb up to 65 kb were reported from West Nigeria (from 0.12 kb to 23 kb), Kathmandu (from 2.05 kb to >33.5 kb) and southwestern Nigeria (from 11.8 kb to 33.5 kb).65,68,69
Different plasmids can co-exist in the same host cell.35 Based on the antibiotic resistance patterns, the high molecular weight plasmid seemed to consist most of the resistance genes as previous studies have also demonstrated that multi-antibiotic resistance is associated with higher molecular weight plasmids.70,71 Comparing the antibiotic pattern with the plasmid profile pattern does not offer much in linking specific antibiotic patterns with specific plasmid profiles. This suggests that the genes resistant to various antibiotic patterns were probably distributed in plasmids of various sizes as well as in the chromosomes.
Strengths and Limitations
This is the first study to explore the plasmid profile in correlation with different β-lactamases of uropathogenic E. coli in patients attending a tertiary hospital in Kathmandu. Constrained by the objective of the study, we did not collect information on socio-demographics of the patients, which could have provided vital information about the source and characteristics of the samples. This study was conducted in one tertiary hospital in Kathmandu; thus, the findings from this study may not be generalizable, although the findings are consistent with previous studies. This study relied on the plasmid profiles of the uropathogens to derive the results, adding genotypic information could have helped to determine the role and distribution of antibiotic resistance. Different buffer solutions used in agarose gel electrophoresis were prepared manually. Therefore, we found a poor resolution of the separated band of plasmid DNA, which is one of the major limitations of this study. Because of logistical region, we used alkaline lysis method for isolation of bacterial plasmid. As a conventional method, it can easily remove chromosomal DNA; however, chances of plasmid sharing could be one of the reasons for small size plasmid in our study. Future studies can provide more information by exploring the plasmid profile together with the genotypic information. In future, studies conducted in multiple sites can be useful in providing more generalizable results.
Conclusions
This study has shown a high prevalence of beta-lactamases-producing E. coli in a tertiary hospital of Kathmandu. This clearly shows the urgency of the emerging resistance. To counteract such an emerging resistance, at first, detection of ESBLs producers should be routinely recommended in all tertiary hospitals including the peripheral laboratories of Nepal. Second, an antibiotic therapy guideline should be developed to scrutinize the use of antibiotics in hospitals including on-the-counter sale of antibiotics for infections such as UTI. In addition, profiling of the plasmid and chromosome of E. coli can be useful to ascertain the evolution and spread of antibiotic resistance among isolates.
Abbreviations
AmpCs BL, AmpC type β-lactamases; AST, antibiotic susceptibility test; ATCC, American Type Culture Collection; CLED, cystine–lactose–electrolyte-deficient agar; CLSI, Clinical and Laboratory Standard Institute; EDTA, ethylene diamine-tetraacetic acid; ESBL, extended-spectrum β-lactamase; IPD, inpatients department; KMH, Kathmandu Model Hospital; KPCs, Klebsiella pneumoniae carbapenemases; MBLss, metallo-β-lactamase; MDR, multidrug-resistant; MHA, Mueller Hinton Agar; MSU, mid-stream urine; NDM, New Delhi metallo β-lactamase; OPD, outpatients department; PBP, phenyl boronic acid; UTI, urinary tract infection; WHO, World Health Organization.
Data Sharing Statement
All data pertaining to this study are within the manuscript.
Ethical Approval and Consent
This study was ethically approved from institutional review committee (IRC); phect-NEPAL. A written informed consent was obtained from each patient before their participation and collection of samples. In the case of illiterate participants, information was provided by reading the consent form in the presence of witnesses.
