Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
Pharmacogene Variation in Thai Plasmodium vivax Relapse Patients Treated with a Combination of Primaquine and Chloroquine
Authors Chamnanphon M , Gaedigk A , Puangpetch A, Pasomsub E, Chantratita W , Longley RJ, Sattabongkot J, Chariyavilaskul P , Sukasem C
Received 9 January 2019
Accepted for publication 3 December 2019
Published 10 January 2020 Volume 2020:13 Pages 1—12
DOI https://doi.org/10.2147/PGPM.S201007
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Monpat Chamnanphon, 1 Andrea Gaedigk, 2 Apichaya Puangpetch, 3, 4 Ekawat Pasomsub, 5 Wasun Chantratita, 6 Rhea J Longley, 7, 8 Jetsumon Sattabongkot, 7 Pajaree Chariyavilaskul, 1 Chonlaphat Sukasem 3, 4
1Clinical Pharmacokinetics and Pharmacogenomics Research Unit, Department of Pharmacology, Faculty of Medicine, Chulalongkorn University, Bangkok 10330, Thailand; 2Division of Clinical Pharmacology, Toxicology & Therapeutic Innovation, Children’s Mercy Kansas City, and School of Medicine, University of Missouri-Kansas City, Kansas City, MO, USA; 3Division of Pharmacogenomics and Personalized Medicine, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand; 4Laboratory for Pharmacogenomics, Somdech Phra Debaratana Medical Center (SDMC), Ramathibodi Hospital, Bangkok, Thailand; 5Division of Virology, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand; 6Center of Genomics Medicine, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand; 7Mahidol Vivax Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand; 8Walter and Eliza Hall Institute of Medical Research, Melbourne, VIC, Australia; Department of Medical Biology, University of Melbourne, Melbourne, VIC, Australia
Correspondence: Chonlaphat Sukasem
Division of Pharmacogenomics and Personalized Medicine, Department of Pathology, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
Tel +66 22004330-2
Fax +66 22004332
Email [email protected]
Purpose: Pharmacogenes have an influence on biotransformation pathway and clinical outcome of primaquine and chloroquine which are often prescribed to treat Plasmodium vivax infection. Genetic variation may impact enzyme activity and/or transporter function and thereby contribute to relapse. The aim of the study was to assess allele, genotype frequencies and the association between pharmacogenes variation and primaquine response in Thai patients infected with Plasmodium vivax.
Patients and Methods: Fifty-one patients were genotyped for 74 variants in 18 genes by Sequenom MassARRAY® and Taqman® SNP Real-Time PCR.
Results: SNP frequencies were not significantly different between relapse (n=4) and non-relapse (n=47) patients. However, the CYP2C19 c.681G>A, the frequency of the A-allele that defines the non-functional CYP2C19*2 haplotype was significantly higher compared to the G-allele (OR=5.14, p=0.021). Patients heterozygous for ABCG2 c.421C>A had a higher odds ratio (OR=8.75, p=0.071) and the frequency of the G-allele of UGT2B7 c.372G>A was higher compared to the A-allele (OR=3.75, p=0.081). CYP2C19, ABCG2 and UGT2B7 emerged as potential high priority genes.
Conclusion: Decreased activity of CYP2C19, ABCG2 and UGT2B7 in combination with CYP2D6 intermediate or poor metabolizer status may expose patients to a higher risk of Plasmodium vivax relapse. Further investigations are warranted to substantiate these findings.
