Back to Journals » OncoTargets and Therapy » Volume 12
Oral microbiota and gastrointestinal cancer
Authors Zhang Y, Niu Q, Fan W, Huang F , He H
Received 10 November 2018
Accepted for publication 12 March 2019
Published 18 June 2019 Volume 2019:12 Pages 4721—4728
DOI https://doi.org/10.2147/OTT.S194153
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Yangyang Zhang,1,2 Qiaoli Niu,2 Wenguo Fan,1 Fang Huang,1 Hongwen He1
1Guanghua School of Stomatology, Institute of Stomatological Research, Guangdong Provincial Key Laboratory of Stomatology, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2The Oral Medicine Clinical Center, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang Uygur Autonomous Region, People’s Republic of China
Abstract: The microbiota inhabiting the oral cavity is a complex ecosystem and responsible for resisting pathogens, maintaining homeostasis, and modulating the immune system. Some components of the oral microbiota contribute to the etiology of some oral diseases. Accumulating evidence suggests that the human oral microbiota is implicated in the development and progression of gastrointestinal cancer. In this review, we described the current understanding of possible roles and mechanisms of oral microbiota in the gastrointestinal cancers studied to date. The perspectives for oral microbiota as the biomarkers for early detection and new therapeutic targets were also discussed.
Keywords: oral microbiota, gastrointestinal cancer, etiology
Introduction
The oral microbiota is perhaps one of the most complex ecosystems in the body. Teeth, gingival sulcus, cheeks, palates, tongue, and tonsils are different oral habitats which are colonized by different oral microorganisms.1 The roles of these commensal microorganisms include resisting pathogens, maintaining homeostasis, and modulating the immune system,2 but they are responsible for a variety of oral diseases3 such as dental caries and periodontal diseases (the two common oral diseases) and oral cancer.4 Mounting evidence suggests that the microbiota may amplify or mitigate carcinogenesis, responsiveness to cancer therapeutics, and cancer-associated complications.5,6 A great deal of evidence indicates that oral microbiota plays important roles in gastrointestinal cancers. The aim of this review is to give an overview of oral microbiota and analyze different lines of evidence for the role of oral microbiota in gastrointestinal cancers studied to date. Possible mechanisms regarding the connection between oral microbiota and gastrointestinal cancers are discussed. The perspectives for future therapeutic and prophylactic modalities based on oral microbiota are also discussed.
Oral microbiota
The human mouth is heavily colonized by microorganisms, including viruses, protozoa, fungi, archaea, and bacteria.7 Up to now, there are 770 taxa in the expanded Human Oral Microbiota Database (eHOMD).1 Among them, 57% are cultivated and officially named, 13% are cultivated but unnamed, and 30% are known as uncultivated phylotypes.1 Bacteria, the most abundant taxonomic group of oral microbiota, have been deeply studied.8 The major phyla of oral bacteria comprise Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria.8 In the oral cavity, there are only a small number of archaea including Methanobrevibacter oralis, Methanobacterium curvum/congolese, and Methanosarcina mazeii.8 The prevalence of archaea seems to increase in patients with periodontitis and endodontic infections.8 And they may play a role in the oral mucosal diseases by favoring the growth of certain bacterial group.8 Fungi represent a small minority within oral microbiota,8 in which Candida species in the oral cavity may serve as a bridge between the oral mucosa and bacteria and present the potential capacities to modulate the oral microbiota.8 Oral protozoa, such as Entamoeba gingivalis and Trichomonas tenax, are usually nonpathogenic commensals.8 Food debris and bacteria are nutritional sources for these protozoa.8 Most oral viruses are bacteriophages which can regulate the microbial diversity and work as reservoirs of pathogenic gene function.8 In addition, there are also some other viruses, such as Herpes Simplex 1 or 2, Epstein–Barr virus, hepatitis A, B, and C, HIV, which are reported in saliva or oral swabs obtained from infected patients.8
Some components of the oral microbiota contribute to the etiology of several oral diseases. Oral microbiota-associated diseases including dental caries and periodontal diseases have been certainly caused by oral bacteria.2 Oral squamous cell carcinoma (OSCC) is the most common cancer of the oral cavity.2 Recently, there has been more and more evidence suggesting that oral bacteria may play a role in oral cancer.8 Some systemic diseases, such as cardiovascular diseases, adverse pregnancy outcomes, are also found to be linked with some oral pathogens.8 In addition, it has been suggested that the composition of the oral microbiota may be linked to carcinogenesis of distant organs, particularly the gastrointestinal tract.
