Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 12
Pharmacogenomics And Hypertension: Current Insights
Authors Oliveira-Paula GH, Pereira SC , Tanus-Santos JE, Lacchini R
Received 7 September 2019
Accepted for publication 5 November 2019
Published 22 November 2019 Volume 2019:12 Pages 341—359
DOI https://doi.org/10.2147/PGPM.S230201
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Gustavo H Oliveira-Paula,1,2 Sherliane C Pereira,2 Jose E Tanus-Santos,2 Riccardo Lacchini3
1Department of Medicine, Division of Cardiology, Wilf Family Cardiovascular Research Institute, Albert Einstein College of Medicine, New York, NY, USA; 2Department of Pharmacology, Ribeirao Preto Medical School, University of Sao Paulo, Ribeirao Preto, SP, Brazil; 3Department of Psychiatric Nursing and Human Sciences, Ribeirao Preto College of Nursing, University of Sao Paulo, Ribeirao Preto, SP, Brazil
Correspondence: Riccardo Lacchini
Department of Psychiatric Nursing and Human Sciences, Ribeirao Preto College of Nursing, University of Sao Paulo, Av. Bandeirantes, 3900, Ribeirao Preto 14040-902, SP, Brazil
Tel +55 16 3315-3447
Email [email protected]
Abstract: Hypertension is a multifactorial disease that affects approximately one billion subjects worldwide and is a major risk factor associated with cardiovascular events, including coronary heart disease and cerebrovascular accidents. Therefore, adequate blood pressure control is important to prevent these events, reducing premature mortality and disability. However, only one third of patients have the effective control of blood pressure, despite several classes of antihypertensive drugs available. These disappointing outcomes may be at least in part explained by interpatient variability in drug response due to genetic polymorphisms. To address the effects of genetic polymorphisms on blood pressure responses to the antihypertensive drug classes, studies have applied candidate genes and genome wide approaches. More recently, a third approach that considers gene-gene interactions has also been applied in hypertension pharmacogenomics. In this article, we carried out a comprehensive review of recent findings on the pharmacogenomics of antihypertensive drugs, including diuretics, β-blockers, angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers, and calcium channel blockers. We also discuss the limitations and inconsistences that have been found in hypertension pharmacogenomics and the challenges to implement this valuable approach in clinical practice.
Keywords: antihypertensive therapy, candidate genes, GWAS, gene-gene interactions, hypertension, pharmacogenomics
Introduction
Cardiovascular disease is the leading global cause of death, accounting for approximately 17 million deaths annually.1 Hypertension is one of the most common cardiovascular diseases and it is a major risk factor for coronary heart disease and cerebrovascular accidents, which can lead to premature mortality, morbidity and significant economic costs.2 Studies indicate that within the next 20 years, the number of individuals affected by hypertension will increase by 60% to a total of more than 1.5 billion subjects.3 Despite the increasing public awareness of hypertension and its complications, the rates of adequate blood pressure control (<140/90 mmHg) among patients receiving antihypertensive therapy remain unsatisfactory.4 The reasons for these disappointing outcomes are complex, but include medication non-adherence, which may be due to adverse effects or treatment costs, and interindividual genetic variability.5
Indeed, genetic factors can affect blood pressure increases by 30–50%.6,7 Therefore, over the past two decades, many genetic studies have aimed to clarify the causal genes of hypertension. In these studies, several genetic polymorphisms, including single nucleotide polymorphisms (SNPs), variable number of tandem repeats, microsatellites and insertions/deletions (I/D), were found associated with hypertension.8 Moreover, these studies have shown that genetic factors are involved not only in the blood pressure elevation, but also contribute to the large interindividual variability in response to antihypertensive treatment, opening a window of opportunity for pharmacogenomic investigation and potential individualization of drug therapy.5 In fact, considering the low rates of blood pressure control, the ability to identify the most effective antihypertensive agent for an individual patient prior to initiation of therapy has potential to be beneficial.9 Successful examples of targeted antihypertensive therapy based on genetics include the personalized treatments available for most forms of monogenic hypertension.10,11 Regarding essential hypertension, the risks of a genetic-guided approach for prescription of antihypertensive drugs would be relatively low, considering that the current method for selection of antihypertensive therapy is predominantly empirical, and frequently involves a trial and error approach to find the optimal regimen for a given patient.9 Thus, the goal of hypertension pharmacogenomics is to use genetic information, in addition to other pertinent clinical or demographic parameters, to select the right antihypertensive therapy and the most favorable dose to maximize the drug efficacy and reduce the risk for adverse effects.5
In order to identify potential genetic predictors of antihypertensive responses, two main approaches have been applied: a hypothesis-driven approach on the candidate genes, encoding proteins involved in signaling pathways affected by antihypertensive drugs, and an unbiased hypothesis-free approach with genome-wide association studies (GWAS), supported by the randomness basis of frequentist statistics.12,13 During the past decade, GWAS have overcome the application of candidate gene approach, resulting in the identification of several previously unknown candidate loci or genes, but the advantages and limitations of this method in hypertension pharmacogenomics compared to the hypothesis-driven approach are still under debate.13 In addition to these strategies that focus on single-locus analysis, a third approach that takes into account gene-gene interactions has been recently applied in pharmacogenomic studies.14 This strategy considers the biological complexity underlying drug response and evaluate potential epistatic interactions that may predict how a patient will respond to a given treatment.15 In this review article, we summarize the recent findings on the pharmacogenomics of antihypertensive drugs and discuss current insights and future directions of this field.
Studies On The Pharmacogenomics Of Hypertension
Here, we review the pharmacogenomics of the most commonly prescribed antihypertensive agents in clinical practice, including diuretics, β-blockers, angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers, and calcium channel blockers. A search was performed in the PubMed database for original articles focusing on the effects of genetic polymorphisms on blood pressure responses to the antihypertensive drug classes described above. The search terms used were: “Antihypertensive therapy”, “Pharmacogenomics and Pharmacogenetics”, “[each antihypertensive drug class] and pharmacogenomics”. The literature search was limited to full-text articles in the English language. In addition, the reference lists of identified articles were searched for further papers.
Diuretics
Diuretics are the first-line drugs of choice for most patients with hypertension.2 Their mechanism of action involves the increases in sodium excretion (natriuresis) and decreases in extracellular volume, leading to a reduction in cardiac output. Although the initial antihypertensive effects of these drugs are in fact due to diuresis, their long-term effects are maintained due to decreases in vascular resistance, possibly resulted from an inhibition of sympathetic nervous and/or renin-angiotensin systems.16 Given the different mechanisms underlying the effects of diuretics, several candidate genes may predict individual responses to these drugs.
The most commonly used diuretic is the thiazide diuretic hydrochlorothiazide, which acts by inhibiting the sodium chloride cotransporter expressed in the distal convoluted tubule of the nephron.17 Considering the substantial inter-individual variation in the antihypertensive responses to hydrochlorothiazide, a large number of studies has evaluated polymorphisms in candidate genes or in GWAS as predictors of blood pressure responses to this drug (Table 1). In this regard, the ADD1 gene was one of the first candidate genes examined for antihypertensive responses to thiazide diuretics.18,19 The ADD1 gene encodes α-adducin, a cytoskeleton-associated protein that modulates ion transport.20 Interestingly, it was found that carriers of the Trp allele for the Gly460Trp (rs4961) polymorphism in the ADD1 gene showed a reduced baseline plasma renin activity and a better antihypertensive response to hydrochlorothiazide treatment compared to Gly/Gly homozygotes.19 A subsequent study found evidence suggesting that the rs4961 polymorphism may modulate renal sodium handling by changing ion transport across the cell membrane.21 While the association between rs4961 polymorphism and the antihypertensive responses to thiazide diuretics has been confirmed by some later studies,22 lack of association was observed in others.23,24
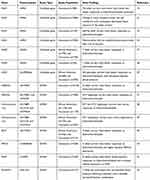 |
Table 1 Summary Of Studies On The Pharmacogenomics Of Diuretics |
Another gene that has been evaluated for hydrochlorothiazide responses is GNB3, which encodes β3-subunit of the G-protein.25 This family of proteins is critical for many physiological and pharmacological responses once they mediate signal transduction from membrane receptors to a wide range of intracellular effectors.25 Interestingly, it was reported that the T allele for C825T (rs5443) polymorphism in the GNB3 gene is related to an RNA splice variant that lacks the nucleotides 498–620 of exon 9, resulting in structural modifications in the β3-subunit of G-protein and potentially affecting signal transduction.26 Indeed, the T allele for this polymorphism was associated with better antihypertensive responses to hydrochlorothiazide with a gene-dose effect,27 and this association was further confirmed by an independent study.28 However, another study with a larger sample size failed to replicate these findings,29 and therefore the association between the rs5443 polymorphism and hydrochlorothiazide responses remains unclear.