Keywords: primaquine, chloroquine, Plasmodium vivax, relapse, pharmacogenes
Introduction
Primaquine (PQ), an 8-amino-6-methoxyquinoline (8AQ) derivative is the only drug approved by the United States Federal Drug Administration to treat acute illness and relapse of Plasmodium vivax (P. vivax) and P. ovale infection caused by hypnozoites that persist in the hepatocytes of infected patients.1,2 Recently, a number of studies reported on the metabolism of PQ showing that biotransformation occurs through three main pathways which are Cytochrome P450 (CYP450) enzymes (CYP2D6, CYP2C19, CYP3A4, CYP1A2), monoamine oxidases (MAO-A and B) and flavin-containing monooxygenase-3 (FMO-3).3–7 PQ is a pro-drug primarily metabolized by MAO-A to PQ aldehyde, which is further oxidized by aldehyde dehydrogenase (ADH) to carboxyprimaquine, the major PQ metabolite found in plasma.4,8 Carboxyprimaquine is further oxidized by FMO to the N-hydroxylated PQ metabolite which can cause hemotoxicity.9 Finally, PQ is metabolized via CYP2D6 to 5-hydroxyprimaquine, 5, 6-orthoquinone, and other phenolic metabolites; other P450 enzymes are also believed to contribute to PQ metabolism.4,5,10 Chloroquine (CQ) is metabolized into N-desethylchloroquine by CYP2C8, CYP3A4, and CYP2D6 by in vitro study.11
The major challenge of elimination of malaria caused by P. vivax and P. ovale in endemic areas is relapse of dormant hypnozoites that survive in the liver of the patient after primary infection. These hypnozoites can persistent in the liver for weeks, months or even years following a primary attack.12,13 Although PQ has been used to treat P. vivax and P. ovale infections for several decades, the exact mechanisms of PQ efficacy and toxicity are still not well understood, neither have the metabolic pathways been fully elucidated. It has been postulated that human host genetics may, at least in part, contribute to the failure of PQ treatment.7 Bennett et al7 first reported a significant association between CYP2D6 metabolizer phenotype and relapsing P. vivax infection. Relapsing CYP2D6 poor (PM) and intermediate metabolizer (IM) patients had a significant higher plasma concentration of the parent drug after 24 hrs compared to non-relapsing patients. These data supported the hypothesis that the CYP2D6-dependent pathway is crucial for the bioactivation of PQ to its phenolic metabolites which are the active metabolites responsible for the elimination of dormant hypnozoites in the liver. Furthermore, these data suggested that patients with impaired CYP2D6 activity caused by genetic variation in the CYP2D6 gene may be at a higher risk of relapse of P. vivax. However, a previous study by our group has also found relapsing infections in patients with normal CYP2D6 metabolism.14 It needs to be noted that our patients have been treated with a combination of PQ and CQ (per standard Thai guidelines). Thus, we are speculating that genetic variation in other genes that contribute to PQ and CQ metabolism may also impact a patient’s response to P. vivax treatment in Thai patients. Moreover, in addition to drug-metabolizing enzymes, transporters have been shown to affect PQ efficacy.15,16 Sortica et al found that SLCO2B1, SLCO1A2 and SLCO1B1 were associated with the clearance of P. vivax in patients treated with PQ and CQ.16 The MRP transporter, for example, can be inhibited by quinoline derivatives,17 and Hayeshi et al demonstrated inhibitory effects of several antimalarial drugs to P-glycoprotein (P-gp) mediated transport and reported that both, PQ and chloroquine, inhibit P-gp.18
This study aimed to investigate genetic variation in drug-metabolizing enzymes and drug transporters and their association with relapse in Thai patients treated with a PQ/CQ combination regimen.
Materials and Methods
This exploratory investigation included 51 Thai patients from a previous study.14 The study was approved by the Internal Ethics Review Committee on Human Research of the Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Thailand (MURA 2016/657) and conducted in accordance with the Declaration of Helsinki. Briefly, symptomatic P. vivax patients from the Tha Song Yang malaria clinic, Tak province, Thailand were recruited from April 2014 to September 2015; all patients gave written informed consent. Patients were diagnosed with P. vivax infection and treated with 25 mg base/kg body weight CQ over 3 days and 0.3 mg/kg PQ daily for 14 days. Finger-prick blood samples were collected before treatment and at 1 and 2 weeks after enrollment, then every 2 weeks for 6 months, then every 4 weeks until 9 months.14 Patients for the current study were selected based on the availability of genomic DNA and clinical data including recurrent status and date of follow up/survival data.
Genes Analyzed with MassARRAY and Real-Time PCR
DNA samples diluted to 10 ng/μL were genotyped using the Sequenom MassARRAY® System (Agena Bioscience™, San Diego, CA, USA). The panel consisted of pre-designed SNPs and indels (referred to SNVs therein), and CNV assays that target the most relevant variants in 11 important pharmacogenes. A total of 53 SNVs were interrogated and five assays were utilized to determine CYP2D6 gene copy number variation (CNV) (Table 1). The iPLEX® PGx 68 Panel (Agena, San Diego, CA) included drug transporters (ABCB1, SLCO1B1, SULT4A1), Phase I enzymes (COMT, CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5). The workflow consisted of five steps: PCR amplification, primer extension or fragmentation, dispensed extension product onto a SpectroCHIP® Array and MassARRAY MALDI-TOF mass spectrometry. Automated software provided diplotype, haplotype, and CNV calls in a single combined report. The overall process was completed in less than 10 hrs.
 |
Table 1 List of Genes and SNPs Detected by MassArray® and Taqman® RT-PCR |
An additional 21 SNVs of 10 genes including CYP2B6, CYP3A4, ABCA1, ABCB1, ABCC2, ABCC4, ABCG2, SLC25A40, SULT1A1, and UGT2B7 were genotyped with commercially available TaqMan® Genotyping Assays (Applied Biosystems™, Carlsbad, CA, USA) following manufacturer’s instructions. Details are provided in Table 1. Allelic variants were designated according to the Pharmacogene Variation Consortium (PharmVar) at www.PharmVar.org.19,20
Activity scores (AS) were assigned as previously described21 and applied in CPIC guidelines.22–25 To assess the combined impact of CYP2D6 and CYP2C19 activity, we assigned a composite AS, which is the sum of the AS assigned to each gene.