Involvement of oral microbiota in gastrointestinal cancers
Increasing number of studies provide evidence that oral microorganisms are associated with gastrointestinal cancers,9–11 such as esophageal cancer, colorectal cancer (CRC), and pancreatic cancer.
Esophageal cancer
Esophageal cancer, affecting 456,000 people each year and leading to around 400,000 death per year, is the eighth most common cancer and the sixth most common cause of cancer-related death in the world.12,13 A recent study, a sub-study of a case–control study on upper gastrointestinal cancers, shows that there is a correlation between altered salivary bacterial microbiota and esophageal adenocarcinoma (ESCC) risk.9 Another study, a sub-study of a case–control study on upper gastrointestinal cancers, demonstrates that the periodontal pathogen Tannerella forsythia may be associated with higher risk of esophageal adenocarcinoma (EAC) and the abundance of the periodontal pathogen Porphyromonas gingivalis trended with higher risk of ESCC.12 Porphyromonas gingivalis is detected immunohistochemically in 61% of the cancerous tissues from ESCC patients, 12% of the adjacent tissues, and it is undetected in normal esophageal mucosa,14 similarly to what is observed by Yuan et al.15 Furthermore, the serum levels of IgG and IgA for Porphyromonas gingivalis are significantly higher in ESCC patients than healthy controls.16 The quantity of Streptococcus anginosus DNA is higher in esophageal cancer tissues than in oral cancer tissues.17 These results suggest that Streptococcus anginosus has a close relationship with esophageal cancer, but is not closely associated with oral cancer.17 Yamamura et al find that there are significantly more Fusobacterium nucleatum DNA in esophageal cancer tissues than matched normal mucosa.18 They also provide the evidence that Fusobacterium nucleatum DNA positivity is significantly associated with tumor stage and cancer-specific survival.18 Although these results indicate that oral microbiota may play an important role in esophageal cancer, case sample sizes remained small, limiting statistical power to detect significant associations. Thus, the causal relationship between them still needs to be clarified.
Colorectal cancer
As one of the most common gastrointestinal cancers worldwide, CRC is affecting 1.4 million people and responsible for approximately 610,000 deaths each year.13,19 The microbial community structure of the tongue coating is different between CRC patients and healthy people.10,20 CRC-associated oral bacteria, such as Peptostreptococcus, Parvimonas, and Fusobacterium are more abundant in CRC than in healthy controls.21 Fusobacterium nucleatum, a periodontal pathogen, has been found to be overabundant in CRC. Castellarin et al find that the abundance of Fusobacterium nucleatum DNA was 415 times greater in CRC tissues than adjacent normal tissues.22 There are several similar results that the abundance of Fusobacterium nucleatum are higher in CRC tissues than in normal mucosa.23–32 Furthermore, Fusobacterium nucleatum promoted CRC resistance to chemotherapy.33 Fusobacterium nucleatum in colorectal tissue could induce inflammatory response and promote CRC development.32–34 Introduction of Fusobacterium nucleatum to ApcMin/+mice results in accelerated small intestinal and colonic cancerogenesis.23 However, in a recent study, the difference in Fusobacterium nucleatum expression between CRC and adjacent normal tissues is not statistically significant, while the original study22 reported a significant increase in Fusobacterium nucleatum expression in CRC.35 Moreover, in some more recent studies, Fusobacterium nucleatum may play a role in the early stage of tumorigenesis.36,37 Other groups of oral bacteria, such as Porphyromonas, Peptostreptococcus,21,38 Prevotella, Parvimonas,21,38 and Gemella genera, are also found associated with colon cancer.39 Treponema denticola and Prevotella intermedia are associated with increased CRC risk.40 In addition, a metacommunity on gut mucosa predominated by oral microbiota is primarily related to CRC.6 The relationship between oral microbiota and CRC is deeply investigated. Although these observations show a relationship between oral microbiota and CRC, we still need larger studies to clarify the relationship and additional investigations are needed to determine the possible mechanisms.
