Back to Journals » Cancer Management and Research » Volume 12
Oncogenic Genetic Alterations in Non-Small-Cell Lung Cancer (NSCLC) in Southwestern China
Authors Ma Y, Li Q, Du Y, Chen W, Zhao G, Liu X, Li H, Liu J, Shen Z, Ma L, Zhou Y
Received 6 June 2020
Accepted for publication 1 October 2020
Published 29 October 2020 Volume 2020:12 Pages 10861—10874
DOI https://doi.org/10.2147/CMAR.S266069
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Yuhui Ma,1,* Quan Li,2,* Yaxi Du,2,* Wanlin Chen,1 Guanqiang Zhao,1 Xing Liu,2 Hongsheng Li,3 Junxi Liu,3 Zhenghai Shen,4 Luyao Ma,2 Yongchun Zhou4
1Department of Thoracic Surgery I, The Third Affiliated Hospital of Kunming Medical University (Yunnan Cancer Hospital), Kunming 650118, People’s Republic of China; 2Key Laboratory of Lung Cancer Research of Yunnan Province, The Third Affiliated Hospital of Kunming Medical University, Kunming 650118, People’s Republic of China; 3International Joint Laboratory on High Altitude Regional Cancer of Yunnan Province, The Third Affiliated Hospital of Kunming Medical University, Kunming 650118, People’s Republic of China; 4Yunnan Cancer Center, The Third Affiliated Hospital of Kunming Medical University, Kunming 650118, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongchun Zhou
Yunnan Cancer Center, The Third Affiliated Hospital of Kunming Medical University, Kunming 650118, People’s Republic of China
Tel/Fax +86-087168172772
Email [email protected]
Purpose: To investigate the impact of oncogenic genetic alterations (GAs) on non-small-cell lung cancer (NSCLC) in southwestern China.
Patients and Methods: We first collected 579 pathologically confirmed NSCLC specimens and then used next-generation sequencing (NGS) to evaluate the DNA samples for GAs. Both the tissue and plasma samples were provided by 28 patients. Furthermore, subgroup analyses based on sample type, concordance, and GA type were carried out.
Results: GAs were detected by NGS in 61.8% (358/579) of patients. Two hundred and twenty-nine patients (39.6%) harbored EGFR mutations, 63 (10.9%) harbored KRAS mutations, 13 (2.2%) harbored BRAF mutations, 30 (5.18%) harbored ALK fusions, and 13 (2.2%) had ROS1 fusions. We found that females (p < 0.01), nonsmokers (p < 0.001), adenocarcinoma (p < 0.001), and tissue (p = 0.03) had a relatively high EGFR mutation rate. Notably, NSCLC patients from Xuanwei had a significantly different mutational pattern for EGFR in comparison with that of non-Xuanwei patients (higher G719X + S768I mutations and multiple gene alterations, but fewer exon 19 deletion mutations and single gene alterations). We found that adenocarcinoma (p = 0.02), family history of malignancy (p = 0.03), Xuanwei origin (p < 0.001), and tissue (p = 0.04) were associated with a higher number of KRAS mutations. Subgroup analysis showed that ALK (p < 0.001) and ROS1 (p < 0.05) fusions and rare EGFR mutations (p < 0.001) were associated with non-Han ethnic patients.
Conclusion: Yunnan NSCLC patients from Xuanwei and non-Han ethnic patients had an obviously unique prevalence of GAs.