Statistical Analysis
Deviation from Hardy-Weinberg expectations was assessed using an exact and chi-square test. SNV and genotype frequencies were determined by direct counting. Linkage disequilibrium measures (r2) and haplotype analysis were performed using Haploview version 4.2.26 Comparisons of SNV and genotype frequencies between relapsing and non-relapsing patients were performed using the χ2 test, odds ratio and 95% confidence intervals were calculated as measurements of the strength of association. The probability of significant associations was set at p<0.05. Relapse-free survival (RFS) was estimated using the log-rank test and Kaplan–Meier curves. The Cox proportional hazards model was conducted to assess the impact of candidate SNPs on relapse of P. vivax. All statistical analyses were performed using STATA software version 14 (StataCorp LP, TX, USA).
Results
Demographic Data
The average age of the 51 patients was 26.37 (min–max; 7–71) years with 32 (63%) males and 19 (37%) females. There were four recurrent P. vivax infections, which occurred between 8 and 32-weeks after the initial treatment, the average time of follow-up was 7.7±1.7 months. The clinical status of 45 patients was recorded for at least 6 months.
Association of Pharmacogene Variation and Relapse
The call rate for SNV genotyping was 100% for both testing platforms. Table 1 summarizes the genes and SNVs tested by MassArray® and Taqman RT-PCR®. SNV and genotype frequencies in the relapsing and non-relapsing patients are provided in Tables 2 and 3, respectively. All SNVs were in Hardy-Weinberg equilibrium except CYP2D6 g.4180G>C and ABCA1 c.4760A>G (p<0.05) which were excluded from subsequent analyses. Nine SNVs in nine genes had minor allele frequency (MAF) of less than 5% (Table S1). The frequencies of the most common genotypes are shown in Table 3 which were CYP1A2*1A/*1F (31.4%), CYP2B6*1/*6 (41.2%), CYP2C9*1/*3 (5.9%), CYP2C19*1/*2 (33.3%), CYP2D6*2/*10 (21.6%), CYP3A4*1/*1B (3.9%), CYP3A5*3/*3 (54.9%), and SLCO1B1*1/*5 (7.8%). In addition, frequencies for 0, 1, 2, 3 and 4 CYP2D6 gene copies were 1.96% (n=1), 9.80% (n=5), 45.10% (n=23), 41.18% (n=21), and 1.96% (n=1), respectively. The CYP2D6 CNV status and genotype of the four relapsed patients were three copies (two patients with a CYP2D6*2/*10 and 1 patient with a CYP2D6*10/*10 genotype) and one copy (one patient genotyped as CYP2D6*1/*5). None of these genotypes, or any other genotypes, revealed statistically significant association between relapse and non-relapse (Tables 3 and S2).
 |
Table 2 SNP Frequencies of Drug Metabolizing Genes Between Relapse and Non-Relapse Groups |
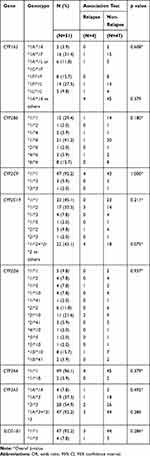 |
Table 3 Genotype Frequencies of Drug Metabolizing Genes Between Relapse and Non-Relapse Groups |
The genotype frequencies of the majority of drug-metabolizing enzyme and transporter genes of the relapsing patients were similar with those previously described for Thai. Frequencies for the candidate genes including CYP2D6, CYP2C9, CYP2C19, CYP3A4, CYP1A2 were not significantly different among non-relapsing and relapsing patients (p>0.05). As shown in Table 3 there was a trend, however, for a significant association between CYP2C19 c.681G>A, and relapse (G/A+A/A genotypes vs G/G genotype; OR=7.25, 95% CI; 0.79–66.84, p=0.082). Furthermore, the c.681A SNV defining the non-functional CYP2C19*2 allele was significantly more common than the c.681G allele in relapse versus non-relapse patients (OR=5.14, 95% CI; 0.90–35.01, p=0.021). Moreover, the ABCG2 c.421C>A transporter SNV had a higher odds ratio, although this did not reach statistical significance (OR=8.75, 95% CI; 0.83–92.32, p=0.071). Lastly, we also explored the relationship between a composite AS for CYP2D6 + CYP2C19 and PQ response. A composite AS, however, was not significantly different (p=0.09) (Table S3).