Pancreatic cancer
Pancreatic cancer, a relatively less common cancer, is the fourth leading causes of cancer-related deaths.11,13,41,42 Poor oral hygiene plays an important role in the development of pancreatic cancer.43 The composition of the salivary microflora is significantly different between pancreatic cancer patients and healthy controls.11,44,45 Neisseria elongata and Streptococcus mitis in saliva can distinguish patients with pancreatic cancer from healthy subjects.46 In a prospective nested case–control study, including 361 pancreatic cancer cases and 371 matched controls, Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans in the oral cavity are positively correlated with pancreatic cancer.47 This study is large enough to provide evidence that the oral microbiota may play a role in the etiology of pancreatic cancer. The abundance ratio of Leptotrichia to Porphyromonas in the saliva of patients with pancreatic cancer is significantly higher than controls.48 The oral microbial diversity of pancreatic ductal adenocarcinoma (PDAC) patients does not differ from that of healthy controls, but the mean relative proportions of Firmicutes are higher in PDAC cases.49 Individuals who have high levels of antibodies against Porphyromonas gingivalis ATTC 53,978 are at higher risk of pancreatic cancer.50 Mitsuhashi et al find that Fusobacterium, an anaerobic, oral bacterium, can be detected in pancreatic cancer tissues and the presence of Fusobacterium colonization means shorter survival.51 But Yamamura et al report an inconsistent result that Fusobacterium nucleatum cannot be detected in pancreatic cancer tissues.52 Oral microbiota may serve as a non-invasive biomarker for early detection of pancreatic cancer, though better characterization of oral bacterial dysbiosis through disease course would be necessary. Because most of these studies are small sample sizes, much larger patient population studies are needed to verify their predictive utility.
Possible oral microbiota mechanisms in gastrointestinal cancers
There are several potential ways that bacteria may induce carcinogenesis (shown in Figure 1): induction of chronic inflammation, immune regulation, interference with signaling pathways and cell cycles, and local metabolism of carcinogens.3,53,54
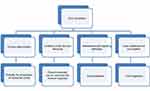 | Figure 1 Possible Oral microbiota mechanisms in gastrointestinal cancer.Abbreviation: CRC, colorectal cancer. |
Chronic inflammation
Chronic inflammation has been verified as the most important preventable cause of cancer.3,41,53,55–57 Almost 20% of the human malignancies can be related to infectious agents.57 On the one hand, chronic infection is believed to contribute to the initiation of several cancer types. Some inflammatory cytokines can activate oncogenes.53 On the other hand, inflammation can promote cancer progression and accelerate the process of invasion and metastasis.53,57 Chronic infection contributes to cancer progression by activating cancer-promoting signaling pathways which augment the production of anti-apoptotic proteins, growth factors, and cytokines that foster cancer growth and dissemination.57 The inflammation evoked by periodontitis could result in low-grade systemic inflammation.58 Porphyromonas gingivalis, and other oral bacteria, may have a significant role in the diseases of distant organs by causing inflammation and promoting tissue degenerative processes.59 Porphyromonas gingivalis can enhance local inflammation that contributes to carcinogenesis.59,60 Lipopolysaccharide (LPS) of Porphyromonas gingivalis can specifically activate host response through Toll-like receptors (TLRs), including TLR2 and TLR4, which can inhibit apoptosis and promote tumor growth.59 TLR signaling plays an important role in pancreatic tumors, thereby providing a potential mechanistic link between direct microbial stimulation of Porphyromonas gingivalis and pancreatic carcinogenesis.59 LPS and cell extracts of Fusobacterium nucleatum can increase inflammatory cytokines and chemokine and generate a proinflammatory microenvironment that promotes cancer progression.54 Therefore, one way that we can prevent CRC is to reduce periodontal pathogen including Porphyromonas gingivalis and Fusobacterium nucleatum by preventing and treating periodontitis.
Inhibition of the immune response
Fusobacterium nucleatum has been shown to expand myeloid-derived immune cells, which inhibit T-cell proliferation and induce T-cell apoptosis in CRC.30 Fusobacterium nucleatum can also protect tumor cells from NK-mediated killing and immune cell attack by the interaction between its Fap2 protein and the inhibitory immunoreceptor TIGIT (T cell immunoreceptor with Ig and ITIM domains) on natural killer (NK) and T cells.61 In addition, Porphyromonas gingivalis could also induce inhibition of the host’s immune system.60 These findings suggested that reducing Fusobacterium nucleatum and Porphyromonas gingivalis may reduce the inhibition of the immune response.