Keywords: non-small-cell lung cancer, oncogenic genetic alteration, next-generation sequencing
Introduction
Lung cancer is the most pervasive malignancy and causes high mortality worldwide. The most commonly diagnosed cancers in China, in 2018, were lung cancers (774,323 new cases with an incidence of 35.10/100,000) and they were the dominant causes of cancer-related death (690,567 deaths were estimated) in 2018.1 Studies have shown that non-small-cell lung cancer (NSCLC) is the most common subtype of lung cancers.2 The treatment strategy of NSCLC has been revolutionized for the development of molecularly targeted therapy, in genomically defined subsets of patients. The identification of epidermal growth factor receptor (EGFR) mutations3 and anaplastic lymphoma kinase (ALK) fusions4 are classic examples of oncogenic genetic alterations (GAs). Two of these GAs are sensitivity to matched targeted therapies. The subsequent discovery of Ret proto-oncogene (RET) and ROS proto-oncogene 1 receptor tyrosine kinase (ROS1) translocations as potentially treatable targets suggested that several chromosomal translocations and corresponding gene fusions may serve as a driving force for NSCLC.5,6 The National Comprehensive Cancer Network (NCCN) guidelines recommend biomarker testing for GAs, including EGFR, ALK, B-raf proto-oncogene, serine/threonine kinase (BRAF), RET, v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), ROS1, and human epidermal growth factor receptor 2 (HER2).7,8
Yunnan province, with an average altitude of 2000 m, is located in the Yunnan-Guizhou Plateau in southwestern China. Xuanwei, a city of Yunnan region, is 6257 km2 and lies on a highland punctuated by mountain ridges. The total population of Yunnan province is approximately one million people. The rate of tobacco smoking is more than 40% for males but less than 0.1% for females in this region. Xuanwei has some local companies, engaging in industries including coal mining and electric power generation. However, over 90% of the residents are peasants. Therefore, industrial pollution is not a main factor causing lung cancers in this area. Lung cancers have been characterized in Xuanwei city: (1) Relatively high incidence and mortality rates of lung cancers in females who did not smoke; (2) In females, adenocarcinoma is the major type of lung cancer.9,10 Former studies have indicated that the lung cancer rate in Xuanwei area of Yunnan region is higher than that in other places of China. Indoor air pollution caused by polycyclic aromatic hydrocarbon (PAH)-containing smoky coal emissions is the primary reason for the high incidence and mortality of lung cancers.11,12 In Xuanwei city, the mortality rate of some regions has started to decrease; However, the total mortality rate of Xuanwei city remains high or even elevated.9 According to this result, we assume that oncogenic factors are vital to the development of lung cancers in Xuanwei City. Based on a former study, we observed that the prevalence of (EGFR) mutation in Xuanwei was different from that in other populations (higher G719X, G719X+S768I, fewer exon 19 deletions and L858R mutations).13 Therefore, it is important for us to determine whether the oncogenic GAs have local specificity in Xuanwei, as lung cancers have become a major health issue.
Currently, tumor tissue is the gold standard for the detection of oncogenic GAs, and these are usually acquired via surgery or biopsy. However, approximately 70% of NSCLC cases are diagnosed at an advanced stage and became ineligible to receive operation. Additionally, it is not feasible to execute repetitive biopsy, which can cause discomfort to NSCLC patients with recurrent cancer or acquired resistance to EGFR tyrosine kinase inhibitors (TKIs). Therefore, researches have confirmed that as a repeat biopsy of tumor tissue, noninvasive detection of oncogenic GAs in plasma circulating tumor DNA (ctDNA) is a viable method. Moreover, this method provides the similar genetic information as a tissue biopsy. During the course of therapy, it can be performed at any time, and dynamic monitoring of molecular changes is possible.14,15 Thus, the prediction of the clinical outcome of EGFR-TKI treatment and the guide for further treatment of advanced NSCLC patients could be reinforced by continuous monitoring of oncogenic GA dynamics in plasma ctDNA. Here, we use next-generation sequencing (NGS), which is an advanced method with efficient and economical detection of multiple genetic variations simultaneously and oncogenic GAs. High consistency between NGS and qPCR was verified and confirmed by Sanger sequencing, which indicates the clinical application prospects of NGS.16,17
Our center has had established oncogenic GA testing facilities (by NGS) since 2016; the data acquired from these methods can provide guiding information about local patients with NSCLC. The oncogenic GA status in NSCLC patients in Yunnan province is unclear, so we first revealed the oncogenic GAs of patients with NSCLC in Yunnan province and next explored the significance of NSCLC patients’ oncogenic GAs and advanced the effective guidance of clinical treatment.
Patients and Methods
Study Population and Samples
For the primary study population, we recruited NSCLC patients undergoing routine tumor genotyping from the Third Affiliated Hospital of Kunming Medical University between November 2016 and June 2019. All included individuals provided written informed consent. The study was approved by the ethics committee of the Third Affiliated Hospital of Kunming Medical University, complying with the Declaration of Helsinki.
Enrolment criteria are 1) adult (>18 years) residents of Yunnan region and 2) eligible individuals with pathologically confirmed NSCLC and sufficient tissue or plasma for GA analysis.
Exclusive criteria are as follows: any specimens with possible contamination.
In total, 751 specimens from NSCLC patients were collected, but only 579 patient specimens were involved in this research for various reasons (such as substandard quality or incomplete information of some patients). The patient data were subgrouped according to age, sex, TNM stage, smoking history, family history of malignancy, histology, ethnicity, sample type, demographic, and Xuanwei origin.