Survival Analysis
RFS was calculated from enrollment date to the time of P. vivax relapse. The effect of genetic variation in pharmacogenes on RFS was investigated by grouping patients by SNVs and genotype. Kaplan–Meier analysis revealed no statistically significant differences in RFS rates for most of the variable factors. However, a trend toward statistical significance of RFS rates was observed for patients carrying the CYP2C19*2 allele (c.681G>A) (Log-rank test; p=0.077, Figure 1A), and ABCG2 c.421C>A (Log-rank test; p=0.099, Figure 1B). Also, as shown in Figure 1C and D, a sub-analysis of CYP2D6 copy number variation revealed significantly different RFS rates between 1 and 2 gene copies (p=0.007) and between 2 and 3 gene copies (p=0.065), respectively. There was no statistical difference in RFS rates among groups for any of the other variable factors (Table S4).
Relapse-Free Survival Rate by Univariate and Multivariate Analysis
Cox proportional hazards analysis was used for the determination of univariate and multivariate association analysis of the SNVs with RFS. In multivariate analyses, there was no significant difference in RFS rates between SNVs, genotype or other variables (Table 4). However, a sub-analysis showed that CYP1A2 g.-163C>A, CYP2C19*2 (c.681G>A), CYP2D6 copy number, ABCA1 c.2649A>G, ABCB1 c.1236C>T, ABCC2 g.68231A>G, ABCG2 c.421C>A, and SLCO1B1 g.37041T>C had statistically different RFS rates (p<0.05). However, there were no differences in RFS rates in the multivariate analysis.
 |
Table 4 Univariate and Multivariate Analysis of the Effects of Different Variables on P. vivax Relapse Outcome |
Discussion
This study investigated the frequencies of sequence variations in eleven drug-metabolizing enzymes contributing to phase I (CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4 and CYP3A5) and Phase II (COMT, SULT1A1, SULT1A4 and UGT2B7) metabolism of PQ and CQ as well as seven drug transporter genes (ABCB1, ABCA1, ABCC2, ABCC4, ABCG2, SLCO1B1 and SLC25A40) in 51 Thai patients infected with P. vivax. Although the influence of drug-metabolizing enzymes and drug transporters on the efficacy of PQ treatment on P. vivax infection has been studied,7,14,16,27 their contribution to relapse is still unclear. In vitro studies have shown that several phase I and II enzymes strongly relate with PQ metabolisms including CYP450 1A2, 2C19, 2C9, 2D6, 3A4, FMO-1, 3, 5 and MAO-A and B4–6 and CYP2C8, CYP3A4, and CYP2D6 with CQ metabolism.11 Therefore, genetic variation in these enzymes likely impacts the pharmacokinetics of both drugs to various degrees. However, to the best of our knowledge, no studies have assessed the association of pharmacogene variation and relapse other than CYP2D6 after treatment with PQ or a combination of PQ and CQ. It has been reported that CYP2D6 metabolizer status influences PQ efficacy.7,14 Bennette et al7 first described that two study participants (6%), one PM with a CYP2D6*5/*6 genotype and one IM with a CYP2D6*4/*41 genotype signifying considerable decreased activity, had multiple relapses of P. vivax while participants with genotypes predicting normal metabolizer (NM) status were not found to relapse. Furthermore, relapse patients had significantly higher amounts of the parent drug PQ AUCinf (p<0.001) compared with non-relapse patients. These data support their hypothesis that the highly polymorphic CYP2D6 gene contributed to the failure to bioactivate PQ to its active phenol metabolites that are responsible for killing hypnozoites. The clinical study by Nelwan et al28 demonstrated high efficacy of PQ against P. vivax relapse in Indonesia although relatively high relapse rates of 14% were observed among treatment groups while 75% of the patients in the control arm relapsed. Unfortunately, CYP2D6 genotyping was not performed in this study. Subsequent studies by Baird et al29 reported that decreased CYP2D6 activity was associated with increased risk of therapeutic failure. These authors reported that patients with CYP2D6 activity scores of 1.0 or less had a higher risk of relapse compared to patients with scores >1.0 (OR=9.4, P=0.001). Another study conducted by Spring et al30 confirmed that subjects with CYP2D6 IM or PM phenotypes have reduced PQ metabolism compared to those with an NM phenotype. In contrast, Chen et al31 found that CYP2D6 phenotype or activity scores were not significantly different between their relapse and non-relapse groups.