Given that the antihypertensive effects of diuretics are in part due to renin-angiotensin system inhibition,16 some studies have tested whether polymorphisms in the gene encoding the angiotensin converting enzyme (ACE) affect the responses to these drugs. In a study evaluating the I/D polymorphism in intron 16 of ACE gene in 87 never-treated hypertensive patients, Sciarrone et al found that individuals carrying the I/I genotype had better antihypertensive responses to hydrochlorothiazide compared to those carrying the D/D genotype.22 A later study in the Han Chinese population showed that this polymorphism affects hydrochlorothiazide responses in a gender-specific manner, since better antihypertensive effects were observed in men carrying the D/D genotype and women carrying the I/I genotype.30 These associations were not replicated in a study including 208 hypertensive Finnish men.31
The NEDD4L has also been considered a candidate gene for hydrochlorothiazide responses. This gene encodes a ubiquitin ligase that targets the epithelial sodium channel for degradation, therefore affecting sodium reabsorption in the distal nephron.32 Consistent with its function, studies have shown that polymorphisms in NEDD4L gene affect salt sensitivity, plasma renin concentrations and susceptibility to hypertension.33–35 To test the effects of NEDD4L variants on the responses to antihypertensive drugs, the NORDIL (Nordic Diltiazem) Study evaluated Caucasian hypertensive patients randomized to beta-blockers or thiazide diuretics and followed-up for six months.36 Interestingly, it was found that G allele carriers for the rs4149601 polymorphism in NEDD4L gene have better antihypertensive responses to hydrochlorothiazide and β-blockers than patients with the AA genotype.36 These results were significantly replicated in white subjects in subsequent studies.37 In addition, better blood pressure responses to hydrochlorothiazide was observed in white hypertensive patients carrying increasing copies of the G-C haplotype of NEDD4L gene (for the SNPs rs4149601 and rs292449, respectively).37 These findings, however, were not replicated in African Americans.37
The Genetic Epidemiology of Responses to Antihypertensives (GERA) study was the first GWAS on the pharmacogenomics of hypertension therapy.38 While no significant associations were observed in Caucasians, this study identified a region of chromosome 12q associated with the antihypertensive responses to hydrochlorothiazide in African Americans. The authors found that haplotypes on this chromosome composed by SNPs rs317689, rs315135, rs7297610 near lysozyme (LYZ), YEATS domain containing 4 (YEATS4), and fibroblast growth receptor substrate 2 (FRS2) were significantly associated with diastolic blood pressure responses to hydrochlorothiazide. The ATT and ATC haplotypes composed by these SNPs were more frequent in poor responders than in good responders. The SNP rs7297610 had the most significant individual value, and it was suggested to drive the observed association. Consistently, these results were replicated in an independent population of 291 African-Americans and 294 Caucasians,38 therefore validating the findings. Interestingly, these results were also replicated in a later study enrolling 746 hypertensive subjects from Pharmacogenomics Evaluation of Antihypertensive Responses (PEAR) study,39 suggesting that haplotypes composed by SNPs rs317689, rs315135, rs7297610 may be potential predictors of hydrochlorothiazide responses. In addition, the chromosome 12q was recently re-sequenced in participants from GERA and PEAR studies and a novel missense SNP, rs61747221 in the BEST3 gene was significantly associated with blood pressure responses to hydrochlorothiazide treatment.40 In this study, subjects carrying AA+AG genotypes for this polymorphism showed better antihypertensive responses to hydrochlorothiazide compared to GG carriers.
In order to identify novel genes affecting the antihypertensive responses to hydrochlorothiazide, Turner et al conducted genome-wide association meta-analyses combining the results from independent white hypertensive populations: PEAR, GERA, NORDIL, and Genetics of Drug Responsiveness in Essential Hypertension Study (GENRES).23 By using this approach, they provided substantial evidence of associations between the SNP rs16960228 in PRKCA gene and blood pressure responses to hydrochlorothiazide. The A-allele carriers showed consistently better antihypertensive responses to this drug. Moreover, this study provided functional evidence that the A-allele for the SNP rs16960228 is associated with higher baseline PRKCA expression.23
More recently, several novel SNPs have been associated with blood pressure responses to hydrochlorothiazide in GWAS. Hiltunen et al identified four new SNPs affecting hydrochlorothiazide response (rs3825926, rs4867623, rs321329 and rs321320) in subjects from GENRES, PEAR and GERA studies.41 Another GWAS including individuals from PEAR and PEAR-2 studies tested 1082 SNPs, prioritized according to their biological function, using RegulomeDB, haploreg and GWAVA software packages.42 The results from the prioritization analysis showed the SNP rs10995 in the VASP gene (encoding the vasodilator-stimulated phosphoprotein) as the most likely functional SNP, among SNPs tested, that has been associated with hydrochlorothiazide responses. The G allele for this SNP was associated with greater blood pressure responses to hydrochlorothiazide and increased mRNA expression of VASP.42 Consistently, these findings were replicated in an independent cohort treated with the thiazide diuretic chlorthalidone, thus validating the associations.42 Also employing a GWAS approach, Magvanjav et al found that the SNP rs261316 in the ALDH1A2 gene was associated with uncontrolled blood pressure following treatment with a thiazide diuretic/β-blocker combination in white participants of the PEAR study.43 These findings were further validated in a replication study including subjects from INVEST study.43 Taken together, these findings highlight GWAS as an efficient approach to identify novel polymorphisms involved with the antihypertensive responses to hydrochlorothiazide. Further studies are still required to confirm the functional relevance of the SNPs identified in the mentioned GWAS.
β-Blockers
Although β-blockers are no longer considered as first-line antihypertensive pharmacotherapy by the Joint National Committee (JNC8) hypertension guidelines,2 they are still widely prescribed and retain an important place in the management of hypertension for certain subgroups of patients.44 β-blockers decrease myocardial contractility, heart rate and cardiac output.45 In addition to the cardiac effects, the antihypertensive effects of these drugs result from the blockage of their targets on juxtaglomerular cells of the kidney, decreasing renin secretion and thereby leading to a reduced production of circulating angiotensin II.45,46
The primary protein target of all β‑blockers is the β1‑adrenergic receptor, encoded by ADRB1.47 This gene contains two common and widely studied genetic polymorphisms that lead to changes in the encoded amino acids: rs1801252, which leads to a serine to glycine change at the position 49 of the protein (Ser49Gly), and rs1801253, which results in the arginine to glycine change at the position 389 of the protein (Arg389Gly).47 These two polymorphisms show important evidence for functional impact once they affect intracellular signaling mediated by the β1‑adrenergic receptor.48 Indeed, the ancestral alleles (Ser49 and Arg389) for these polymorphisms are both associated with improved intracellular responses to β1‑adrenergic receptor agonists compared to variant alleles.48 Despite the functional relevance, there is no consensus for the association between Ser49Gly or Arg389Gly polymorphisms and antihypertensive responses to β-blockers (Table 2). In a study including white, African American, and Hispanic individuals, carriers of Arg/Arg genotype for Arg389Gly polymorphism had better blood pressure responses to the β‑blocker metoprolol compared to subjects carrying the Gly allele.49 In addition to the genotypic findings, the authors observed that the haplotypes composed by Ser49Gly and Arg389Gly polymorphisms affect the blood pressure responses to metoprolol. Similar results were also found in Chinese hypertensive patients treated with the β‑blocker carvedilol.50 In the opposite direction, Chen et al recently reported that subjects carrying the Gly/Gly genotype for Arg389Gly polymorphism show greater antihypertensive responses to metoprolol.51 Regardless of these positive findings, lack of association between Ser49Gly or Arg389Gly polymorphisms and blood pressure responses to β-blockers was reported in two European prospective studies.52,53
 |
Table 2 Summary Of Studies On The Pharmacogenomics Of β-Blockers |
In addition to polymorphisms in genes that modulate the pharmacodynamics of β‑blockers, variants in genes regulating their pharmacokinetics are also potential candidates to impact the antihypertensive responses to these drugs. One of these candidates is the gene encoding the cytochrome enzyme CYP2D6, which is a major responsible for metabolizing β‑blockers.42 Because there are evidences suggesting that the CYP2D6 polymorphisms affect the pharmacokinetics of β-blockers,54–56 some studies have evaluated whether these variations translate into variability in their effects. In fact, there are studies showing associations of CYP2D6 genotypes and blood pressure responses to β-blockers.57,58 Based on these evidences, the Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association has established therapeutic dose recommendations for metoprolol based on CYP2D6 genotypes.59 However, other studies have suggested that there is no sufficient evidence for the clinical utility of CYP2D6 genotyping to guide metoprolol therapy in hypertension,60 since lack of association between CYP2D6 variants and blood pressure responses to this drug and other β-blockers has been observed in some studies.56,60–62
Recently, several GWAS have found polymorphisms affecting blood pressure responses to β-blockers (Table 2). Using this approach, Gong et al evaluated 318 African-American hypertensive participants from PEAR and PEAR-2 studies treated with β-blockers and observed that individuals carrying one variant allele (a deletion in the intronic region) for the SNP rs201279313 in SLC25A31 gene (which encodes ADP/ATP translocase 4) have better antihypertensive responses to β-blockers compared to subjects carrying two wild-type alleles.63 Moreover, they also found that subjects carrying the deletion allele of rs11313667, located in the intronic region of LRRC15 gene (encoding leucine rich repeat containing 15) show greater blood pressure responses to β-blockers compared to individuals carrying two wild-type alleles. Importantly, these associations were replicated in an independent cohort of PEAR study,63 thereby validating these findings.