DNA Extraction
Tissue samples were collected from surgical resection specimens and biopsy specimens, and cytology was performed on pleural fluids. The AmoyDx tissue/pleural fluid DNA Kit (Amoy Diagnostics, Xiamen, China) was used to extract the tissue and cytological DNA, based on the manufacturer’s instructions.18 The patients’ blood samples were collected in tubes with 10 mL EDTA anticoagulant and centrifuged at 2000 g and 8000 g for 10 min at 4°C within 2 hrs. The plasma supernatant was isolated, and DNA was immediately obtained. The Circulating DNA Kit (AmoyDx, Xiamen, China) was used to extract DNA from the plasma supernatant samples according to the manufacturer’s instructions.18
GA Analysis by NGS
For the NGS assay, the AmoyDx Essential NGS panel (reversible terminator sequencing) was used for qualitative detection of single nucleotide variants, copy number variants, indels, and gene rearrangements in the lung panel (including EGFR, KRAS, RET, ROS1, ALK, NRAS, BRAF, MET, PIK3CA, and HER2) in NSCLC in accordance with the manufacturer’s instructions.16,17 Each result was qualified only when the sample coverage was more than 98%, the average original depth (Mean Depth) was more than 10,000× and the average effective depth (SSBCDepth) was more than 500×. The threshold of mutation frequency for mutation was 0.4%. The mutant copy number (DSBCDepth) was not less than 2.
Statistical Analysis
The correlation between GAs and clinical factors were analyzed by χ2 tests. Statistical data were used to calculate sample concordance regarding sensitivity, specificity, negative predictive value (NPV) and positive predictive value (PPV). All the statistics were performed by using SPSS 19.0 software (SPSS IBM Corp., Armonk, NY, USA); two-side, p < 0.05 was considered statistically significant.
Results
Patient Characteristics
There were 579 consecutive patients, including 246 (42.5%) females and 333 (57.5%) males, enrolled in our study. These patients were between ages 29 and 89, with a median age of 58 years. The most common histology was adenocarcinoma (n=524, 90.5%), and the others were squamous carcinoma (n=46, 8.0%), large cell carcinoma (n=1, 0.2%), and other kinds of NSCLC (n=8, 1.3%). Additionally, 62.3% of the patients were diagnosed with stage IV disease. Half of the patients (61.2%, 354 of 579) were non-smokers. The difference in smoking status is based on sex and pathological type. Patients who were males (p < 0.01) or who had squamous cell carcinoma (p < 0.05) appeared to be more likely to smoke. Twenty-eight (4.8%) patients had a family history of malignancy, and 119 (20.6%) patients were from Xuanwei city. Of the 36 young patients diagnosed with adenocarcinoma at age <40 years, 18 (50.0%) were women, with a median age of 38 years (range 23–40 years), and 21 (58.4%) never smoked. In the patient population, Stage IV cancer was more frequent in younger group than in older age (age > 40 years old, p = 0.02). However, there were no significant differences in the characteristics, such as sex distribution, histology, smoking, malignant family history or Xuanwei origin, between the young and the elder groups. Table 1 summarizes the primary patients’ characteristics.
 |
Table 1 Characteristics of 579 Patients with NSCLC |
Prevalence of Driver Alterations in Patients
The NGS assay detected GAs in 61.8% (358/579) of the patients. Through NGS genotyping, we identified a wide range of tumor-related changes (Figure 1). In addition, we identified driver mutations in 96.0% (344/358) of the patients; these mutations included common changes such as EGFR mutations, secondary EGFR resistance alterations (20 insertions, MET amplification and T790M), rare mutations in EGFR, ALK fusion, MET exon 14 skip change, BRAF alteration, ROS1 fusion and RET translocation, and nontransformable changes such as KRAS, HER2 and PIK3CA mutations (Figure 2).
 |
Figure 1 Plot of all alterations detected by next-generation sequencing (n =579). |
 |
Figure 2 Genetic driver alteration identified by next-generation sequencing (n = 358). |
Incidence of EGFR, KRAS, ALK, ROS1 and BRAF Alterations and Their Association with Demographic and Clinical Factors
The frequency of EGFR mutations in Yunnan patients with NSCLC and their relationship with clinicopathological parameters are similar to those in other East Asian countries. We detected EGFR mutations in 229 (39.6%) NSCLC patients. The observed differences in EGFR mutation rates were based on smoking situation, sex, pathological type, and sample type. Females (p < 0.01), never-smokers (p < 0.001), adenocarcinoma status (p < 0.001), and tissue type (p < 0.03) appeared to be associated with higher EGFR mutations. We detected KRAS mutations in 63 (10.9%) NSCLC patients. The differences in KRAS mutation rates were based on pathology type, TNM stage, family history of malignancy, region distribution, Xuanwei origin and sample type. It seemed that adenocarcinoma (p = 0.02), family history of malignancy (p = 0.03), northeast region (p < 0.001), Xuanwei origin (p < 0.001) and tissue as the specimen source (p < 0.04) were associated with a high frequency of KRAS mutations. Table 2 summarizes the incidence of EGFR, KRAS, ALK, ROS1 and BRAF alterations and their association with demographic and clinical factors.
 |
Table 2 Frequency of EGFR, KRAS, ALK, ROS1 and BRAF Alterations According to Clinical Characteristics |
ALK fusions are identified in 5.1% (30/579) of patient samples. The difference in the rate of ALK fusions is associated with patient age ≤ 40 years (p < 0.001) and non-Han ethnicity (p < 0.001). It seems that younger non-Han patients have a high likelihood of ALK fusions.