One explanation of why the present study did not find a relationship between CYP2D6 genotype or metabolizer status and PQ response may be due to the co-administration of CQ. Based on in-vitro studies, CQ is metabolized into N-desethylchloroquine by CYP2C8, CYP3A4, and CYP2D6. CYP2C8 and CYP3A4 constitute low-affinity, high-capacity systems while CYP2D6 may play a more important role at low CQ concentrations contributing to low-affinity, high-capacity systems11 of metabolism explaining why CQ may cause modest inhibition of CYP2D6 activity in humans when co-administered with debrisoquine.32 Thus, it cannot be excluded that CQ inhibits CYP2D6 activity to a certain extent and thereby reduce the enzyme’s capacity to efficiently bioactivate PQ. Furthermore, our study is limited by the small number of relapsed patients decreasing statistical power; small number size may also be a source of misclassification bias. Therefore, the findings of this study need to be viewed as preliminary. Although the patients were extensively genotyped for CYP2D6 and CNVs, we cannot exclude the possibility that the relapsing patients possess additional rare, or novel sequence variants and/or structural variants that decrease or obliterate CYP2D6 activity and drug response.33 We also like to stress that a lack of hypnozoites in the controls at the time of treatment initiation with PQ may have misclassified these patients as treatment successes, in fact over 22% of the relapse controls in clinical trial treated without PQ did not relapse.28,34 Thus, the recruitment of controls lacking hypnozoites would bias the statistical analysis for patients with decreased or no CYP2D6 activity. In addition, yet another explanation needs to be entertained, ie hypnozoite resistance to PQ,35,36 which may occur especially in some relapsing patients with normal and ultrarapid CYP2D6 activity.
Finally, there is sparse information, regarding the level of activity towards PQ that is conferred by alleles classified as decreased function alleles. For example, the CYP2D6*10 allele, which is the most common decreased function allele in Asians and has a frequency of 34.31% in our population, appears to have considerably less activity towards tamoxifen compared to the probe drug dextromethorphan. This observation triggered specific recommendations in the tamoxifen/CYP2D6 drug/gene pair guideline recently published by the Clinical Pharmacogenetic Implementation Consortium (CPIC) for patients carrying the CYP2D6*10 allele.37 It is not inconceivable that CYP2D6*10 and other decreased function alleles metabolize PQ and/or CQ at rates that are lower than expected from their current function classifications (see variation and functionality tables available at https://www.pharmgkb.org/page/pgxGeneRef) and put carriers at risk of relapse due to the failure of producing sufficiently high levels of exposure to the metabolites that are responsible for eradicating hypnozoites.
Regarding CYP2C19, the non-functional CYP2C19*2 allele (c.681G>A; rs4244285) was found at a statistically significantly higher allele frequency among the relapse patients (62.5%, 5/8) compared with 24.5% (23/94) among the non-relapse patients. The minor allele frequency was 26% in our study cohort making this a rather common non-functional allele in the Thai population.38–40 Although Pybus et al4 describe CYP2C19 as a minor contributing pathway for PQ metabolisms into its active metabolites, its contribution may assume a more prominent role in patients with compromised CYP2D6 activity. This is exemplified by one of our relapsing patients who was genotyped as CYP2D6*2/*10 and CYP2C19*2/*2. A composite activity score taking CYP2D6 and CYP2C19 into account may therefore be more informative to predict the risk of P. vivax relapse than either of these genes alone as suggested by our data (Table S3). The use of a combined metabolism index has been proposed previously by Villagra et al41 concluding that a
combinatory approach represents an improvement over the current gene-by-gene reporting by providing greater scope while still allowing for the resolution of a single-gene index when needed, especially when drugs are metabolized or activated by multiple pathways
which is clearly the case for the treatment with a combined PQ/CQ regimen.