Given that black subjects have different antihypertensive responses to β-blockers compared to Caucasians, recent GWAS have focused on this last population.41,64,65 Hiltunen et al identified the SNPs rs2514036, rs948445, and rs2514037 in the ACY3 gene (coding for aminoacylase III) associated with blood pressure responses to the β‑blocker bisoprolol in Caucasians.41 In a later replication study, only the SNP rs2514036 was found associated with blood pressure responses to atenolol.65 Another GWAS evaluating European-Americans participants from PEAR-2 study found that the A allele carriers for rs294610 polymorphism in the FGD5 gene (which encodes for FYVE, RhoGEF and PH Domain Containing 5) show better blood pressure responses to β-blockers. Consistently, these findings were replicated in European-Americans participants from PEAR study. In a secondary replication approach, the authors performed a meta-analysis between PEAR-2 and PEAR studies with an independent cohort from PEAR and identified an additional single nucleotide polymorphism (rs45545233) in the SLC4A1 gene (encoding for Solute Carrier Family 4 Member 1) associated with poor responses to β-blockers.64 More recently, Singh et al carried out a GWAS involving 5 randomized clinical trials consisting of 1254 patients with hypertension of European ancestry and identified A allele for the SNP rs28404156 in the BST1 gene associated with better blood pressure responses to β‐blockers.66 Consistently, these findings were successfully replicated in 3 additional randomized clinical trials.66
In addition to African Americans and European-Americans, recent studies have used GWAS approach to identify genetic polymorphisms associated with blood pressure responses to atenolol in other populations. This is the case of a study performed by Sy et al, which investigated the association of genetic variants with antihypertensive responses to β-blockers among Filipinos.67 Interestingly, they found that the deletion of at least one copy of allele A for rs36217263 polymorphism in the Klotho gene is associated with poor response to β-blockers. Further studies are required to test if these findings can be replicated in independent cohorts of Filipino subjects.
Angiotensin-Converting Enzyme Inhibitors And Angiotensin II Receptor Blockers
The renin-angiotensin system is well known for its modulation of the blood pressure and sodium homeostasis.68 These effects are coordinated through integrated mechanisms in the kidney, cardiovascular system and the central nervous system.68 The cascade that results in the physiologic effects of renin-angiotensin system involves the renin-mediated conversion of angiotensinogen into angiotensin I, which is further cleaved by angiotensin-converting enzyme (ACE), to produce angiotensin II, the final effector of the system.69 Angiotensin II directly regulates blood pressure by stimulating angiotensin II type 1 receptor (AT1R) receptors present in the vasculature, kidney and central nervous system, leading to vasoconstriction, sodium reabsorption, and increases in sympathetic tone.69
Two different classes of drugs targeting the renin-angiotensin system are widely prescribed to treat hypertension: ACE inhibitors, which preclude the formation of angiotensin II, and angiotensin II receptor blockers (ARB), which bind to AT1R, thereby antagonizing the effect of angiotensin II. Not surprisingly, genes encoding components of the renin-angiotensin system are the most likely candidate genes for a pharmacogenomic approach on these drugs (Table 3). The most studied polymorphism of these genes is the I/D in intron 16 of ACE.70 Indeed, D allele of this polymorphism seems to be associated with higher levels of ACE in Caucasian71 and Asian72,73 populations, but not in African-Americans.74 Accordingly, this allele has also been associated with cardiovascular75–78 and renal79,80 disease. However, conflicting results have been found regarding to the antihypertensive responses to ACE inhibitors or ARB. This is because the D allele was associated with better antihypertensive responses to these drugs in some studies,81,82 while the I allele was associated with those effects in others;83–85 in addition, several studies have shown no effects of this polymorphism on blood pressure responses to ACE inhibitors or ARB.86–89 Many factors, including study population, sample size, and possible interethnic differences in the distribution of I/D ACE polymorphism could explain these discrepancies.
 |
Table 3 Summary Of Studies On The Pharmacogenomics Of Angiotensin-Converting Enzyme Inhibitors And Angiotensin II Receptor Blockers |
Other candidate genes that are part of the classical cascade of the renin-angiotensin system and therefore could affect the antihypertensive responses to ACE inhibitors and ARB include AGT and CYP11b2 genes. The AGT gene encodes angiotensinogen, and the most studied polymorphism of this gene is the SNP Met235Thr (rs699), which results in a threonine instead of a methionine in codon 235 of the protein.90 However, similar to the I/D polymorphism in ACE gene, there is no consensus regarding the effects of this polymorphism on the antihypertensive responses to ACE inhibitors or ARB.85,87–89,91,92 The CYP11b2 gene encodes aldosterone synthase, the enzyme that catalyzes the final step of aldosterone synthesis in juxtaglomerular cells.93 One important polymorphism in this gene is the SNP −344C/T (rs179998), which was shown to affect aldosterone levels and hypertension susceptibility.94–97 Interestingly, the −344C/T polymorphism has been found associated with blood pressure responses to ARB,92,98–100 although it is still not clear which variant contributes to greater responses. In fact, while this effect was attributed to the C allele of this SNP in some studies,99,100 the same effect was associated with the T allele in others.92,98 Studies with larger populations are required to interpret the findings more conclusively.
In addition to the renin-angiotensin system inhibition, the beneficial effects of ACE inhibitors and ARB are associated with several pleiotropic effects.101–103 Indeed, many of these effects are related to the vasodilation produced by nitric oxide (NO) as a result of endothelial NO synthase (NOS3) activation.104,105 NOS3 is the dominant NO synthase in the vasculature and polymorphisms in the gene encoding this enzyme have been associated with hypertension and other cardiovascular alterations.106–108 The pharmacogenomic relevance of NOS3 for ACE inhibitor and ARB responses has been shown in different studies. One of these studies focused on hypertensive patients treated with the ACE inhibitor enalapril.109 This study observed higher frequencies of the C allele for the NOS3 SNP −786T/C (rs2070744) in patients with good responses to this drug as compared with those patients classified as poor responders.109 In line with these findings, another study showed that the ARB olmesartan promotes increased NO formation in endothelial cells that are homozygous for C allele of this polymorphism compared to heterozygous cells.110 Therefore, both enalapril and olmesartan seem to have more significant effects in the presence of C allele for −786T/C polymorphism, suggesting that hypertensive subjects carrying this allele may have better responses to these drugs. More recently, the T allele for the NOS3 SNP −665C/T (rs3918226) was associated with greater responses to enalapril, whereas the A allele for the NOS3 tagSNP rs3918188 and the CAG haplotype involving NOS3 tagSNPs were associated with the opposite effect.111 Additionally, different proteins have been described to contribute to NOS3 activation promoted by ACE inhibitors, including bradykinin receptor B2 (BDKRB2), protein kinase C (PKC) and vascular endothelial growth factor (VEGF).105,112,113 Taken this into consideration, we have recently shown that polymorphisms in genes encoding BDKRB2, PKC and VEGF affect the antihypertensive responses to the enalapril109,114,115 (Table 3). Taken together, these findings clearly suggest that genetic variability in NOS3 gene or in genes that contribute to NOS3 activation may affect the responses to ACE inhibitors and ARB.
In addition to the candidate gene approach, GWAS have also identified genetic predictor of response to drugs targeting the renin-angiotensin system (Table 3). This is the case of a study enrolling 372 Italian hypertensive subjects treated with the ARB losartan and further replicated in two independent populations.116 In this GWAS, the authors found the GG genotype for the SNP rs10752271 in CAMK1D gene (encoding calcium/calmodulin dependent protein kinase 1D, involved in aldosterone synthesis) associated with better blood pressure responses to losartan.116 Another GWAS identified multiple loci influencing antihypertensive response to the ARB candesartan in subjects from GERA study.117 The associations were observed with the SNPs rs11020821 in FUT4 gene (encoding fucosyltransferase 4), rs3758785 in GPR83 gene (encoding G protein-coupled receptor 83), and rs11649420 in SCNN1G gene (encoding sodium channel, non-voltage-gated 1, gamma subunit).117 Replication in independent populations is required to make these findings more robust. Additionally, a further GWAS involving GENRES participants found the SNP rs3814995 in NPHS1 gene (encoding nephrin, a transmembrane protein that is a structural component of the slit diaphragm in the kidney and an important contributor to blood pressure regulation) associated with improved blood pressure responses to losartan.41 Consistently, this association was also replicated in GERA2 and Italian hypertensive patients treated with the Angiotensin receptor blocker losartan (SOPHIA) populations.
Calcium Channel Blockers
Calcium channel blockers (CCB) constitute a heterogeneous class of drugs commonly prescribed to treat a wide array of cardiovascular conditions, including hypertension.118 Although all agents in this class act by blocking calcium channels, each subclass binds at a specific location. While the subclass of dihydropyridines, such as nifedipine and amlodipine, has vascular selectivity, verapamil has cardiac selectivity, and diltiazem can act both in the heart and blood vessels.45 Given that blood pressure responses to CCB are widely variable,119 studies have investigated potential genetic polymorphisms that could contribute to this variability. Consistent with CCB mechanism of action, studies have shown that polymorphisms in genes encoding different ion channels, such as voltage-gated calcium channel α1C (CACNA1C), α1D (CACNA1D) and β2 (CACNB2), large-conductance calcium and voltage-dependent potassium channel β1 (KCNMB1), and ERG potassium channel (KCNH2) affect the antihypertensive responses to CCB or the risk of adverse cardiovascular outcomes120–124 (Table 4).
 |
Table 4 Summary Of Studies On The Pharmacogenomics Of Calcium Channel Blockers |
In addition to explore polymorphisms in genes implicated in CCB pharmacodynamics, studies have also tested the effects of variants in genes regulating CCB pharmacokinetics on the antihypertensive responses to these drugs. Calcium channel blockers are largely metabolized in the liver via the cytochrome P450 3A5 (CYP3A5),125,126 and therefore, studies have examined if polymorphisms in the gene encoding this enzyme affect CCB responses. Contrary to the functional CYP3A5*1 allele, the CYP3A5*3 variant has a 6986A>G mutation in intron 3 that results in a splicing defect in CYP3A5 mRNA and a nonfunctional protein.127 Another mutation (14690G>A) in exon 7 of CYP3A5 gene (CYP3A5*6) also leads to a splicing defect and deletion of exon 7, thereby resulting in a protein truncation.127 Despite the role of CYP3A5 in CCB metabolism, the effects of these CYP3A5 variants on CCB responses are not clear. Indeed, Zhang et al and Huang et al found the CYP3A5*3 allele associated with better antihypertensive responses to the CCB amlodipine in the Chinese population.128,129 However, no associations between CYP3A5 variants and CCB effects were observed in Koreans130 and African-Americans.131 These discrepancies may be explained by the fact that amlodipine responses can be influenced by ethnicity as a result of genetic factors, environmental factors and their interaction.