Interference with signaling pathways
Fusobacterium nucleatum can bind to both normal and cancerous epithelial cells via FadA binding to epithelial (E)-cadherin.62 But this binding can only lead to growth stimulation of human CRC cells.62,63 Furthermore, this binding activates β-catenin-regulated transcription, resulting in increased expression of oncogenes cyclin D1 and c-Myc, Wnt (wingless-related integration site) signaling genes Wnt7a, Wnt7b, and Wnt9a, and inflammatory genes nuclear factor-κB, interleukin-6 (IL-6), IL-8, and IL-18, all of which are responsible for carcinogenesis.62,63 Thus, we can block the oncogenic, Wnt, and inflammatory gene expression by preventing Fusobacterium nucleatum from binding and invasion of CRC cells.62
Local metabolism of carcinogens
Oral microbiota may affect gastrointestinal cancer risk by activating alcohol and smoking-related carcinogens.56 High salivary levels of acetaldehyde have been found in alcohol drinkers and smokers.64 This carcinogenic compound has been validated as a major causing factor in the upper digestive tract cancer.64 Although acetaldehyde can be produced from alcohol by mucosal alcohol dehydrogenases (ADH) in the upper digestive tract, much higher levels derive from the bacterial oxidation of alcohol by the oral microflora.64 Some species of oral microbiota have been shown to be capable of converting ethanol to acetaldehyde.55,65–67 Genus Neisseria has extremely high ADH activity.66 The ability of Neisseria to produce acetaldehyde is extremely higher than other genera. Furthermore, alcohol can increase the proportion of Neisseria in oral microflora. These findings suggest that this microbe can be a source of acetaldehyde and thus potentially play an important role in alcohol-related carcinogenesis.66 Candida glabrata (a non-Candida albicans species) which can produce carcinogenic amounts of acetaldehyde from both ethanol and glucose may be another source of acetaldehyde in oral cavity and gastrointestinal tract.3 In addition, oral bacteria may play a role in increased activation of carcinogenic nitrosamines from tobacco smoke.56,68 Smoking also contributes to alcohol–tobacco interactions in carcinogenesis by increasing the alcohol-related acetaldehyde production of oral bacteria.56 Taken together, these findings suggest that oral microbiota may play a role in oral and gastrointestinal carcinogenesis by local metabolism of alcohol and smoking-related carcinogens.3,56
Oral microbiota as a novel biomarker or therapeutic target
Studies of the relationship between oral microbiota and gastrointestinal cancer seem to be important not only for better understanding of cancer growth regulation but also for clinical practice. The mechanistic studies identified novel diagnostic and therapeutic targets. As a unique protein of Fusobacterium nucleatum, fadA may be an ideal diagnostic marker for early detection of CRC.62 The inhibitory peptide against fadA may be used to treat CRC or reduce CRC risk by specifically eradicating Fusobacterium nucleatum.62 Fusobacterial elimination might improve treatment outcome of CRC.69 There is a correlation between the shift of salivary bacterial microbiota and ESCC risk.9 Furthermore, the composition of tongue coating was different between gastrointestinal cancer patients and healthy people.9,10,20,70 These results suggested that the detection of samples derived from oral cavity may be a simple method to screen gastrointestinal cancer. Flemer et al assess the suitability of oral microbiota as a screening tool for identifying subjects with CRC and find that some oral microbiota operational taxonomic units (OTUs) can distinguish individuals with CRC from healthy controls.21 However, the causal relationship between oral microbiota and gastrointestinal cancer was not clear enough. The role and mechanisms that the oral microbiota involved in gastrointestinal cancers remain to be elucidated.
CRCs arise with genomic and epigenomic alterations through interactions between neoplastic cells, immune cells, and microbiota.71 Bacterial microbiota contribute to carcinogenesis in many ways, and the complete understanding of it will provide new ways for diagnosis, prevention, and treatment.72 Molecular pathological epidemiology (MPE) is an integrative transdisciplinary field that addresses heterogeneous effects of exogenous and endogenous factors, including microorganisms, on disease occurrence and consequence utilising molecular pathological signatures of the disease.73 It addresses etiologic heterogeneity according to subgroups of CRC classified by tumor tissue microbial profiling.74 Using this approach, we can examine how lifestyle factors, dietary patterns, medications, environmental exposures, and germline genetics influence cancer development and progression through impacting the microbial communities in the human body.73 Prudent diets rich in whole grains and dietary fiber are associated with a lower risk for Fusobacterium nucleatum-positive CRC but not Fusobacterium nucleatum-negative cancer, supporting a potential role for intestinal microbiota in mediating the association between diet and colorectal neoplasms.75 Dietary interventions could be useful for cancer prevention and precision medicine.74 MPE research combined with oral microbiota analyses might play a role in providing rationales and discovering insights into precision medicine for gastrointestinal cancer.