Acknowledgments
The authors would like thank Kantipur College of Medical Science and Kathmandu Model Hospital for providing an opportunity to conduct this study. The authors are also grateful to all the patients and technical staff for their help during the study. We would like to express our gratitude to Mr. Gordon Tambellini, USA, for proof reading and edits.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
1. Pokharel S, Raut S, Adhikari B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob Health. 2019;4(6):e002104. doi:10.1136/bmjgh-2019-002104
2. Parajuli NP, Acharya SP, Mishra SK, Parajuli K, Rijal BP, Pokharel BM. High burden of antimicrobial resistance among Gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob Resist Infect Control. 2017;6(1):67. doi:10.1186/s13756-017-0222-z
3. Gajdacs M. The concept of an Ideal antibiotics: implications for drug design. Molecules. 2019;24(5):892. doi:10.3390/molecules24050892
4. Raut S, Adhikari B. ESBL and their identification in peripheral laboratories of Nepal. Nepal Med Coll J. 2015;17(3–4):176–181.
5. Guragin N, Pradhan A, Dhungel B, Banjara MR, Rijal KR, Ghimire P. Extended spectrum B-lactamase producing Gram negative bacterial isolates from urine of patients visiting Everest Hospital, Kathmandu, Nepal. TUJM. 2019;6(1):26–31.
6. Kayastha K, Dhungel B, Karki S, et al. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella species in pediatric patients visiting International Friendship Children’s Hospital, Kathmandu, Nepal. Infect Dis. 2020;13:1–7.
7. Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum b-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26(4):744–758. doi:10.1128/CMR.00023-13
8. Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev. 2009;22(9):161–82.
9. Deshmukh DG, Damle AS, Bajaj JK, Bhakre JB. The metallo β-lactamase producing clinical isolates from the patients of a tertiary care hospital. J Lab Physicians. 2011;3(2):93–97. doi:10.4103/0974-2727.86841
10. Bush K. Alarming beta-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010;13:558–564. doi:10.1016/j.mib.2010.09.006
11. Nordmann P, Poirel L, Toleman MA, Walsh TR. Does broad spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? Antimicrob Agents Chemother. 2011;66(4):689–692. doi:10.1093/jac/dkq520
12. Pitout JD. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol. 2012;3:9. doi:10.3389/fmicb.2012.00009
13. Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of extended spectrum-ß-lactamase (ESBL) producing Gram-negative bacilli and further molecular characterization of ESBL producing E. coli and Klebsiella spp. J Clin & Diag Res. 2013;7(10):2173–2177.
14. Saeed A, Khatoon H, Ansari FA. Multidrug resistant Gram-negative bacteria in clinical isolates from Karachi. Pak J Pharm Sci. 2009;22:44–48.
15. Motayo BO, Ogiogwa IJ, Okerentugba PO, et al. Antimicrobial resistance profile of Extra-intestinal E. coli infections in a Southwestern Nigerian City. J Microbiol Res. 2012;2(5):141–144. doi:10.5923/j.microbiology.20120205.05
16. Munita JM, Arias CA. Mechanism of antibiotic resistance. Microbial Spectra. 2016;4(2):10.
17. Adelberg EA, Pittard J. Chromosome transfer in bacterial conjugation. Bacteriol Rev. 1965;29(2):161–172. doi:10.1128/MMBR.29.2.161-172.1965
18. Rozwandowicz M, Brouwer MSM, Fischer J, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrobial Chemother. 2018;73(5):1121–1137. doi:10.1093/jac/dkx488
19. Tayfour MA, Eris FN, Alanazi AR. Comparison of antibiotic susceptibility tests, plasmids profiles and restriction enzyme analysis of plasmid DNA of methicillin susceptible and resistant Staphylococcus aureus strains isolated from intensive care units. Saudi Med J. 2005;26(1):57–63.
20. Pradhan B, Pradhan SB. Prevalence of urinary tract infection and antibiotic susceptibility pattern to urinary pathogens in Kathmandu Medical College and Teaching Hospital, Duwakot. BJHS. 2017;2(1):134–137.