We detected ROS1 fusions in 13 (2.2%) of patient samples. The differences in the rate of ROS1 fusions were associated with non-Han ethnicity (p < 0.05). It seemed that non-Han ethnic patients had a high likelihood of ROS1 fusions.
BRAF mutations were identified in 2.2% (13/579) of patient samples. Differences in the rate of BRAF mutations were associated with the sex of the patient (p = 0.01). It seemed that being female was associated with a high likelihood of BRAF mutations.
Prevalence of HER2, RET, PIK3CA, MET, NRAS and ERBB2 Alterations in Patients with NSCLC
Figure 1 indicates that 24 patients have HER2, PIK3CA, NRAS, ERBB2, MET and RET alterations. We identified HER2 mutations in 5 patients and PIK3CA mutations in 2 patients. One patient carried PIK3CA mutations plus EGFR L858R and KRAS mutations. NRAS mutations were identified in 3 patients. Two patients carried NRAS mutations plus EGFR L858R, EGFR T790M and MET exon 14 skipping alterations. ERBB2 mutations were identified in 1 patient. MET exon 14 skipping alterations were identified in 6 patients. Two patients carried MET exon 14 skipping alterations plus EGFR L858R, EGFR T790M and NRAS mutations. RET alterations were identified in 7 patients. Seven patients carried RET translocations, and one patient carried RET translocations plus EGFR G719X and EGFR S768I mutations. One patient carried RET translocations plus the EGFR G719X mutation and ALK fusion.
Tissue and Plasma Concordance
We provided plasma-based NGS to 28 patients who received tissue NGS genotyping at the center to assess tissue and plasma consistency. They are part of the 579 samples. GAs were detected in 24 (82.7%) samples, of which three harbored the EGFR exon 19 deletion, six harbored EGFR L858R, one harbored EGFR G719X, one harbored EGFR L861Q, two harbored EGFR G719X plus S768I, one harbored G719X plus L861Q, three harbored KRAS mutations, one harbored BRAF mutation, two harbored ALK fusions, three harbored ROS1 fusions, and one harbored RET translocations. Some GAs (EGFR, ALK, ROS1, KRAS, MET, RET, BRAF and HER2) have been recognized among patients who were with plasma NGS-positive under the NCCN guidelines.18 The oncogenic driver alteration statuses of matched tissue and plasma are concordant for 21 patients (Table 3). Three inconsistent cases include KRAS mutations found in tissue NGS that were not identified in plasma NGS, RET translocations found in tissues that were not identified by plasma NGS, and KRAS mutations found in plasma NGS that were not identified by tissue NGS. The sensitivity of plasma-based ctDNA NGS is 91.3% (21/23), the specificity is 80.0% (4/5), the PPV is 95.45% (21/22), and the NPV is 66.7% (4/6).
 |
Table 3 GAs Rates in Patients Who Provided Both Samples |
GA Status Among Patients with NSCLC
The EGFR mutation pattern in Yunnan region is consistent with that in other East Asian populations. In the Xuanwei subgroup, we found that the prevalence of EGFR mutation differs from that of the other population (G719X + S768I was higher, but exon 19 deletion mutations were lower) (multiple EGFR mutations were higher, but a single EGFR mutation was lower). Overall, we detected GAs in 358 patients, and EGFR mutations in 229 patients. The most common EGFR mutations are the exon 19 deletion and the L858R point mutation observed in 67 (29.2%) and 60 (26.2%), respectively. A single EGFR mutation was observed in 146 patients (63.7%), an EGFR combined mutation was detected in 57 patients (24.9%), multiple gene alterations (EGFR mutation plus other GAs) were found in 7 patients (3.1%), and a rare EGFR mutation was detected in 19 patients (8.3%). Among 229 patients, 145 patients (63.3%) had sensitizing mutations, 7 patients (3.0%) had resistant mutations, and 51 patients harbored both sensitizing and resistant mutations. Twenty-four patients had a single T790M mutation or its combined mutations (one with T790M, fourteen with 19 del + T790M, three with L858R + T790M, one with L858R + T790M + 20ins, two with L858R + T790M + NRAS + MET, and three with the L858R + T790M + rare mutation). Fifteen had never received EGFR-TKI therapy in these patients.