Conclusion
Oral microbiota may play an important role in different gastrointestinal cancers. Validating the association of the oral microbiota with gastrointestinal cancers may lead to significant advances in understanding the etiology of gastrointestinal cancers. Some species of oral microbiota or the shift of the oral ecosystem may also serve as readily accessible, noninvasive biomarkers for the identification of high risk for gastrointestinal cancers. Therefore, a comprehensive understanding of the underlying mechanisms will be necessary for the prevention and/or treatment of gastrointestinal cancers.
Acknowledgments
This work was supported partly by the National Natural Science Foundation of China (No. 81470760).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. In J Bacteriol. 2010;192(19):5002–5017. doi:10.1128/JB.00542-10
2. Zhang Y, Wang X, Li H, Ni C, Du Z, Yan F. Human oral microbiota and its modulation for oral health. Biomed Pharmacother. 2018;99:883–893. doi:10.1016/j.biopha.2018.01.146
3. Srinivasprasad V, Dineshshankar J, Sathiyajeeva J, Karthikeyan M, Sunitha J, Ragunathan R. Liaison between micro-organisms and oral cancer. J Pharm Bioallied Sci. 2015;7(Suppl 2):S354–S360. doi:10.4103/0975-7406.163451
4. Meurman JH. Oral microbiota and cancer. J Oral Microbiol. 2010;2. doi:10.3402/jom.v2i0.5195
5. Thomas LV, Suzuki K, Zhao J. Probiotics: a proactive approach to health. A symposium report. Br J Nutr. 2015;114(Suppl 1):S1–S15. doi:10.1017/S0007114515004043
6. Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi:10.1038/ncomms9727
7. Nieboer P, Roodenburg JL, van der Laan BF, de Vries EG, Mulder NH. van der Graaf WT. Screening for infectious foci in breast cancer patients prior to high-dose chemotherapy and stem cell transplantation. Anticancer Res. 2003;23(2c):1779–1783.
8. Sampaio-Maia B, Caldas IM, Pereira ML, Perez-Mongiovi D, Araujo R. The oral microbiome in health and its implication in oral and systemic diseases. Adv Appl Microbiol. 2016;97:171–210. doi:10.1016/bs.aambs.2016.08.002
9. Chen X, Winckler B, Lu M, et al. oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS One. 2015;10(12):e0143603. doi:10.1371/journal.pone.0143603
10. Han S, Yang X, Qi Q, et al. Potential screening and early diagnosis method for cancer: tongue diagnosis. Int J Oncol. 2016;48(6):2257–2264. doi:10.3892/ijo.2016.3466
11. Garcia-Castillo V, Sanhueza E, McNerney E, Onate SA, Garcia A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65(12):1347–1362. doi:10.1099/jmm.0.000371
12. Peters BA, Wu J, Pei Z, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77(23):6777. doi:10.1158/0008-5472.CAN-17-1296
13. Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: the current state and clinical perspectives. Semin Cancer Biol. 2018;51:36–49.
14. Gao S, Li S, Ma Z, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer. 2016;11:3. doi:10.1186/s13027-016-0049-x
15. Yuan X, Liu Y, Kong J, et al. Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. 2017;404:1–7. doi:10.1016/j.canlet.2017.07.003
16. Gao SG, Yang JQ, Ma ZK, et al. Preoperative serum immunoglobulin G and A antibodies to Porphyromonas gingivalis are potential serum biomarkers for the diagnosis and prognosis of esophageal squamous cell carcinoma. BMC Cancer. 2018;18(1):17. doi:10.1186/s12885-018-4242-8
17. Morita E, Narikiyo M, Yano A, et al. Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer Sci. 2010;94(6):492–496. doi:10.1111/cas.2003.94.issue-6g
18. Yamamura K, Baba Y, Nakagawa S, et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22(22):5574–5581. doi:10.1158/1078-0432.CCR-16-1786
19. Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Junior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46(4):1135–1140. doi:10.1590/S1517-838246420140665
20. Han S, Chen Y, Hu J, Ji Z. Tongue images and tongue coating microbiome in patients with colorectal cancer. Microb Pathog. 2014;77:1–6. doi:10.1016/j.micpath.2014.10.003
21. Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018:67(8):1454–1463.
22. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi:10.1101/gr.126516.111
23. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi:10.1016/j.chom.2013.07.007
24. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi:10.1101/gr.126573.111
25. Mccoy AN, Araújopérez F, Azcárateperil A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PLoS One. 2013;8(1):e53653. doi:10.1371/journal.pone.0053653
26. Warren RL, Freeman DJ, Pleasance S, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1(1):16. doi:10.1186/2049-2618-1-16
27. Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–1318. doi:10.1158/0008-5472.CAN-13-1865
28. Mira-Pascual L, Cabrera-Rubio R, Ocon S, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50(2):167–179. doi:10.1007/s00535-014-0963-x
29. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi:10.1016/j.chom.2013.07.012
30. Nosho K, Sukawa Y, Adachi Y, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22(2):557–566. doi:10.3748/wjg.v22.i2.557
31. Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33(8):1381–1390. doi:10.1007/s10096-014-2081-3
32. Li YY, Ge QX, Cao J, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22(11):3227–3233. doi:10.3748/wjg.v22.i11.3227
33. Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappab, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–866.e824. doi:10.1053/j.gastro.2016.11.018
34. Ye X, Wang R, Bhattacharya R, et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res. 2017;10(7):398–409. doi:10.1158/1940-6207.CAPR-16-0178
35. Repass J, Iorns E, Denis A, et al. Replication study: fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Elife. 2018;7. doi:10.7554/eLife.42270
36. Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137(6):1258–1268. doi:10.1002/ijc.29488
37. Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci Rep. 2016;6:25271. doi:10.1038/srep25271
38. Drewes JL, White JR, Dejea CM, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017;3:34. doi:10.1038/s41522-017-0040-3
39. Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: evidence of association and causality. Semin Immunol. 2017;32:25–34. doi:10.1016/j.smim.2017.08.001
40. Yang Y, Cai Q, Shu XO, et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer. 2018;78(1):176–181.
41. Jia G, Zhi A, Lai PFH, et al. The oral microbiota - a mechanistic role for systemic diseases. Br Dent J. 2018;224(6):447–455. doi:10.1038/sj.bdj.2018.217
42. Stolzenberg-Solomon RZ, Dodd KW, Blaser MJ, Virtamo J, Taylor PR, Albanes D. Tooth loss, pancreatic cancer, and Helicobacter pylori. Am J Clin Nutr. 2003;78(1):176–181. doi:10.1093/ajcn/78.1.176
43. Huang J, Roosaar A, Axéll T, Ye W. A prospective cohort study on poor oral hygiene and pancreatic cancer risk. Int J Cancer. 2016;138(2):340–347. doi:10.1002/ijc.29710
44. Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35(2):249–255. doi:10.1093/carcin/bgt392
45. Ertz-Archambault N, Keim P, Von Hoff D. Microbiome and pancreatic cancer: A comprehensive topic review of literature. World J Gastroenterol. 2017;23(10):1899–1908. doi:10.3748/wjg.v23.i10.1899
46. Anderson RS. Risks. J Miss State Med Assoc. 2008;49(12):385–386.
47. Fan X, Alekseyenko AV, Wu J, Peters BA. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–127. doi:10.1136/gutjnl-2016-312580
48. Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. doi:10.7717/peerj.1373
49. Olson SH, Satagopan J, Xu Y, et al. The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: a pilot study. Cancer Causes Control. 2017;28(9):959–969. doi:10.1007/s10552-017-0933-8
50. Michaud DS, Izard J, Wilhelm-Benartzi CS, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62(12):1764–1770. doi:10.1136/gutjnl-2012-303006
51. Mitsuhashi K, Nosho K, Sukawa Y, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6(9):7209–7220. doi:10.18632/oncotarget.3109
52. Yamamura K, Baba Y, Miyake K, et al. Fusobacterium nucleatum in gastroenterological cancer: evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol Lett. 2017;14(6):6373–6378. doi:10.3892/ol.2017.7001
53. Meng C, Bai C, Brown TD, Hood LE, Human Gut TQ. Microbiota and gastrointestinal cancer. Genomics Proteomics Bioinformatics. 2018;16(1):33–49. doi:10.1016/j.gpb.2017.06.002
54. Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother. 2017;89:918–925. doi:10.1016/j.biopha.2017.02.102
55. Meurman JH. Infectious and dietary risk factors of oral cancer. Oral Oncol. 2010;46(6):411–413. doi:10.1016/j.oraloncology.2010.03.003
56. Kiyabu MT, Shibata D, Arnheim N, Martin WJ, Fitzgibbons PL. Detection of human papillomavirus in formalin-fixed, invasive squamous carcinomas using the polymerase chain reaction. Am J Surg Pathol. 1989;13(3):221–224.