Besides amlodipine, studies have also investigated the influence of CYP3A5 polymorphisms on blood pressure responses to the CCB verapamil. Langaee et al addressed this question by using a haplotype approach, with any allele containing either (*3) or (*6) designated as nonfunctional.132 The authors observed that the number of CYP3A5 functional alleles was marginally associated with blood pressure responses to verapamil in blacks and Hispanics, but not in whites, with the effect being largely driven by worst responses in the carriers of two functional alleles.
Along with the candidate gene approach, some GWAS have shown SNPs associated with blood pressure responses to CCB (Table 4). Using this strategy, Kamide et al133 found that the alleles C for rs588076, G for rs2429427, C for rs10898815 and C for rs564991 of the PICALM, TANC2, NUMA1 and APCDD1 genes, respectively, are associated with antihypertensive responses to CCB in Japanese hypertensive patients. Consistently, these findings were further replicated in an independent Japanese population.133 Taking advantage of previous GWAS findings, another study identified the SNP rs12946454 of PLCD3 gene (which encodes phospholipase C, delta 3) associated with blood pressure responses to the CCB diltiazem.134 Interestingly, the authors found an additive effect for increasing copies of T allele for this SNP and more intense blood pressure reductions (1.53 mmHg per allele for systolic blood pressure and 0.73 mmHg per allele for diastolic blood pressure) after diltiazem treatment. However, replication of these findings in independent populations is still pending.
Gene-Gene Interactions In The Pharmacogenomics Of Hypertension
The two main approaches frequently used in pharmacogenomic studies (candidate gene and GWAS) evaluate associations between individual genes and the drug response. However, these approaches may overlook the associations that can only be detected when the combinations of multiple genomic regions are examined.135,136 Indeed, a lot of the genetic predisposition to drug response phenotypes seems to be hidden in multigenic and multifactorial complex traits.137 Therefore, novel approaches that consider the interactions among polymorphisms from different genes within drug response pathways should be considered in pharmacogenomic studies.
One of these approaches involves the use of the multifactor dimensionality reduction, a machine learning method that detects the high-order gene-gene interactions with the use of relatively small sample sizes.138–140 Using this method, Silva et al showed that interactions between NOS3 and BDKRB2 polymorphisms affect the antihypertensive responses to enalapril.109 Interactions between these genes are expected, as both proteins encoded by them are involved in the same signaling pathway modulated by ACE inhibitors. Interestingly, it was found that, although the CC genotype for the rs1799722 BDKRB2 polymorphism was not associated with the antihypertensive response to enalapril at single-locus analysis, gene-gene interaction analysis showed that the CC genotype for this polymorphism combined with the TT genotype for rs2070744 NOS3 polymorphism was more frequent in poor responders to enalapril, while the combination of BDKRB2 CC genotype with NOS3 TC genotype was more frequent among good responders.109 These findings are underestimated when single BDKRB2 genotypes alone are analyzed, thus highlighting the relevance of gene-gene interaction analysis.
Following up on this study, we have recently investigated the involvement of PRKCA gene (encoding PKCα) in the interactions described above.114 Interestingly, we described that such combinations between BDKRB2 and NOS3 genes found to be associated with antihypertensive responses to enalapril were significant only when combined with the GG genotype for the rs16960228 PRKCA polymorphism.114 These gene-gene interactions are in line with the signaling cascade that, at least in part, contributes to the antihypertensive effects of ACE inhibitors.105 The inhibition of ACE leads to enhanced bradykinin levels, which activate bradykinin receptors on endothelial cells.105 The activation of these receptors stimulates PKCα, which then activates NOS3, resulting in vasodilation due to increased NO production.113,141,142 The evaluation of circulating nitrite levels (a marker of endogenous NO formation) may be a possible approach to clarify the cellular mechanisms underlying the effects of BDKRB2, PRKCA and NOS3 interactions on enalapril responses. Indeed, NOS3 polymorphisms were shown to affect plasma and blood nitrite concentrations.106,107 However, additional studies are required to test the effects of gene interactions within this pathway on circulating nitrite levels in patients treated with ACE inhibitors. Taken together, these findings suggest that gene-gene interaction analysis is a promising approach that should be considered in the pharmacogenomics of hypertension.
Concluding Remarks And Future Directions
In this article, we have reviewed the recent pharmacogenomic literature for the main classes of antihypertensive drugs currently in use (i.e., diuretics, β-blockers, ACE inhibitors, ARB, and CCB). While a number of studies support an important contribution of genetic polymorphisms on blood pressure responses to antihypertensive therapy, some studies have failed to detect significant effects or replicate previous findings. These inconsistencies may be related to interethnic differences in the distributions of polymorphisms tested/identified, or to epigenomic alterations that could mask the contribution of DNA sequence variants.143 Moreover, the inconsistencies may be attributed to heterogeneous phenotypes. In this case, variability in the etiology and mechanisms involved may explain the differences in observed phenotype, thereby reducing the possibility of successfully detecting associations between genetic polymorphisms and antihypertensive responses. Therefore, it is critical that subjects are carefully phenotyped and maybe surrogate markers of disease should also be examined. Another issue that can lead to inconsistent results is the effect of the previous treatments. The persisting antihypertensive effect after stopping therapy may preclude pharmacogenomic studies enrolling previously treated patients without an adequate washout period from getting reliable findings. To avoid this issue, studies should be preferentially performed in recently diagnosed patients that have never been treated before.
The inconsistencies and limitations observed in studies of pharmacogenomics of hypertension may be also dependent on the type of strategy applied. In fact, vast majority of studies discussed here applied two different approaches: gene candidate and GWAS. During the past decade, the shift from gene candidate approach to GWAS has been noteworthy. However, the large and inevitably heterogeneous sample size required for GWAS may preclude these studies from disentangling the complex genetics basis of hypertension, which consists of dynamic interactions among genetic/environmental aspects that may change with aging. This could help to explain the fact that some significant results found in candidate-gene studies, usually performed in more homogeneous cohorts, are not necessarily found in GWAS. On the other hand, the candidate gene approach prevents the identification of novel candidate loci or genes that could impact hypertension therapy. Taking this into consideration, Manunta et al propose to integrate the candidate gene with GWAS approach, in order to minimize their respective limitations.13 A possible strategy to fully exploit the pharmacogenetic relevance of a given polymorphism and therefore increase the chances of defining clinically significant benefits would be testing the significant results found in GWAS in more homogeneous cohorts, in parallel with mechanistic studies to validate the biological significance of the pharmacogenetic findings.
Other than improving approaches that focus on single-locus analysis, another promising strategy to overcome the limitations of hypertension pharmacogenomics is the investigation of interactions among polymorphisms from different genes within drug response pathways.14 Indeed, the polygenic nature of hypertension indicates that single loci may not be the best clinical target for all individuals.144 The studies discussed in this article highlight the relevance of evaluating the interactions among multiple loci when studying complex traits, which is the case of antihypertensive response phenotypes. Given that these ideas are relatively new in the pharmacogenomics of hypertension, further studies in different populations are required to replicate the findings reported by the studies presented here. Another strategy that also takes into account the polygenic nature of hypertension is the use of genetic risk score (GRS). This approach has recently been developed to evaluate the impact of multiple blood pressure-associated variants on blood pressure levels, risk of hypertension, and other cardiovascular diseases.145,146 Interestingly, GRS analysis showed that all blood pressure-elevating alleles combined could increase systolic blood pressure by 10 mm Hg and increase the risk of cardiovascular events.145,146 Therefore, this seems to be a promising strategy to be used in future studies on hypertension pharmacogenomics.
Despite the issues described above, it is reasonable to suggest that data from several genes shown in the studies discussed in this review provide valuable genetic information for hypertension pharmacogenomics research. In order to provide a realistic estimate on how much variation of responses to antihypertensive therapy may be explained by genetic markers, a systematic review and meta-analysis involving the recent studies on hypertension pharmacogenomics would be required. This would help to translate the research findings into clinical practice to improve hypertension therapy, which remains a challenge. In addition, given that a significant number of hypertensive patients require more than one drug to achieve the therapeutic goals, increasing the number of studies focusing on drug combinations, although not an easy task, would also help to approximate hypertension pharmacogenomics to the clinical practice. In fact, while a lot of progress was achieved in the clinical application of pharmacogenomics in oncology and anticoagulation therapy, the same has not been seen in hypertension therapy.147 In this regard, antihypertensive agents do not have serious or lethal side effects such as those of anticancer drugs or warfarin. In addition, cancer is a condition where drug efficacy may have immediate influence on short-term survival. All these reasons may explain the lack of motivation in personalizing medicine for hypertension therapy, resulting in patients often taking an antihypertensive drug that is less effective than required. To start changing this paradigm, several academic medical centers are already piloting programs to implement hypertension pharmacogenomics in clinical practice.148,149
Indeed, an efficient pharmacogenomic strategy to get hypertensive patients on the most effective and well-tolerated drug regimen would be extremely valuable. This would result in less patient visits to readjust the treatment, fewer drugs per patient, and a more cost-effective approach than the trial and error in current practice. An adequate blood pressure control would prevent cardiovascular and renal events and improve the quality of life and longevity of hypertensive patients.