We also conducted a subgroup analysis to investigate whether the sample type, Xuanwei origin, age or ethnicity affected the distribution of GA types. Our study shows that the EGFR mutation type distribution originating from Xuanwei is different from that of other populations in Yunnan region. The prevalence of G719X + S768I mutations and multiple gene alterations is more common in the Xuanwei population than in other populations in Yunnan region. However, compared with other populations in Yunnan region, the mutation rate of exon 19 deletion in NSCLC patients with Xuanwei origin is lower (Table 4 and Figure 1). According to ethnicity, we found that a rare EGFR mutation was significantly more frequent in non-Han patients compared with Han patients, and the frequency of the G719X + S768I mutation in Han patients was higher than that in non-Han patients. There was no difference in the distribution of GA types depending on the sample type and age.
 |
Table 4 GA Type in Our Analysis |
Discussion
The EGFR mutation rate in NSCLC patients is 39.6%, but it is much higher in tissue samples (44.7%, 109/244). According to the report of East Asian countries, the frequency is within the normal range (3150%).19–21 Additionally, our studies showed the correlation between the female, never-smoker and adenocarcinoma characteristics and a higher rate of NSCLC. These observations are consistent with previous studies.19–21 In our study, exon 19 deletion and L858R of EGFR are the most frequent mutation patterns, which is similar to reports performed in East Asian countries.19–21
It has been reported that the EGFR exon 20 S769I mutation is a rare mutation, but in our studies, a total of 36 (6.2%) NSCLC patients with this mutation could be detected. Interestingly, we also found high frequencies of S768I in multiple mutation cases, and approximately 27 cases of G719X/S768I cases came from Xuanwei-origin patients. Chen et al indicated that in contrast to patients from non-Xuanwei areas, a lower frequency of the exon 19 deletion (7.8% versus 49.3%, p < 0.0001) but a higher frequency of G719X + S768I in exons 18 and 20 (45.1% versus 4.1%, p < 0.0001) could be observed in NSCLC patients from the Xuanwei area.22 According to our study, we noticed that there were more G719X + S768I mutations but fewer exon 19 deletion mutations in NSCLC patients in the Xuanwei population than in the non-Xuanwei population (Table 4 and Figure 3). Our analysis also supports previous positive findings in Xuanwei patients. Moreover, our subgroup analysis also indicates that the pattern of the prevalence of multiple EGFR alterations is more generalized in those of Xuanwei origin than in other populations in Yunnan region (Table 4). The function of the S768I mutation in TKI therapy is still unclear. Masago et al23 reported that one patient with the S768I mutation showed a good clinical response to TKI therapy; However, entirely opposite results were found in Asahina et al.24 It is crucial to investigate the role of S768I in NSCLC oncogenesis because S768I showed high frequencies of multiple mutations. There is no doubt that more reports of multiple mutations will emerge and that clinical detection of multiple oncogene mutations is useful in determining the optimal therapeutic method.
 |
Figure 3 Genetic driver alterations identified by next-generation sequencing in the Xuanwei population (n = 79). |
For the treatment of advanced NSCLCs with EGFR-activating mutations, EGFR-TKIs have been a successful option. Somatic mutations in the EGFR gene have been found in NSCLC patients with rapid, durable, complete or partial responses to TKI therapy. Many patients who receive the first-generation TKI treatment for 6 to 12 months develop resistance to TKIs. Additionally, it has been reported that EGFR-mutated patients with TKI resistance account for 50% of secondary T790M mutations.25 Here, we demonstrate that EGFR T790M plus other GAs accounts for 23 (28.8%) of our 80 cases of multiple mutations. Fifteen of those cases might be caused by TKI therapy. However, there were 8 cases in which patients had no TKI therapy treatment but harbored a secondary T790M mutation. This may explain why TKI therapy fails in some NSCLC patients who harbor EGFR-activating mutations.26 It may suggest that multiple mutations with a second resistant mutation play a vital role in resistance to TKI therapy in NSCLCs carrying EGFR-sensitizing mutations, which provides a promising recommendation for designing drugs, more effective strategies against specific subtypes of NSCLC with both EGFR-TKIs and anti-T790M therapies.