This study is the first to examine the impact of variants in the ABCG2 efflux drug transporter on PQ response. This gene, also known as the Breast Cancer Resistance Protein (BRCP) is expressed in intestine, liver, kidney, placenta, and brain capillaries and plays an important role in the absorption, distribution, and removal of drugs across the cell membrane.42 This transporter is believed to play a protective role by blocking drug absorption at the apical membrane of the intestine and the blood–brain barrier among other sites. At the apical membranes of the liver and kidney, it facilitates efflux of xenobiotics lowering intracellular drug levels.43,44 Interestingly, although not significant, more subjects were heterozygous for ABCG2 c.421C/A (rs2231142) than homozygous for the reference allele (C/C; p=0.071). The mechanism of this variant, however, is unknown. ABCB1 is a member of the superfamily of ATP-binding cassette (ABC) transporters like ABCG2. Sortica et al16 reported that ABCB1 (MDR1, P-gp) T/nonG/T haplotype carriers (3435C>T, 2677G>A/T and 1236C>T) were associated with lower parasitemia clearance rates over treatment time in a model adjusted for other clinical factors; however, there was no longer statistically significant difference after false discovery rate analysis. The absence of an association between relapse and ABCG2 in this study may be due to the small number of relapse patients and/or inhibitory effects of CQ that seems to be an inhibitor to some ABC transporters.18 Two previous studies suggested that MAO-A/B enzyme activity relates with PQ metabolism,4,6 however, we did not test the relationships between MAO variants and RFS rates.
In addition to the study limitations mentioned above, we acknowledge that adherence was assessed retrospectively by reviewing a data registry. Although a risk of P. vivax reinfection could not be ruled out, the risk in our region is less than 5% during the 42-day period, assuming that most P. vivax infection recurrences were indeed relapses,45,46 and parasite genotyping for two polymorphic markers suggested a high probability of late relapsing infections in these volunteers.14 Nonetheless, further work is warranted to assess the impact of CYP2D6, CYP2C19, ABCG2 and potentially other drug-metabolizing enzymes and transporters on relapse in larger patient populations.
Conclusion
Although we did not find an association between CYP2D6 genotype and relapse, sequence variations in other pharmacogenes emerged as additional candidate genes that may contribute to variability in PQ/CQ drug response. These findings warrant further investigation, however, in larger study populations.
Abbreviations
PQ, Primaquine; CQ, Chloroquine; CYP, Cytochrome P450; ABCB1, ATP-Binding Cassette Subfamily B Member 1; ABCC2, ATP-Binding Cassette Subfamily C Member 2; MAO, Monoamine oxidase; ABCG2, ATP-Binding Cassette Subfamily G Member 2; COMT, catechol-O-methyltransferase; SLCO1B1, solute carrier organic anion transporter family member 1B1; SULT4A1, Sulfotransferase Family 4A Member 1; SLC25A40, Solute Carrier Family 25 Member 40, UGT2B7, UDP Glucuronosyltransferase Family 2 Member B7; RFS, Relapse-free survival; CPQ, Carboxy primaquine; OR, odds ratio; HR, hazards ratio; MALDI-TOF, matrix-assisted laser desorption ionization- time-of-flight.
Acknowledgments
This research is supported by Rachadapisek Sompote Fund for Postdoctoral Fellowship, Chulalongkorn University.
Disclosure
Rhea J Longley reports a patent PCT/US17/67926 pending. The authors declare that they have no other competing interests.
References
1. Krotoski WA, Krotoski DM, Garnham PC, et al. Relapses in primate malaria: discovery of two populations of exoerythrocytic stages. Preliminary note. Br Med J. 1980;280(6208):153–154. doi:10.1136/bmj.280.6208.153-a
2. Baird JK, Hoffman SL. Primaquine therapy for malaria. Clin Infect Dis. 2004;39(9):1336–1345. doi:10.1086/424663
3. Jacobson AR, Coffin SH, Shearson CM, Sayre LM. Beta, beta’-Iminodipropionitrile (IDPN) neurotoxicity: a mechanistic hypothesis for toxic activation. Mol Toxicol. 1987;1(1):17–34.