Acknowledgments
This study was supported by Fundaçao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP – grant number: 2018/18312-2), Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq – grant number: 302241/2017-5) and Coordenaçao de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES – student’s scholarships).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pagidipati NJ, Gaziano TA. Estimating deaths from cardiovascular disease: a review of global methodologies of mortality measurement. Circulation. 2013;127(6):749–756. doi:10.1161/CIRCULATIONAHA.112.128413
2. James PA, Oparil S, Carter BL, et al. evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the eighth Joint National Committee (JNC 8). JAMA. 2014;311(5):507–520. doi:10.1001/jama.2013.284427
3. Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365(9455):217–223. doi:10.1016/S0140-6736(05)17741-1
4. Chobanian AV; Shattuck Lecture. The hypertension paradox–more uncontrolled disease despite improved therapy. N Engl J Med. 2009;361(9):878–887. doi:10.1056/NEJMsa0903829
5. Johnson JA. Advancing management of hypertension through pharmacogenomics. Ann Med. 2012;44(Suppl 1):S17–22. doi:10.3109/07853890.2011.653399
6. Menni C, Mangino M, Zhang F, et al. Heritability analyses show visit-to-visit blood pressure variability reflects different pathological phenotypes in younger and older adults: evidence from UK twins. J Hypertens. 2013;31(12):2356–2361. doi:10.1097/HJH.0b013e32836523c1
7. Dominiczak AF, Negrin DC, Clark JS, Brosnan MJ, McBride MW, Alexander MY. Genes and hypertension: from gene mapping in experimental models to vascular gene transfer strategies. Hypertension. 2000;35(1 Pt 2):164–172. doi:10.1161/01.HYP.35.1.164
8. Tabara Y, Kohara K, Miki T; Millennium Genome Project for H. Hunting for genes for hypertension: the millennium genome project for hypertension. Hypertens Res. 2012;35(6):567–573. doi:10.1038/hr.2012.41
9. Cooper-Dehoff RM, Johnson JA. Hypertension pharmacogenomics: in search of personalized treatment approaches. Nat Rev Nephrol. 2016;12(2):110–122. doi:10.1038/nrneph.2015.176
10. Lee WK, Padmanabhan S, Dominiczak AF. Genetics of hypertension: from experimental models to clinical applications. J Hum Hypertens. 2000;14(10–11):631–647. doi:10.1038/sj.jhh.1001043
11. Padmanabhan S, Paul L, Dominczak AF. The pharmacogenomics of anti-hypertensive therapy. Pharmaceuticals (Basel). 2010;3(6):1779–1791. doi:10.3390/ph3061779
12. Kamide K, Kawano Y, Rakugi H. Pharmacogenomic approaches to study the effects of antihypertensive drugs. Hypertens Res. 2012;35(8):796–799. doi:10.1038/hr.2012.82
13. Manunta P, Ferrandi M, Cusi D, Ferrari P, Staessen J, Bianchi G. Personalized therapy of hypertension: the past and the future. Curr Hypertens Rep. 2016;18(3):24. doi:10.1007/s11906-016-0632-y
14. Luizon MR, Pereira DA, Sandrim VC. Pharmacogenomics of hypertension and preeclampsia: focus on gene-gene interactions. Front Pharmacol. 2018;9:168. doi:10.3389/fphar.2018.00168
15. Motsinger AA, Ritchie MD, Reif DM. Novel methods for detecting epistasis in pharmacogenomics studies. Pharmacogenomics. 2007;8(9):1229–1241. doi:10.2217/14622416.8.9.1229
16. Arnett DK, Claas SA, Glasser SP. Pharmacogenetics of antihypertensive treatment. Vascul Pharmacol. 2006;44(2):107–118. doi:10.1016/j.vph.2005.09.010
17. Duarte JD, Cooper-Dehoff RM. Mechanisms for blood pressure lowering and metabolic effects of thiazide and thiazide-like diuretics. Expert Rev Cardiovasc Ther. 2010;8(6):793–802. doi:10.1586/erc.10.27
18. Glorioso N, Manunta P, Filigheddu F, et al. The role of alpha-adducin polymorphism in blood pressure and sodium handling regulation may not be excluded by a negative association study. Hypertension. 1999;34(4 Pt 1):649–654. doi:10.1161/01.HYP.34.4.649
19. Cusi D, Barlassina C, Azzani T, et al. Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349(9062):1353–1357. doi:10.1016/S0140-6736(97)01029-5
20. Matsuoka Y, Li X, Bennett V. Adducin: structure, function and regulation. Cell Mol Life Sci. 2000;57(6):884–895. doi:10.1007/PL00000731
21. Glorioso N, Filigheddu F, Cusi D, et al. alpha-adducin 460Trp allele is associated with erythrocyte Na transport rate in North Sardinian primary hypertensives. Hypertension. 2002;39(2 Pt 2):357–362. doi:10.1161/hy0202.103065
22. Sciarrone MT, Stella P, Barlassina C, et al. ACE and alpha-adducin polymorphism as markers of individual response to diuretic therapy. Hypertension. 2003;41(3):398–403. doi:10.1161/01.HYP.0000057010.27011.2C
23. Turner ST, Boerwinkle E, O’Connell JR, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62(2):391–397. doi:10.1161/HYPERTENSIONAHA.111.00436
24. Schelleman H, Klungel OH, Witteman JC, et al. The influence of the alpha-adducin G460W polymorphism and angiotensinogen M235T polymorphism on antihypertensive medication and blood pressure. Eur J Hum Genet. 2006;14(7):860–866. doi:10.1038/sj.ejhg.5201632
25. Klenke S, Siffert W. SNPs in genes encoding G proteins in pharmacogenetics. Pharmacogenomics. 2011;12(5):633–654. doi:10.2217/pgs.10.203
26. Siffert W, Rosskopf D, Siffert G, et al. Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet. 1998;18(1):45–48. doi:10.1038/ng0198-45
27. Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. C825T polymorphism of the G protein beta(3)-subunit and antihypertensive response to a thiazide diuretic. Hypertension. 2001;37(2 Pt 2):739–743. doi:10.1161/01.HYP.37.2.739
28. Schelleman H, Stricker BH, Verschuren WM, et al. Interactions between five candidate genes and antihypertensive drug therapy on blood pressure. Pharmacogenomics J. 2006;6(1):22–26. doi:10.1038/sj.tpj.6500339
29. Turner ST, Chapman AB, Schwartz GL, Boerwinkle E. Effects of endothelial nitric oxide synthase, alpha-adducin, and other candidate gene polymorphisms on blood pressure response to hydrochlorothiazide. Am J Hypertens. 2003;16(10):834–839. doi:10.1016/S0895-7061(03)01011-2
30. Li Y, Yang P, Wu S, et al. Gender-specific association between ACE gene I/D polymorphism and blood pressure response to hydrochlorothiazide in Han Chinese hypertensive patients. Biochem Genet. 2011;49(11–12):704–714. doi:10.1007/s10528-011-9444-6
31. Suonsyrja T, Hannila-Handelberg T, Fodstad H, Donner K, Kontula K, Hiltunen TP. Renin-angiotensin system and alpha-adducin gene polymorphisms and their relation to responses to antihypertensive drugs: results from the GENRES study. Am J Hypertens. 2009;22(2):169–175. doi:10.1038/ajh.2008.343
32. Rotin D, Schild L. ENaC and its regulatory proteins as drug targets for blood pressure control. Curr Drug Targets. 2008;9(8):709–716. doi:10.2174/138945008785132367
33. Luo F, Wang Y, Wang X, Sun K, Zhou X, Hui R. A functional variant of NEDD4L is associated with hypertension, antihypertensive response, and orthostatic hypotension. Hypertension. 2009;54(4):796–801. doi:10.1161/HYPERTENSIONAHA.109.135103
34. Russo CJ, Melista E, Cui J, et al. Association of NEDD4L ubiquitin ligase with essential hypertension. Hypertension. 2005;46(3):488–491. doi:10.1161/01.HYP.0000178594.63193.c0
35. Dahlberg J, Nilsson LO, von Wowern F, Melander O. Polymorphism in NEDD4L is associated with increased salt sensitivity, reduced levels of P-renin and increased levels of Nt-proANP. PLoS One. 2007;2(5):e432. doi:10.1371/journal.pone.0000432
36. Svensson-Farbom P, Wahlstrand B, Almgren P, et al. A functional variant of the NEDD4L gene is associated with beneficial treatment response with beta-blockers and diuretics in hypertensive patients. J Hypertens. 2011;29(2):388–395. doi:10.1097/HJH.0b013e3283410390
37. McDonough CW, Burbage SE, Duarte JD, et al. Association of variants in NEDD4L with blood pressure response and adverse cardiovascular outcomes in hypertensive patients treated with thiazide diuretics. J Hypertens. 2013;31(4):698–704. doi:10.1097/HJH.0b013e32835e2a71
38. Turner ST, Bailey KR, Fridley BL, et al. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52(2):359–365. doi:10.1161/HYPERTENSIONAHA.107.104273
39. Duarte JD, Turner ST, Tran B, et al. Association of chromosome 12 locus with antihypertensive response to hydrochlorothiazide may involve differential YEATS4 expression. Pharmacogenomics J. 2013;13(3):257–263. doi:10.1038/tpj.2012.4
40. Singh S, Wang Z, Shahin MH, et al. Targeted sequencing identifies a missense variant in the BEST3 gene associated with antihypertensive response to hydrochlorothiazide. Pharmacogenet Genomics. 2018;28(11):251–255. doi:10.1097/FPC.0000000000000353