In contrast to EGFR mutations, KRAS mutations related to a poor NSCLC prognosis were typically found in smokers and patients resistant to TKIs. In our study, the KRAS mutation rate of NSCLC patients is 10.9%, and it is much higher (54.0%, 34/63) in the tissue samples. The frequency is similar to that in other reports (710.9%).27–30 Additionally, a correlation between adenocarcinoma, family history of malignancy, northeast region, Xuanwei origin characteristics and a higher KRAS mutation rate was observed in NSCLC patients in our studies. These results show partial similarity with other previous studies.27–30 It is remarkable that a KRAS mutation is the second most common driver oncogene identified in Xuanwei-origin adenocarcinoma patients (Table 2 and Figure 3). According to our data, in the 579 NSCLC cases, only 4.5% of smokers and 6.3% of nonsmokers had KRAS mutations, which is much lower than that formerly reported in the United States (34% in smokers and 6% in nonsmokers).31 Originally, Rodenhuis et al reported that KRAS mutations are uncommon in NSCLC patients with a history of nonsmoking.32 However, we found KRAS mutations in 6.3% of nonsmokers, as well as mutations in the United States population (Riely et al 2008),33 which suggested the significance of exploring other factors, such as environmental or genetic inheritance, on KRAS mutations. We discuss two reasons for this issue. First, previous studies showed that the distribution of EGFR, KRAS and rare mutations originating from Xuanwei is different from that of other populations in Yunnan Province,13,22,34 and the genetic spectrum of lung cancers in Xuanwei is unique. The unique genetic spectrum might be related to the mechanism of lung cancer in Xuanwei. Therefore, the difference in KRAS mutations in nonsmokers might be related to the unique genetic spectrum of lung cancer in Xuanwei. Second, since our study recruited patients in a single center and the number of Xuanwei NSCLC patient samples was not large enough, our sample data may not accurately reflect the status of KRAS mutations in Xuanwei area.
Interestingly, we found that the KRAS mutation spectrum in Xuanwei city is different compared to that in other populations in China. DeMarini et al suggested that exposure to indoor air pollution from local smoky coal may be the primary reason.35 A high concentration of PAHs can be released through burning coal, and PAH-DNA adducts have been found in bronchoalveolar lavage of Xuanwei residents.36 Similarly, the EGFR mutation spectra in Xuanwei were in accordance with PAH exposure.27 These results show that the KRAS and EGFR gene mutations could be a symbol of specific environmental exposure. In summary, the unique KRAS and EGFR mutation spectrum in Xuanwei region might be related to the exposure of air pollution from local coal burning. Moreover, our study also showed that a family history of malignancy is correlated with a higher KRAS mutation rate. These studies present several questions regarding which component plays the dominant role: PAHs, inherited factors, or particulate matter. In addition, further investigation is needed to determine the potential mechanism involved.
Interestingly, KRAS and BRAF mutations are mutually exclusive, reported by Rajagopalan et al.37 The KRAS mutation occurs only in tumors without BRAF mutations, as KRAS and BRAF provide an equivalent or at least redundant oncogenic stimulus in cancer pathogenesis. De Roock et al has also reported this result in colorectal cancer.38 However, we found one case of coexisting BRAF and KRAS mutations, which have been formerly considered mutually exclusive mutations.
Kosuke et al showed that ALK fusions were significantly less frequent in elder patients compared with younger patients (4% vs 41%; p < 0.001), however EGFR and KRAS mutations were more frequent in elder patients (EGFR: 45% vs 30%; p = 0.006; KRAS: 10% vs 2%; p = 0.026).39 According to our data, younger NSCLC patients had a higher rate of ALK fusions than elder patients, but there were no differences in EGFR and KRAS mutations (Table 2). We propose that the main reason may be the source limitation of recruited patients and quantity limitation of samples, making it unfeasible to reflect the younger population based on our samples. Surprisingly, for the first time, we discovered that non-Han ethnicity is associated with an increased likelihood of harboring rare oncogenic GAs in NSCLC. ALK fusions, ROS1 fusions and EGFR rare mutations were significantly more frequent in non-Han patients compared with Han patients (ALK: 13.7% vs 3.7%, p < 0.001; ROS1: 5.7% vs 1.7%, p < 0.05; EGFR rare: 16.0% vs 1.0%, p < 0.001) (Table 2, Table 4 and Figure 4). The Yunnan Plateau is one of the four plateaus in China, with an average altitude of 2000 m. There are two geographical characteristics of the Yunnan Plateau: hypoxia and high ultraviolet radiation. These two characteristics may account for the discrepancies. First, most of the non-Han ethnic patients live in mountainous areas with an average altitude of more than 2000 m, and living in a hypoxic environment for a long time may lead to an increase in the incidence of rare GAs. Second, high altitude areas are also high ultraviolet radiation areas. Living under high ultraviolet radiation for a long time may also lead to an increase in the incidence of rare GAs. However, it is unclear which component plays the dominant role: hypoxia, high ultraviolet radiation, or an interaction between the two. Further studies are expected.