21. Gajdacs M, Urban E. Resistance Trends and epidemiology of Citrobacter-Enterobacter- Serratia in urinary tract infections of inpatients and outpatients (RECESUTI): a 10-year survey. Medicina. 2019;55(6):285. doi:10.3390/medicina55060285
22. Gajdacs M, Abrok M, Lazar A, Burain K. Comparative epidemiology and resistance trends of common urinary pathogens in a tertiary care hospital: a 10-year surveillance study. Medicina. 2019;55(7):356. doi:10.3390/medicina55070356
23. Raut S, Gokhale S, Adhikari B. Prevalence of extended spectrum beta-lactamases among Escherichia coli and Klebsiella spp. Isolates in Manipal Teaching Hospital, Pokhara, Nepal J Microbiol and Infect Dis. 2015;5(2):69–75.
24. Raut S, Adhikari B. Global leadership against antimicrobial resistance ought to include developing countries. Lancet Infect Dis. 2016;16(7):775. doi:10.1016/S1473-3099(16)30078-0
25. Sharma AR, Bhatta DR, Shrestha J, Banjara MR. Antimicrobial susceptibility pattern of Escherichia coli isolated from urinary tract infected patients attending Bir Hospital. Nep J Sci and Tech. 2013;14(1):177–184. doi:10.3126/njst.v14i1.8938
26. Shrestha A, Manandhar S, Pokharel P, Panthi P, Chaudhry DK. Prevalence of extended spectrum beta-lactamase (ESBL) producing multidrug resistance Gram-negative isolates causing urinary tract infection. EC Microbiol. 2016;4(5):749–755.
27. Sharma KR, Bhandari P, Adhikari N, Tripathi P, Khanal S, Tiwari BR. Extended spectrum ß-lactamase (ESBL) producing multi-drug resistant urinary pathogens in a children hospital from Nepal. Kathmandu Univ Med J (KUMS). 2018;16(62):151–155.
28. Nepal K, Pant ND, Neupane B, et al. Extended spectrum beta-lactamase and metallo-beta-lactamase producing E. coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care- hospital in Kathmandu. Ann Clin Microbiol Antimicrob. 2017;19(16):62. doi:10.1186/s12941-017-0236-7
29. Rai S, Pant ND, Bhandari R, et al. AmpC and extended spectrum beta-lactamase production urinary isolates from a tertiary care hospital in Lalitpur, Nepal. BMC Res Notes. 2017;10(1):467. doi:10.1186/s13104-017-2784-5
30. Hiroshi N. Multidrug resistance in Bacteria. Annu Rev Biochem. 2009;78:119–146. doi:10.1146/annurev.biochem.78.082907.145923
31. Cheesbrough M. District Laboratory Practice in Tropical Countries, Part II.
32. Isenberg HD. Clinical Microbiology Procedures Handbook.
33. Clinical Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty Second Informational Supplement Document. Wayne, PA: CLSI; 2012:M100–S20.
34. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pan drug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
35. Polsfuss S, Bloemberg GV, Giger J, Meyer V, Böttger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clinic Microbiol. 2011;49(8):2798–2803. doi:10.1128/JCM.00404-11
36. Tan TY, Yong-Ng LS, He J, Koh TH, Hsu LY. Evaluation of screening methods to detect plasmid mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. Antimicrob Agents Chemother. 2009;53(1):146–149. doi:10.1128/AAC.00862-08
37. Tsakris A, Poulou A, Pournaras S, et al. A simple phenotypic method for the differentiation of metallo-β-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. Antimicrob Agents Chemother. 2010;65(8):1664–1671. doi:10.1093/jac/dkq210
38. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual.
39. Daini OA, Adesemowo A. Antimicrobial susceptibility patterns and R-plasmids of clinical strains of Escherichia coli. Aust J Basic Appl Sci. 2008;2:397–400.
40. Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7(12):653–660. doi:10.1038/nrurol.2010.190
41. Al-Tawfiq JA. Increasing antibiotic resistance among isolates of Escherichia coli recovered from inpatients and outpatients in a Saudi Arabian hospital. Infect Control Hosp Epidemiol. 2006;27:748–753. doi:10.1086/505336
42. Salem MM, Muharram M, Alhosiny IM. Distribution of classes 1 and 2 integrons among multi drug resistant E. coli isolated from hospitalized patients with urinary tract infection in Cairo, Egypt. Aus J Basic and Appl Sci. 2010;4:398–407.