41. Hiltunen TP, Donner KM, Sarin AP, et al. Pharmacogenomics of hypertension: a genome-wide, placebo-controlled cross-over study, using four classes of antihypertensive drugs. J Am Heart Assoc. 2015;4(1):e001521. doi:10.1161/JAHA.114.001521
42. Shahin MH, Sa AC, Webb A, et al. Genome-wide prioritization and transcriptomics reveal novel signatures associated with thiazide diuretics blood pressure response. Circ Cardiovasc Genet. 2017;10(1):e001404. doi:10.1161/CIRCGENETICS.116.001404
43. Magvanjav O, Gong Y, McDonough CW, et al. Genetic variants associated with uncontrolled blood pressure on thiazide diuretic/beta-blocker combination therapy in the PEAR (Pharmacogenomic Evaluation of Antihypertensive Responses) and INVEST (International Verapamil-SR Trandolapril Study) trials. J Am Heart Assoc. 2017;6(11):e006522. doi:10.1161/JAHA.117.006522
44. Mann SJ. Redefining beta-blocker use in hypertension: selecting the right beta-blocker and the right patient. J Am Soc Hypertens. 2017;11(1):54–65. doi:10.1016/j.jash.2016.11.007
45. Laurent S. Antihypertensive drugs. Pharmacol Res. 2017;124:116–125. doi:10.1016/j.phrs.2017.07.026
46. Blumenfeld JD, Sealey JE, Mann SJ, et al. Beta-adrenergic receptor blockade as a therapeutic approach for suppressing the renin-angiotensin-aldosterone system in normotensive and hypertensive subjects. Am J Hypertens. 1999;12(5):451–459. doi:10.1016/S0895-7061(99)00005-9
47. Johnson JA, Liggett SB. Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. Clin Pharmacol Ther. 2011;89(3):366–378. doi:10.1038/clpt.2010.315
48. Zhang F, Steinberg SF. S49G and R389G polymorphisms of the beta(1)-adrenergic receptor influence signaling via the cAMP-PKA and ERK pathways. Physiol Genomics. 2013;45(23):1186–1192. doi:10.1152/physiolgenomics.00087.2013
49. Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74(1):44–52. doi:10.1016/S0009-9236(03)00068-7
50. Si D, Wang J, Xu Y, Chen X, Zhang M, Zhou H. Association of common polymorphisms in beta1-adrenergic receptor with antihypertensive response to carvedilol. J Cardiovasc Pharmacol. 2014;64(4):306–309. doi:10.1097/FJC.0000000000000119
51. Chen L, Xiao T, Chen L, Xie S, Deng M, Wu D. The association of ADRB1 and CYP2D6 polymorphisms with antihypertensive effects and analysis of their contribution to hypertension risk. Am J Med Sci. 2018;355(3):235–239. doi:10.1016/j.amjms.2017.11.002
52. Suonsyrja T, Donner K, Hannila-Handelberg T, Fodstad H, Kontula K, Hiltunen TP. Common genetic variation of beta1- and beta2-adrenergic receptor and response to four classes of antihypertensive treatment. Pharmacogenet Genomics. 2010;20(5):342–345. doi:10.1097/FPC.0b013e328338e1b8
53. Filigheddu F, Argiolas G, Degortes S, et al. Haplotypes of the adrenergic system predict the blood pressure response to beta-blockers in women with essential hypertension. Pharmacogenomics. 2010;11(3):319–325. doi:10.2217/pgs.09.158
54. Rau T, Heide R, Bergmann K, et al. Effect of the CYP2D6 genotype on metoprolol metabolism persists during long-term treatment. Pharmacogenetics. 2002;12(6):465–472. doi:10.1097/00008571-200208000-00007
55. Nozawa T, Taguchi M, Tahara K, et al. Influence of CYP2D6 genotype on metoprolol plasma concentration and beta-adrenergic inhibition during long-term treatment: a comparison with bisoprolol. J Cardiovasc Pharmacol. 2005;46(5):713–720. doi:10.1097/01.fjc.0000184117.76188.68
56. Lefebvre J, Poirier L, Poirier P, Turgeon J, Lacourciere Y. The influence of CYP2D6 phenotype on the clinical response of nebivolol in patients with essential hypertension. Br J Clin Pharmacol. 2007;63(5):575–582. doi:10.1111/bcp.2007.63.issue-5
57. Bijl MJ, Visser LE, van Schaik RH, et al. Genetic variation in the CYP2D6 gene is associated with a lower heart rate and blood pressure in beta-blocker users. Clin Pharmacol Ther. 2009;85(1):45–50. doi:10.1038/clpt.2008.172
58. Rau T, Wuttke H, Michels LM, et al. Impact of the CYP2D6 genotype on the clinical effects of metoprolol: a prospective longitudinal study. Clin Pharmacol Ther. 2009;85(3):269–272. doi:10.1038/clpt.2008.218
59. Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. doi:10.1038/clpt.2011.34
60. Hamadeh IS, Langaee TY, Dwivedi R, et al. Impact of CYP2D6 polymorphisms on clinical efficacy and tolerability of metoprolol tartrate. Clin Pharmacol Ther. 2014;96(2):175–181. doi:10.1038/clpt.2014.62
61. Wu D, Li G, Deng M, et al. Associations between ADRB1 and CYP2D6 gene polymorphisms and the response to beta-blocker therapy in hypertension. J Int Med Res. 2015;43(3):424–434. doi:10.1177/0300060514563151
62. Zineh I, Beitelshees AL, Gaedigk A, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther. 2004;76(6):536–544. doi:10.1016/j.clpt.2004.08.020
63. Gong Y, Wang Z, Beitelshees AL, et al. Pharmacogenomic genome-wide meta-analysis of blood pressure response to beta-blockers in hypertensive African Americans. Hypertension. 2016;67(3):556–563. doi:10.1161/HYPERTENSIONAHA.115.06345
64. Singh S, El Rouby N, McDonough CW, et al. Genomic association analysis reveals variants associated with blood pressure response to beta-blockers in European Americans. Clin Transl Sci. 2019. doi:10.1111/cts.12643
65. Rimpela JM, Kontula KK, Fyhrquist F, et al. Replicated evidence for aminoacylase 3 and nephrin gene variations to predict antihypertensive drug responses. Pharmacogenomics. 2017;18(5):445–458. doi:10.2217/pgs-2016-0204
66. Singh S, Warren HR, Hiltunen TP, et al. Genome-wide meta-analysis of blood pressure response to beta1-blockers: results from ICAPS (International Consortium of Antihypertensive Pharmacogenomics Studies). J Am Heart Assoc. 2019;8(16):e013115. doi:10.1161/JAHA.119.013115
67. Sy RG, Nevado JB
68. Sparks MA, Crowley SD, Gurley SB, Mirotsou M, Coffman TM. Classical renin-angiotensin system in kidney physiology. Compr Physiol. 2014;4(3):1201–1228.
69. van Thiel BS, van der Pluijm I, te Riet L, Essers J, Danser AH. The renin-angiotensin system and its involvement in vascular disease. Eur J Pharmacol. 2015;763(Pt A):3–14. doi:10.1016/j.ejphar.2015.03.090
70. Lupoli S, Salvi E, Barcella M, Barlassina C. Pharmacogenomics considerations in the control of hypertension. Pharmacogenomics. 2015;16(17):1951–1964. doi:10.2217/pgs.15.131
71. Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–1346. doi:10.1172/JCI114844
72. Chiang FT, Lai ZP, Chern TH, et al. Lack of association of the angiotensin converting enzyme polymorphism with essential hypertension in a Chinese population. Am J Hypertens. 1997;10(2):197–201. doi:10.1016/S0895-7061(96)00345-7
73. Nakai K, Itoh C, Miura Y, et al. Deletion polymorphism of the angiotensin I-converting enzyme gene is associated with serum ACE concentration and increased risk for CAD in the Japanese. Circulation. 1994;90(5):2199–2202. doi:10.1161/01.CIR.90.5.2199
74. Bloem LJ, Manatunga AK, Pratt JH. Racial difference in the relationship of an angiotensin I-converting enzyme gene polymorphism to serum angiotensin I-converting enzyme activity. Hypertension. 1996;27(1):62–66. doi:10.1161/01.HYP.27.1.62
75. Iwai N, Ohmichi N, Nakamura Y, Kinoshita M. DD genotype of the angiotensin-converting enzyme gene is a risk factor for left ventricular hypertrophy. Circulation. 1994;90(6):2622–2628. doi:10.1161/01.CIR.90.6.2622
76. Schunkert H, Hense HW, Holmer SR, et al. Association between a deletion polymorphism of the angiotensin-converting-enzyme gene and left ventricular hypertrophy. N Engl J Med. 1994;330(23):1634–1638. doi:10.1056/NEJM199406093302302
77. Cambien F, Poirier O, Lecerf L, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature. 1992;359(6396):641–644. doi:10.1038/359641a0
78. Abouelfath R, Habbal R, Laaraj A, Khay K, Harraka M, Nadifi S. ACE insertion/deletion polymorphism is positively associated with resistant hypertension in Morocco. Gene. 2018;658:178–183. doi:10.1016/j.gene.2018.03.028
79. Fernandez-Llama P, Poch E, Oriola J, et al. Angiotensin converting enzyme gene I/D polymorphism in essential hypertension and nephroangiosclerosis. Kidney Int. 1998;53(6):1743–1747. doi:10.1046/j.1523-1755.1998.00946.x
80. Pontremoli R, Sofia A, Tirotta A, et al. The deletion polymorphism of the angiotensin I-converting enzyme gene is associated with target organ damage in essential hypertension. J Am Soc Nephrol. 1996;7(12):2550–2558.