57. Binder GA, Stuart F, Brurya R, et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatumpromote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6(26):22613–22623. doi:10.18632/oncotarget.4209
58. Lira-Junior R, Figueredo CM. Periodontal and inflammatory bowel diseases: is there evidence of complex pathogenic interactions? World J Gastroenterol. 2016;22(35):7963–7972. doi:10.3748/wjg.v22.i35.7963
59. Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34(10):2193–2197. doi:10.1093/carcin/bgt249
60. Le Bars P, Matamoros S, Montassier E, et al. The oral cavity microbiota: between health, oral disease, and cancers of the aerodigestive tract. Can J Microbiol. 2017;63(6):475–492. doi:10.1139/cjm-2016-0603
61. Colucci F. An oral commensal associates with disease: chicken, egg, or red herring? Immunity. 2015;42(2):208–210. doi:10.1016/j.immuni.2015.01.024
62. Han YW. Oral bacteria as drivers for colorectal cancer. J Periodontol. 2014;85(9):1155–1157. doi:10.1902/jop.2014.140039
63. Lam SY, Yu J, Wong SH, Peppelenbosch MP, Fuhler GM. The gastrointestinal microbiota and its role in oncogenesis. Best Pract Res Clin Gastroenterol. 2017;31(6):607–618. doi:10.1016/j.bpg.2017.09.010
64. Rota MT, Poggi P. Reduction of oral acetaldehyde levels using a controlled-release chlorhexidine chip as a prevention strategy against upper digestive tract cancer. Med Hypotheses. 2003;60(6):856–858.
65. Marttila E, Uittamo J, Rusanen P, Lindqvist C, Salaspuro M, Rautemaa R. Acetaldehyde production and microbial colonization in oral squamous cell carcinoma and oral lichenoid disease. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;116(1):61–68. doi:10.1016/j.oooo.2013.02.009
66. Muto M, Hitomi Y, Ohtsu A, et al. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: implications for carcinogenesis in upper aerodigestive tract. Int J Cancer. 2015;88(3):342–350. doi:10.1002/1097-0215(20001101)88:3<342::AID-IJC4>3.0.CO;2-I
67. Zhang M, Fan X, Fang B, Zhu C, Zhu J, Ren F. Effects of Lactobacillus salivarius Ren on cancer prevention and intestinal microbiota in 1, 2-dimethylhydrazine-induced rat model. J Microbiol. 2015;53(6):398–405. doi:10.1007/s12275-015-5046-z
68. Yang L, Ganly I, Morris L, et al. Relevance of microbiome to cigarette smoking and oral cancer. Proc Iadr General Session. 2011.
69. Abed J, Maalouf N, Parhi L, Chaushu S, Mandelboim O, Bachrach G. Tumor targeting by fusobacterium nucleatum: a pilot study and future perspectives. Front Cell Infect Microbiol. 2017;7:295. doi:10.3389/fcimb.2017.00517
70. Hu J, Han S, Chen Y, Ji Z. variations of tongue coating microbiota in patients with gastric cancer. Biomed Res Int. 2015;2015:173729. doi:10.1155/2015/173729
71. Mima K, Nowak JA, Qian ZR, et al. Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget. 2016;7(34):55098–55109. doi:10.18632/oncotarget.10398
72. Rajpoot M, Sharma AK, Sharma A, Gupta GK. Understanding the microbiome: emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol. 2018;52(Pt 1):1–8. doi:10.1016/j.semcancer.2018.02.003
73. Hamada T, Nowak JA, Milner DA
74. Kosumi K, Mima K, Baba H, Ogino S. Dysbiosis of the gut microbiota and colorectal cancer: the key target of molecular pathological epidemiology. J Lab Precis Med. 2018;3. doi:10.21037/jlpm.2018.09.05
75. Mehta RS, Nishihara R, Cao Y, et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017;3(7):921–927. doi:10.1001/jamaoncol.2016.6374
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