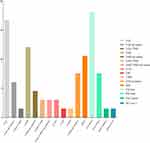 |
Figure 4 Genetic driver alterations identified by next-generation sequencing in the non-Han ethnic population (n = 66). |
NGS has provided us with abundant genetic information, and meanwhile it greatly decreased the cost and time of sequencing with high output and high resolution. In this study, we demonstrate that it is feasible, rapid, and useful to apply NGS genotyping for NSCLC patients in a real-world clinical practice setting. Direct clinical care can be carried out by a driver alteration identified through NGS. However, a negative result needs further exploration. The NCCN guidelines now recommend routine mutation testing for EGFR, BRAF, ERBB2, and MET; Rearrangements in ALK, ROS1, and RET; MET amplification in all patients diagnosed with NSCLC. For patients and clinicians, the NGS platform offers a single test that can capture point mutations, indels, copy number variants, and gene rearrangements across many cancer-related genes. Generally, NGS outperforms each individual molecular test, and NGS is a vital tool in the effective diagnosis and treatment of NSCLC patients.40 It was able to analyze both tissue and plasma in 28 patients in our study. The patients who tested plasma NGS-positive for GAs had a tissue NGS concordance of 95.5%. The sensitivity was 91.3%, the specificity was 80.0%, the PPV was 95.5%, and the NPV was 66.7%. These data are consistent with other previous studies.22,37 However, the smaller sample size may have led to bias. Nevertheless, our study showed that, with rapid and noninvasive genotyping-related plasma-based ctDNA NGS assays, precision therapy could immediately be guided, and a crucial supplement to tissue NGS could be provided.
However, one of the fundamental limitations of plasma NGS genotyping is the low concentration of ctDNA shed into the peripheral circulation.41 The plasma NGS was incapable of detecting drivers in some cases in this study, which means a negative result on ctDNA does not exclude the presence of a targetable driver. Plasma sequencing depth as a technical factor may affect results, and for further plasma detection, ultradeep sequencing methods have to develop. The function of biological factors, including tumor shedding and ctDNA kinetics, in the field of liquid biopsy needs to be explored. However, plasma NGS may be a beneficial method for evaluating the genomic landscape and incorporating tumor heterogeneity, especially in patients with multiple metastatic sites and this strategy is also important for the clinical setting as acquired resistance to targeted therapies occurs with the emergence of subclonal resistance mutations.42 Promising evidence offered by our study supported the incorporation of plasma NGS into lung cancer guidelines. Future plasma-based NGS applications, such as detection in earlier stages or minimal residual disease, may provide neoadjuvant or adjuvant therapies and early detection of cancers.43
Conclusion
In conclusion, our data show a clear panoramic perspective of GA frequencies from 579 NSCLC patients and their associations with demographic and clinical parameters in Yunnan Province. For the first time, we reported that Yunnan NSCLC patients from Xuanwei have a distinctly unique pattern of oncogenic GAs. In addition, we also reported here, for the first time, that Yunnan non-Han ethnic patients with NSCLC had a uniquely high incidence of rare GAs. Finally, NGS is one of the most vital improvements in NSCLC oncology for the detection of GAs. Considering the limitation of our study, unlike a positive finding, a negative finding of an oncogenic driver in plasma may still require additional testing, which cannot immediately guide treatment. In addition, exploring an enough sample size from multiple cancer centers in Yunnan region could improve our study, making it more representative of the Yunnan region population.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81860513), the Project of Basic Applied Research in Yunnan Province (2016FB145) and the Project of Basic Applied Research in Yunnan Province (2017FA037).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 global cancer statistics? Cancer Commun. 2019;39(1):22.
2. Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(760774).
3. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi:10.1056/NEJMoa040938
4. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi:10.1038/nature05945
5. Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(378381). doi:10.1038/nm.2658.
6. Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi:10.1038/nm.2673
7. Clinical Lung Cancer Genome Project (CLCGP); Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153.
8. Serizawa M, Koh Y, Kenmotsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: a prospective, single-institute study. Cancer. 2014;120:1471–1481. doi:10.1002/cncr.28604
9. Chen G, Sun X, Ren H, et al. The mortality patterns of lung cancer between 1990 and 2013 in Xuanwei, China. Lung Cancer. 2015;90:155–160. doi:10.1016/j.lungcan.2015.08.006
10. Xiao Y, Shao Y, Yu X, Zhou G. The epidemic status and risk factors of lung cancer in Xuanwei City, Yunnan Province, China. Front Med. 2012;6:388–394. doi:10.1007/s11684-012-0233-3
11. Mumford JL, He XZ, Chapman RS, et al. Lung cancer and indoor air pollution in Xuan Wei, China. Science. 1987;235:217–220. doi:10.1126/science.3798109
12. Lan Q, He X, Shen M, et al. Variation in lung cancer risk by smoky coal subtype in Xuanwei, China. Int J Cancer. 2008;123:2164–2169. doi:10.1002/ijc.23748
13. Zhou YC, Yang YL, Yang CG, et al. Epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) of Yunnan in southwestern china. Oncotarget. 2016.