81. Heidari F, Vasudevan R, Mohd Ali SZ, et al. Association of insertion/deletion polymorphism of angiotensin-converting enzyme gene among Malay male hypertensive subjects in response to ACE inhibitors. J Renin Angiotensin Aldosterone Syst. 2015;16(4):872–879. doi:10.1177/1470320314538878
82. Stavroulakis GA, Makris TK, Krespi PG, et al. Predicting response to chronic antihypertensive treatment with fosinopril: the role of angiotensin-converting enzyme gene polymorphism. Cardiovasc Drugs Ther. 2000;14(4):427–432. doi:10.1023/A:1007820401377
83. Kurland L, Melhus H, Karlsson J, et al. Angiotensin converting enzyme gene polymorphism predicts blood pressure response to angiotensin II receptor type 1 antagonist treatment in hypertensive patients. J Hypertens. 2001;19(10):1783–1787. doi:10.1097/00004872-200110000-00012
84. Ohmichi N, Iwai N, Uchida Y, Shichiri G, Nakamura Y, Kinoshita M. Relationship between the response to the angiotensin converting enzyme inhibitor imidapril and the angiotensin converting enzyme genotype. Am J Hypertens. 1997;10(8):951–955. doi:10.1016/S0895-7061(97)00121-0
85. Heidari F, Vasudevan R, Mohd Ali SZ, Ismail P, Arkani M. RAS genetic variants in interaction with ACE inhibitors drugs influences essential hypertension control. Arch Med Res. 2017;48(1):88–95. doi:10.1016/j.arcmed.2017.03.003
86. Sasaki M, Oki T, Iuchi A, et al. Relationship between the angiotensin converting enzyme gene polymorphism and the effects of enalapril on left ventricular hypertrophy and impaired diastolic filling in essential hypertension: M-mode and pulsed doppler echocardiographic studies. J Hypertens. 1996;14(12):1403–1408. doi:10.1097/00004872-199612000-00003
87. Hingorani AD, Jia H, Stevens PA, Hopper R, Dickerson JE, Brown MJ. Renin-angiotensin system gene polymorphisms influence blood pressure and the response to angiotensin converting enzyme inhibition. J Hypertens. 1995;13(12 Pt 2):1602–1609.
88. Dudley C, Keavney B, Casadei B, Conway J, Bird R, Ratcliffe P. Prediction of patient responses to antihypertensive drugs using genetic polymorphisms: investigation of renin-angiotensin system genes. J Hypertens. 1996;14(2):259–262. doi:10.1097/00004872-199602000-00016
89. Konoshita T; Genomic Disease Outcome Consortium Study I. Do genetic variants of the renin-angiotensin system predict blood pressure response to renin-angiotensin system-blocking drugs? A systematic review of pharmacogenomics in the renin-angiotensin system. Curr Hypertens Rep. 2011;13(5):356–361. doi:10.1007/s11906-011-0212-0
90. Joshi PH, Xu H, Lestrange R, et al. The M235T single nucleotide polymorphism in the angiotensinogen gene is associated with coronary artery calcium in patients with a family history of coronary artery disease. Atherosclerosis. 2013;226(2):433–439. doi:10.1016/j.atherosclerosis.2012.10.039
91. Mondorf UF, Russ A, Wiesemann A, Herrero M, Oremek G, Lenz T. Contribution of angiotensin I converting enzyme gene polymorphism and angiotensinogen gene polymorphism to blood pressure regulation in essential hypertension. Am J Hypertens. 1998;11(2):174–183. doi:10.1016/S0895-7061(97)00402-0
92. Kurland L, Liljedahl U, Karlsson J, et al. Angiotensinogen gene polymorphisms: relationship to blood pressure response to antihypertensive treatment. results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation vs Atenolol (SILVHIA) trial. Am J Hypertens. 2004;17(1):8–13. doi:10.1016/j.amjhyper.2003.09.009
93. Freel EM, Ingram M, Friel EC, et al. Phenotypic consequences of variation across the aldosterone synthase and 11-beta hydroxylase locus in a hypertensive cohort: data from the MRC BRIGHT study. Clin Endocrinol (Oxf). 2007;67(6):832–838. doi:10.1111/cen.2007.67.issue-6
94. Hautanena A, Lankinen L, Kupari M, et al. Associations between aldosterone synthase gene polymorphism and the adrenocortical function in males. J Intern Med. 1998;244(1):11–18. doi:10.1046/j.1365-2796.1998.00308.x
95. Lim PO, Macdonald TM, Holloway C, et al. Variation at the aldosterone synthase (CYP11B2) locus contributes to hypertension in subjects with a raised aldosterone-to-renin ratio. J Clin Endocrinol Metab. 2002;87(9):4398–4402. doi:10.1210/jc.2001-012070
96. Davies E, Holloway CD, Ingram MC, et al. Aldosterone excretion rate and blood pressure in essential hypertension are related to polymorphic differences in the aldosterone synthase gene CYP11B2. Hypertension. 1999;33(2):703–707. doi:10.1161/01.HYP.33.2.703
97. Lacchini R, Sabha M, Coeli FB, et al. T allele of −344 C/T polymorphism in aldosterone synthase gene is not associated with resistant hypertension. Hypertens Res. 2009;32(2):159–162. doi:10.1038/hr.2008.36
98. Kurland L, Melhus H, Karlsson J, et al. Aldosterone synthase (CYP11B2) −344 C/T polymorphism is related to antihypertensive response: result from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation versus Atenolol (SILVHIA) trial. Am J Hypertens. 2002;15(5):389–393. doi:10.1016/S0895-7061(02)02256-2
99. Ji X, Qi H, Li DB, et al. Associations between human aldosterone synthase CYP11B2 (−344T/C) gene polymorphism and antihypertensive response to valsartan in Chinese patients with essential hypertension. Int J Clin Exp Med. 2015;8(1):1173–1177.
100. Ortlepp JR, Hanrath P, Mevissen V, Kiel G, Borggrefe M, Hoffmann R. Variants of the CYP11B2 gene predict response to therapy with candesartan. Eur J Pharmacol. 2002;445(1–2):151–152. doi:10.1016/S0014-2999(02)01766-1
101. Jankowski P, Safar ME, Benetos A. Pleiotropic effects of drugs inhibiting the renin-angiotensin-aldosterone system. Curr Pharm Des. 2009;15(5):571–584. doi:10.2174/138161209787315747
102. Ismail H, Mitchell R, McFarlane SI, Makaryus AN. Pleiotropic effects of inhibitors of the RAAS in the diabetic population: above and beyond blood pressure lowering. Curr Diab Rep. 2010;10(1):32–36. doi:10.1007/s11892-009-0081-y
103. Fontana V, de Faria AP, Oliveira-Paula GH, et al. Effects of angiotensin-converting enzyme inhibition on leptin and adiponectin levels in essential hypertension. Basic Clin Pharmacol Toxicol. 2014;114(6):472–475. doi:10.1111/bcpt.12195
104. Oliveira-Paula GH, Pinheiro LC, Ferreira GC, et al. Angiotensin converting enzyme inhibitors enhance the hypotensive effects of propofol by increasing nitric oxide production. Free Radic Biol Med. 2018;115:10–17. doi:10.1016/j.freeradbiomed.2017.11.010
105. Linz W, Wohlfart P, Scholkens BA, Malinski T, Wiemer G. Interactions among ACE, kinins and NO. Cardiovasc Res. 1999;43(3):549–561. doi:10.1016/S0008-6363(99)00091-7
106. Oliveira-Paula GH, Lacchini R, Tanus-Santos JE. Endothelial nitric oxide synthase: from biochemistry and gene structure to clinical implications of NOS3 polymorphisms. Gene. 2016;575(2 Pt 3):584–599. doi:10.1016/j.gene.2015.09.061
107. Oliveira-Paula GH, Lacchini R, Tanus-Santos JE. Clinical and pharmacogenetic impact of endothelial nitric oxide synthase polymorphisms on cardiovascular diseases. Nitric Oxide. 2017;63:39–51. doi:10.1016/j.niox.2016.08.004
108. Oliveira-Paula GH, Lacchini R, Pinheiro LC, et al. Endothelial nitric oxide synthase polymorphisms affect the changes in blood pressure and nitric oxide bioavailability induced by propofol. Nitric Oxide. 2018;75:77–84. doi:10.1016/j.niox.2018.02.007
109. Silva PS, Fontana V, Luizon MR, et al. eNOS and BDKRB2 genotypes affect the antihypertensive responses to enalapril. Eur J Clin Pharmacol. 2013;69(2):167–177. doi:10.1007/s00228-012-1326-2
110. Mason RP, Jacob RF, Kubant R, et al. Effects of angiotensin receptor blockers on endothelial nitric oxide release: the role of eNOS variants. Br J Clin Pharmacol. 2012;74(1):141–146. doi:10.1111/bcp.2012.74.issue-1
111. Oliveira-Paula GH, Lacchini R, Luizon MR, et al. Endothelial nitric oxide synthase tagSNPs influence the effects of enalapril in essential hypertension. Nitric Oxide. 2016;55-56:62–69. doi:10.1016/j.niox.2016.03.006
112. Li P, Kondo T, Numaguchi Y, et al. Role of bradykinin, nitric oxide, and angiotensin II type 2 receptor in imidapril-induced angiogenesis. Hypertension. 2008;51(2):252–258. doi:10.1161/HYPERTENSIONAHA.107.097394
113. Yang X, Zhu Q, Fong J, et al. Enalaprilat, an angiotensin-converting enzyme inhibitor, enhances functional preservation during long-term cardiac preservation. Possible involvement of bradykinin and PKC. J Mol Cell Cardiol. 1996;28(7):1445–1452. doi:10.1006/jmcc.1996.0135
114. Oliveira-Paula GH, Luizon MR, Lacchini R, et al. Gene-gene interactions among PRKCA, NOS3 and BDKRB2 polymorphisms affect the antihypertensive effects of enalapril. Basic Clin Pharmacol Toxicol. 2017;120(3):284–291. doi:10.1111/bcpt.12682
115. Oliveira-Paula GH, Lacchini R, Fontana V, Silva PS, Biagi C, Tanus-Santos JE. Polymorphisms in VEGFA gene affect the antihypertensive responses to enalapril. Eur J Clin Pharmacol. 2015;71(8):949–957. doi:10.1007/s00228-015-1872-5
116. Frau F, Zaninello R, Salvi E, et al. Genome-wide association study identifies CAMKID variants involved in blood pressure response to losartan: the SOPHIA study. Pharmacogenomics. 2014;15(13):1643–1652. doi:10.2217/pgs.14.119
117. Turner ST, Bailey KR, Schwartz GL, Chapman AB, Chai HS, Boerwinkle E. Genomic association analysis identifies multiple loci influencing antihypertensive response to an angiotensin II receptor blocker. Hypertension. 2012;59(6):1204–1211. doi:10.1161/HYP.0b013e31825b30f8
118. Elliott WJ, Ram CV. Calcium channel blockers. J Clin Hypertens (Greenwich). 2011;13(9):687–689. doi:10.1111/j.1751-7176.2011.00513.x
119. Nguyen BN, Parker RB, Noujedehi M, Sullivan JM, Johnson JA. Effects of COER-verapamil on circadian pattern of forearm vascular resistance and blood pressure. J Clin Pharmacol. 2000;40(12 Pt 2):1480–1487.