4. Pybus BS, Sousa JC, Jin X, et al. CYP450 phenotyping and accurate mass identification of metabolites of the 8-aminoquinoline, anti-malarial drug primaquine. Malar J. 2012;11:259. doi:10.1186/1475-2875-11-259
5. Pybus BS, Marcsisin SR, Jin X, et al. The metabolism of primaquine to its active metabolite is dependent on CYP 2D6. Malar J. 2013;12:212. doi:10.1186/1475-2875-12-212
6. Jin X, Pybus BS, Marcsisin R, et al. An LC-MS based study of the metabolic profile of primaquine, an 8-aminoquinoline antiparasitic drug, with an in vitro primary human hepatocyte culture model. Eur J Drug Metab Pharmacokinet. 2014;39(2):139–146. doi:10.1007/s13318-013-0139-8
7. Bennett JW, Pybus BS, Yadava A, et al. Primaquine failure and cytochrome P-450 2D6 in Plasmodium vivax malaria. N Engl J Med. 2013;369(14):1381–1382. doi:10.1056/NEJMc1301936
8. Baker JK, Yarber RH, Nanayakkara NP, McChesney JD, Homo F, Landau I. Effect of aliphatic side-chain substituents on the antimalarial activity and on the metabolism of primaquine studied using mitochondria and microsome preparations. Pharm Res. 1990;7(1):91–95. doi:10.1023/A:1015899928897
9. Cashman JR. Structural and catalytic properties of the mammalian flavin-containing monooxygenase. Chem Res Toxicol. 1995;8(2):166–181. doi:10.1021/tx00044a001
10. Potter BM, Xie LH, Vuong C, et al. Differential CYP 2D6 metabolism alters primaquine pharmacokinetics. Antimicrob Agents Chemother. 2015;59(4):2380–2387. doi:10.1128/AAC.00015-15
11. Projean D, Baune B, Farinotti R, et al. In vitro metabolism of chloroquine: identification of CYP2C8, CYP3A4, and CYP2D6 as the main isoforms catalyzing N-desethylchloroquine formation. Drug Metab Dispos. 2003;31(6):748–754. doi:10.1124/dmd.31.6.748
12. White NJ. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 2011;10:297. doi:10.1186/1475-2875-10-297
13. Eyles DE, Young MD. Studies on imported malarias; the parasitological pattern of relapsing Plasmodium vivax in military patients. J Natl Malar Soc. 1948;7(1):23–37.
14. Longley RJ, Sripoorote P, Chobson P, et al. High efficacy of primaquine treatment for Plasmodium vivax in Western Thailand. Am J Trop Med Hyg. 2016;95(5):1086–1089. doi:10.4269/ajtmh.16-0410
15. Jin X, Luong TL, Reese N, et al. Comparison of MDCK-MDR1 and Caco-2 cell based permeability assays for anti-malarial drug screening and drug investigations. J Pharmacol Toxicol Methods. 2014;70(2):188–194. doi:10.1016/j.vascn.2014.08.002
16. Sortica VA, Lindenau JD, Cunha MG, et al. SLCO1A2, SLCO1B1 and SLCO2B1 polymorphisms influences chloroquine and primaquine treatment in Plasmodium vivax malaria. Pharmacogenomics. 2017;18(15):1393–1400.
17. Wu CP, Klokouzas A, Hladky SB, Ambudkar SV, Barrand MA. Interactions of mefloquine with ABC proteins, MRP1 (ABCC1) and MRP4 (ABCC4) that are present in human red cell membranes. Biochem Pharmacol. 2005;70(4):500–510. doi:10.1016/j.bcp.2005.05.022
18. Hayeshi R, Masimirembwa C, Mukanganyama S, Ungell AL. The potential inhibitory effect of antiparasitic drugs and natural products on P-glycoprotein mediated efflux. Eur J Pharm Sci. 2006;29(1):70–81. doi:10.1016/j.ejps.2006.05.009
19. Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE. The pharmacogene variation (PharmVar) consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin Pharmacol Ther. 2018;103(3):399–401. doi:10.1002/cpt.910
20. Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Twist GP, Klein TE, Miller NA. The evolution of PharmVar. Clin Pharmacol Ther. 2019;105(1):29–32. doi:10.1002/cpt.1275
21. Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234–242. doi:10.1038/sj.clpt.6100406
22. Bell GC, Caudle KE, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther. 2017;102(2):213–218. doi:10.1002/cpt.v102.2
23. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2016;102:37–44.
24. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134. doi:10.1002/cpt.147
25. Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95(4):376–382. doi:10.1038/clpt.2013.254
26. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi:10.1093/bioinformatics/bth457
27. Fasinu PS, Tekwani BL, Nanayakkara NP, et al. Enantioselective metabolism of primaquine by human CYP2D6. Malar J. 2014;13:507. doi:10.1186/1475-2875-13-507
28. Nelwan EJ, Ekawati LL, Tjahjono B, et al. Randomized trial of primaquine hypnozoitocidal efficacy when administered with artemisinin-combined blood schizontocides for radical cure of Plasmodium vivax in Indonesia. BMC Med. 2015;13:294. doi:10.1186/s12916-015-0535-9
29. Baird JK, Louisa M, Noviyanti R, et al. Association of impaired cytochrome P450 2D6 activity genotype and phenotype with therapeutic efficacy of primaquine treatment for latent Plasmodium vivax malaria. JAMA Netw Open. 2018;1(4):e181449. doi:10.1001/jamanetworkopen.2018.1449
30. Spring MD, Sousa JC, Li Q, et al. Determination of cytochrome P450 isoenzyme 2D6 (CYP2D6) genotypes and pharmacogenomic impact on primaquine metabolism in an active-duty US military population. J Infect Dis. 2019;220:1761–1770. doi:10.1093/infdis/jiz386
31. Chen N, Dowd S, Gatton ML, Auliff A, Edstein MD, Cheng Q. Cytochrome P450 2D6 profiles and their relationship with outcomes of primaquine anti-relapse therapy in Australian Defence Force personnel deployed to Papua New Guinea and East Timor. Malar J. 2019;18(1):140. doi:10.1186/s12936-019-2774-2
32. Simooya OO, Sijumbil G, Lennard MS, Tucker GT. Halofantrine and chloroquine inhibit CYP2D6 activity in healthy Zambians. Br J Clin Pharmacol. 1998;45(3):315–317. doi:10.1046/j.1365-2125.1998.00671.x
33. Willyard C. Copy number variations’ effect on drug response still overlooked. Nat Med. 2015;21(3):206. doi:10.1038/nm0315-206
34. Sutanto I, Tjahjono B, Basri H, et al. Randomized, open-label trial of primaquine against vivax malaria relapse in Indonesia. Antimicrob Agents Chemother. 2013;57(3):1128–1135. doi:10.1128/AAC.01879-12
35. Spudick JM, Garcia LS, Graham DM, Haake DA. Diagnostic and therapeutic pitfalls associated with primaquine-tolerant Plasmodium vivax. J Clin Microbiol. 2005;43(2):978–981. doi:10.1128/JCM.43.2.978-981.2005
36. Langholz Kristensen K, Dragsted UB. Recurrent Plasmodium vivax malaria due to dose-dependent primaquine resistance: a case report. Scand J Infect Dis. 2014;46(1):63–65. doi:10.3109/00365548.2013.822093
37. Goetz MP, Sangkuhl K, Guchelaar HJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther. 2018;103(5):770–777. doi:10.1002/cpt.1007
38. Sukasem C, Tunthong R, Chamnanphon M, et al. CYP2C19 polymorphisms in the Thai population and the clinical response to clopidogrel in patients with atherothrombotic-risk factors. Pharmgenomics Pers Med. 2013;6:85–91. doi:10.2147/PGPM.S42332
39. Tassaneeyakul W, Mahatthanatrakul W, Niwatananun K, et al. CYP2C19 genetic polymorphism in Thai, Burmese and Karen populations. Drug Metab Pharmacokinet. 2006;21(4):286–290. doi:10.2133/dmpk.21.286
40. Chuwongwattana S, Jantararoungtong T, Chitasombat MN, et al. A prospective observational study of CYP2C19 polymorphisms and voriconazole plasma level in adult Thai patients with invasive aspergillosis. Drug Metab Pharmacokinet. 2016;31(2):117–122. doi:10.1016/j.dmpk.2015.12.005
41. Villagra D, Goethe J, Schwartz HI, et al. Novel drug metabolism indices for pharmacogenetic functional status based on combinatory genotyping of CYP2C9, CYP2C19 and CYP2D6 genes. Biomark Med. 2005;70(4):427–438. doi:10.2217/bmm.11.32
42. Russel FGM. Transporters: importance in drug absorption, distribution, and removal. In: Pang KS, Rodrigues AD, Peter RM, editors. Enzyme- and Transporter-Based Drug-Drug Interactions: Progress and Future Challenges. New York: Springer New York; 2010. 27–49.
43. Vlaming ML, Lagas JS, Schinkel AH. Physiological and pharmacological roles of ABCG2 (BCRP): recent findings in Abcg2 knockout mice. Adv Drug Deliv Rev. 2009;61(1):14–25. doi:10.1016/j.addr.2008.08.007
44. Huls M, Russel FG, Masereeuw R. The role of ATP binding cassette transporters in tissue defense and organ regeneration. J Pharmacol Exp Ther. 2009;328(1):3–9.
45. Luxemburger C, Thwai KL, White NJ, et al. The epidemiology of malaria in a Karen population on the western border of Thailand. Trans R Soc Trop Med Hyg. 1996;90(2):105–111. doi:10.1016/S0035-9203(96)90102-9
46. Price RN, Nosten F, Luxemburger C, et al. Artesunate versus artemether in combination with mefloquine for the treatment of multidrug-resistant falciparum malaria. Trans R Soc Trop Med Hyg. 1995;89(5):523–527. doi:10.1016/0035-9203(95)90094-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