43. Christopher AF, Hora S, Ali Z. Investigation of plasmid profile, antibiotic susceptibility pattern multiple antibiotic resistance index calculation of Escherichia coli isolates obtained from different human clinical specimens at tertiary care hospital in Bareilly-India. Ann Trop Med Public Health. 2013;6(3):285–289. doi:10.4103/1755-6783.120985
44. Zhanel GG, Simor AE, Vercaigne L, Mandell L. Imipenem and meropenem: comparison of in vitro activity, pharmacokinetics, clinical trials and adverse effects. Can J Infect Dis. 1998;9(4):215–228. doi:10.1155/1998/831425
45. Piller CM, Torres MK, Brown NP, Shah D, Sahm DF. In vitro activity of doripenem, a carbapenem for the treatment of challenging infections caused by Gram-negative bacteria, against recent clinical isolates from the United States. Antimicrob Agents Chemother. 2008;52(12):4388–4399. doi:10.1128/AAC.00381-08
46. Padmini SB, Appalaraju B. Extended spectrum β-lactamases in urinary isolates of E. coli and Klebsiella pneumoniae, prevalence and susceptibility pattern in tertiary care hospital. Ind J Med Microbiol. 2004;223:172–174.
47. Wachtler DA, Joshi MP, Rimal B. Antibiotic dispensing by drug retailers in Kathmandu, Nepal. Trop Med Int Health. 1999;4(11):782–788. doi:10.1046/j.1365-3156.1999.00476.x
48. Nepal A, Hendrie D, Robinsons S, LA S. Survey of the pattern of antibiotic dispensing in private pharmacies in Nepal. BMJ Open. 2019;9(10):e032422. doi:10.1136/bmjopen-2019-032422
49. Khajuria A, Praharaj AK, Kumar M, Grover N. Emergence of Escherichia coli, co-producing NDM-1 and OXA-48 carbapenemases, in urinary isolates, at a tertiary care centre at Central India.. J Clin Diagn Res. 2014;8(6):DC01–4. doi:10.7860/JCDR/2014/7952.4413
50. Hammer DA, Dongol S, Anderson TP, Wong JS, Werno AM, Murdoch DR. High prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Nepal. Int J Antimicrob Agents. 2007;30(5):471–472. doi:10.1016/j.ijantimicag.2007.07.004
51. Baral P, Neupane S, Marasini BP, Ghimire KR, Lekhak B, Shrestha B. High prevalence of multidrug resistance in bacterial uropathogens from Kathmandu, Nepal. BMC Res Notes. 2012;5(1):38. doi:10.1186/1756-0500-5-38
52. Shrestha S, Mali NM, Tiwari KB, Adhikari N, Shrestha UT, Basnyat SR. Antibiogram and plasmid profiling of clinical multidrug resistant Escherichia coli. J Inst Med. 2014;35(2):21–26.
53. Bora A, Ahmed GU, Hazarika NK. Antibiotic resistance pattern and prevalence of ESBL producing Escherichia coli isolates in urinary tract infection from a tertiary care hospital of Guwahati, Assam. IJBPAS. 2012;1(11):1659–1668.
54. Manoharan A, Sugumar M, Kumar A, et al. Phenotypic and molecular characterization of AmpC β lactamases among Escherichia coli, Klebsiella spp and Enterobacter spp from five Indian medical centers. Indian J Med Res. 2012;135:359–364.
55. Dhungana K, Awal BK, Dhungel B, Sharma S, Banjara MR, Rijal KR. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo betalactamae (MBL) producing Gram negative bacteria isolated from different clinical samples in a Transplant Center, Kathmandu, Nepal. ASMI. 2019;2(12):60–69.
56. Sasirekha B. Prevalence of ESBL, AmpC β- lactamases and MRSA among uropathogens and its antibiogram. EXCLI J. 2013;12:81–88.