120. Niu Y, Gong Y, Langaee TY, et al. Genetic variation in the beta2 subunit of the voltage-gated calcium channel and pharmacogenetic association with adverse cardiovascular outcomes in the INternational VErapamil SR-Trandolapril STudy GENEtic Substudy (INVEST-GENES). Circ Cardiovasc Genet. 2010;3(6):548–555. doi:10.1161/CIRCGENETICS.110.957654
121. Beitelshees AL, Gong Y, Wang D, et al. KCNMB1 genotype influences response to verapamil SR and adverse outcomes in the INternational VErapamil SR/Trandolapril STudy (INVEST). Pharmacogenet Genomics. 2007;17(9):719–729. doi:10.1097/FPC.0b013e32810f2e3c
122. Kamide K, Yang J, Matayoshi T, et al. Genetic polymorphisms of L-type calcium channel alpha1C and alpha1D subunit genes are associated with sensitivity to the antihypertensive effects of L-type dihydropyridine calcium-channel blockers. Circ J. 2009;73(4):732–740. doi:10.1253/circj.CJ-08-0761
123. Bremer T, Man A, Kask K, Diamond C. CACNA1C polymorphisms are associated with the efficacy of calcium channel blockers in the treatment of hypertension. Pharmacogenomics. 2006;7(3):271–279. doi:10.2217/14622416.7.3.271
124. He F, Luo J, Luo Z, et al. The KCNH2 genetic polymorphism (1956, C>T) is a novel biomarker that is associated with CCB and alpha,beta-ADR blocker response in EH patients in China. PLoS One. 2013;8(4):e61317. doi:10.1371/journal.pone.0061317
125. Zhu Y, Wang F, Li Q, et al. Amlodipine metabolism in human liver microsomes and roles of CYP3A4/5 in the dihydropyridine dehydrogenation. Drug Metab Dispos. 2014;42(2):245–249. doi:10.1124/dmd.113.055400
126. Tracy TS, Korzekwa KR, Gonzalez FJ, Wainer IW. Cytochrome P450 isoforms involved in metabolism of the enantiomers of verapamil and norverapamil. Br J Clin Pharmacol. 1999;47(5):545–552. doi:10.1046/j.1365-2125.1999.00923.x
127. Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–391. doi:10.1038/86882
128. Huang Y, Wen G, Lu Y, et al. CYP3A4*1G and CYP3A5*3 genetic polymorphisms alter the antihypertensive efficacy of amlodipine in patients with hypertension following renal transplantation. Int J Clin Pharmacol Ther. 2017;55(2):109–118. doi:10.5414/CP202559
129. Zhang YP, Zuo XC, Huang ZJ, et al. CYP3A5 polymorphism, amlodipine and hypertension. J Hum Hypertens. 2014;28(3):145–149. doi:10.1038/jhh.2013.67
130. Kim KA, Park PW, Lee OJ, et al. Effect of CYP3A5*3 genotype on the pharmacokinetics and pharmacodynamics of amlodipine in healthy Korean subjects. Clin Pharmacol Ther. 2006;80(6):646–656. doi:10.1016/j.clpt.2006.09.009
131. Bhatnagar V, Garcia EP, O’Connor DT, et al. CYP3A4 and CYP3A5 polymorphisms and blood pressure response to amlodipine among African-American men and women with early hypertensive renal disease. Am J Nephrol. 2010;31(2):95–103. doi:10.1159/000258688
132. Langaee TY, Gong Y, Yarandi HN, et al. Association of CYP3A5 polymorphisms with hypertension and antihypertensive response to verapamil. Clin Pharmacol Ther. 2007;81(3):386–391. doi:10.1038/sj.clpt.6100090
133. Kamide K, Asayama K, Katsuya T, et al. Genome-wide response to antihypertensive medication using home blood pressure measurements: a pilot study nested within the HOMED-BP study. Pharmacogenomics. 2013;14(14):1709–1721. doi:10.2217/pgs.13.161
134. Hamrefors V, Sjogren M, Almgren P, et al. Pharmacogenetic implications for eight common blood pressure-associated single-nucleotide polymorphisms. J Hypertens. 2012;30(6):1151–1160. doi:10.1097/HJH.0b013e3283536338
135. Lin E, Hsu SY. A bayesian approach to gene-gene and gene-environment interactions in chronic fatigue syndrome. Pharmacogenomics. 2009;10(1):35–42. doi:10.2217/14622416.10.1.35
136. Lin E, Hwang Y, Liang KH, Chen EY. Pattern-recognition techniques with haplotype analysis in pharmacogenomics. Pharmacogenomics. 2007;8(1):75–83. doi:10.2217/14622416.8.1.75
137. Zanger UM. Pharmacogenetics - challenges and opportunities ahead. Front Pharmacol. 2010;1:112.
138. Ritchie MD, Motsinger AA. Multifactor dimensionality reduction for detecting gene-gene and gene-environment interactions in pharmacogenomics studies. Pharmacogenomics. 2005;6(8):823–834. doi:10.2217/14622416.6.8.823
139. Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–147. doi:10.1086/321276
140. Lane HY, Tsai GE, Lin E. Assessing gene-gene interactions in pharmacogenomics. Mol Diagn Ther. 2012;16(1):15–27. doi:10.1007/BF03256426
141. Li H, Oehrlein SA, Wallerath T, et al. Activation of protein kinase C alpha and/or epsilon enhances transcription of the human endothelial nitric oxide synthase gene. Mol Pharmacol. 1998;53(4):630–637. doi:10.1124/mol.53.4.630
142. Amiri F, Garcia R. Renal angiotensin II receptors and protein kinase C in diabetic rats: effects of insulin and ACE inhibition. Am J Physiol Renal Physiol. 2000;278(4):F603–F612. doi:10.1152/ajprenal.2000.278.4.F603
143. Zhang W, Huang RS, Dolan ME. Integrating epigenomics into pharmacogenomic studies. Pharmgenomics Pers Med. 2008;2008(1):7–14. doi:10.2147/pgpm.s4341
144. Padmanabhan S, Joe B. Towards precision medicine for hypertension: a review of genomic, epigenomic, and microbiomic effects on blood pressure in experimental rat models and humans. Physiol Rev. 2017;97(4):1469–1528. doi:10.1152/physrev.00035.2016
145. Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–1425. doi:10.1038/s41588-018-0205-x
146. Warren HR, Evangelou E, Cabrera CP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49(3):403–415. doi:10.1038/ng.3768
147. Cunningham PN, Chapman AB. The future of pharmacogenetics in the treatment of hypertension. Pharmacogenomics. 2019;20(3):129–132. doi:10.2217/pgs-2018-0191
148. O’Donnell PH, Wadhwa N, Danahey K, et al. Pharmacogenomics-based point-of-care clinical decision support significantly alters drug prescribing. Clin Pharmacol Ther. 2017;102(5):859–869. doi:10.1002/cpt.709
149. Rosenman MB, Decker B, Levy KD, Holmes AM, Pratt VM, Eadon MT. Lessons learned when introducing pharmacogenomic panel testing into clinical practice. Value Health. 2017;20(1):54–59. doi:10.1016/j.jval.2016.08.727
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