14. Gill GN, Kawamoto T, Cochet C, et al. Monoclonal anti-EGF receptor antibodies which are inhibitors of growth factor binding and antagonists of EGF-stimulated tyrosine protein kinase activity. J Biol Chem. 1984;259:77557760.
15. Zhu G, Ye X, Dong Z, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015;17:265–272. doi:10.1016/j.jmoldx.2015.01.004
16. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. doi:10.1158/2159-8290.CD-15-0285
17. Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–849. doi:10.1158/2159-8290.CD-14-1467
18. Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;(15):504–535. doi:10.6004/jnccn.2017.0050
19. Liam CK, Wahid MI, Rajadurai P, Cheah YK, Ng TS. Epidermal growth factor receptor mutations in lung adenocarcinoma in Malaysian patients. J Thorac Oncol. 2013;8:766–772. doi:10.1097/JTO.0b013e31828b5228
20. Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154–162. doi:10.1097/JTO.0000000000000033
21. Shi Y, Li J, Zhang S, et al. Molecular epidemiology of EGFR mutations in asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology - mainland china subset analysis of the PIONEER study. PLoS One. 2015;10:e0143515. doi:10.1371/journal.pone.0143515
22. Chen Y, Ye L, Stanford RR, Zhang D, Zhang X, Wei W. Distinct epithelial growth factor receptor mutation profile in non-small-cell lung cancer patients from the Xuanwei area of China. Mol Clin Oncol. 2016;4:749–755. doi:10.3892/mco.2016.805
23. Masago K, Fujita S, Irisa K, et al. Good clinical response to gefitinib in a non-small cell lung cancer patient harboring a rare somatic epidermal growth factor gene point mutation; codon 768 AGC4ATC in exon 20 (S768I). Jpn J Clin Oncol. 2010;40:1105–1109. doi:10.1093/jjco/hyq087
24. Asahina H, Yamazaki K, Kinoshita I, Yokouchi H, Dosaka-Akita H, Nishimura M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer. 2006;54:419–422. doi:10.1016/j.lungcan.2006.09.005
25. Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci USA. 2012;109:E2127–E2133. doi:10.1073/pnas.1203530109
26. Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2010;352:786–792. doi:10.1056/NEJMoa044238
27. Jing C, Mao X, Wang Z, et al. Next generation sequencing‑based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her2 and TP53 mutations in patients with non‑small cell lung cancer. Mol Med Rep. 2018;2191–2197.
28. Wen YS, Cai L, Zhang XW, et al. Concurrent oncogene mutation profile in Chinese patients with stage Ib lung adenocarcinoma. Medicine. 2014;93(e296). doi:10.1097/MD.0000000000000296
29. Sabari JK, Offin M, Stephens D, et al. A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J Natl Cancer Inst. 2018.
30. Sim WC, Loh CH, Toh GL, et al. Non-invasive detection of actionable mutations in advanced non-small-cell lung cancer using targeted sequencing of circulating tumor DNA. Lung Cancer. 2018;124:154–159. doi:10.1016/j.lungcan.2018.08.007
31. Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi:10.1158/1078-0432.CCR-11-3265
32. Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–935. doi:10.1056/NEJM198710083171504
33. Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi:10.1158/1078-0432.CCR-08-0646
34. Hosgood HD
35. DeMarini DM, Landi S, Tian D, et al. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61:6679–6681.
36. Mumford JL, Lee X, Lewtas J, Young TL, Santella RM. DNA adducts as biomarkers for assessing exposure to polycyclic aromatic hydrocarbons in tissues from Xuan Wei women with high exposure to coal combustion emissions and high lung cancer mortality. Environ Health Perspect. 1993;99:83–87. doi:10.1289/ehp.939983
37. Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B. Velculescu VE Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi:10.1038/418934a
38. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi:10.1016/S1470-2045(10)70130-3
39. Tanaka K, Hida T, Oya Y, et al. Unique prevalence of oncogenic genetic alterations in young patients with lung adenocarcinoma. Cancer. 2017;123:1731–1740.
40. Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21:3631–3639. doi:10.1158/1078-0432.CCR-14-2683
41. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi:10.1038/nm.1789
42. Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi:10.1038/nature12065
43. Li BT, Stephens D, Chaft JE, et al. Liquid biopsy for ctDNA to revolutionize the care of patients with early stage lung cancers. Ann Transl Med. 2017;5:479. doi:10.21037/atm.2017.09.02
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