57. Yusuf AH, Arzai M, Haruna AA, Sharif GMI. Detection of multi drug resistant bacteria in major hospitals in Kano, North-West, Nigeria. Braz J Microbiol. 2014;45(3):791–798. doi:10.1590/S1517-83822014000300005
58. Shayan S, Bokaeian M. Detection of ESBL- and AmpC-producing E. coli isolates from urinary tract infections. Adv Biomed Res. 2015;4(1):220. doi:10.4103/2277-9175.166643
59. Gajdacs M. Intravenous or oral antibiotic therapy: Sophie’s choice? Gen Int Med Clin Innov. 2019;4(2):1–2. doi:10.15761/GIMCI.1000176
60. Zavascki AP, Barth AL, Gonçalves AL, et al. The influence of metallo-beta-lactamase production on mortality in nosocomial Pseudomonas aeruginosa infections. J Antimicrob Chemother. 2006;58:387–392. doi:10.1093/jac/dkl239
61. Rawat V, Singhai M, Verma PK. Detection of different β-lactamases and their co-existence by using various discs combination methods in clinical isolates of Enterobacteriaceae and Pseudomonas spp. J Lab Physicians. 2013;5:21–25. doi:10.4103/0974-2727.115918
62. Wadekar MD, Anuradha K, Venkatesha D. Phenotypic detection of ESBL and MBL in clinical isolates of Enterobacteriaceae. Int J Curr Res Acad Rev. 2013;1:89–95.
63. Pandya NP, Prajapati SP, Mehta SJ, Kikani KM, Joshi PJ. Evaluation of various methods for detection of metallo-betalactamase (MBL) production in Gram-negative bacilli. Int J Biol Med Res. 2011;2(3):775–777.
64. Moayednia R, Shokri D, Mobasherizadeh S, Baradaran A, Fatemi SM, Merrikhi A. Frequency assessment of β-lactamase enzymes in Escherichia coli and Klebsiella isolates in patients with urinary tract infection. J Isfahan Uni Med Sci. 2014;19(1):S41–S45.
65. Enwuru NV, Enwuru CA, Ogbonnia SO. Adepoju-Bello metallo-beta-lactamase production by Escherichia coli and Klebsiella species isolated from hospital and community subjects in Lagos, Nigeria Nature and Science. 2011;9(11):1–5.
66. Oberoi L, Singh N, Sharma P, Aggarwal A. ESBL, MBL, AmpC β-lactamases producing superbugs – havoc in the intensive care units of Punjab, India. J Clin Diag Res. 2013;7(1):70–73.
67. Urban C, Bradford PA, Tuckman M, et al. Carbapenem-resistant Escherichia coli harboring Klebsiella pneumoniae carbapenemase β-lactamases associated with long-term care facilities. Clin Infect Dis. 2008;46(1):e127–30. doi:10.1086/588048
68. Shrestha B, Shrestha S, Mishra SK, et al. Phenotypic characterization of multidrug-resistant Escherichia coli with special reference to extended-spectrum-beta-lactamases and metallo-beta-lactamases in a tertiary care center. J Nepal Med Assoc. 2015;53(198):89–95. doi:10.31729/jnma.2768
69. Motayo BO, Akinduti PA, Adeyakinu FA, et al. Antibiogram and plasmid profiling of carbapenemase and extended spectrum beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae in Abeokuta, South western, Nigeria. Afr Health Sci. 2013;13(4):1091–1097. doi:10.4314/ahs.v13i4.33
70. McPherson P, Gealt M. Isolation of indigenous wastewater bacterial strains capable of mobilizing plasmid pBR325. Appl Environ Microbiol. 1986;51(5):904–909. doi:10.1128/AEM.51.5.904-909.1986
71. Zulkifli Y, Alitheen NB, Raha AR, Yeap SK, Marlina SR, Nishibuchi M. Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from cockles in Padang, Indonesia. Inter Food Res J. 2009;16:53–58.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

