Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 13
Omics-Driven Biomarkers of Psoriasis: Recent Insights, Current Challenges, and Future Prospects
Authors Aydin B , Arga KY , Karadag AS
Received 20 June 2020
Accepted for publication 7 August 2020
Published 25 August 2020 Volume 2020:13 Pages 611—625
DOI https://doi.org/10.2147/CCID.S227896
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Busra Aydin,1 Kazim Yalcin Arga,1 Ayse Serap Karadag2
1Department of Bioengineering, Faculty of Engineering, Marmara University, Istanbul, Turkey; 2Department of Dermatology, Istanbul Medeniyet University, School of Medicine, Goztepe Research and Training Hospital, Istanbul, Turkey
Correspondence: Ayse Serap Karadag
Department of Dermatology, Istanbul Medeniyet University, School of Medicine, Goztepe Research and Training Hospital, Istanbul, Turkey
Tel +90 5336552260
Fax +90 216 566 66 14
Email [email protected]
Abstract: Advances in omics technologies have made it possible to unravel biomarkers from different biological levels. Intensive studies have been carried out to uncover the dysregulations in psoriasis and to identify molecular signatures associated with the pathogenesis of psoriasis. In this review, we presented an overview of the current status of the omics-driven biomarker research and emphasized the transcriptomic, epigenomic, proteomic, metabolomic, and glycomic signatures proposed as psoriasis biomarkers. Furthermore, insights on the limitations and future directions of the current biomarker discovery strategies were discussed, which will continue to comprehend broader visions of psoriasis research, diagnosis, and therapy especially in the context of personalized medicine.
Keywords: psoriasis, biomarker, omics, inflammatory skin diseases, personalized medicine, psoriasis biomarkers, omics biomarkers
Introduction
Psoriasis is a chronic, recurrent, and immune-mediated cutaneous disease with variable morphology and severity.1–3 A systematic analysis of worldwide epidemiology of psoriasis estimated varying prevalence ranging from 0.5% to 11.4% in adults and 0% to 1.4% in children.4 The manifestation of the disease is characterized by well-demarcated, scaly, reddish and/or silvery, and oval plaques, which are arisen from hyperplastic regenerative epidermal growth, hyperproliferative keratinocytes, infiltration of the inflammatory cells, and elevated numbers of tortuous capillaries through thinned epithelium.5 Psoriasis affects primarily the skin, however, it can affect nails, peripheral and axial joints, and emotional situation of the patients, as well. The etiopathogenesis of psoriasis hinged on the chaotic reciprocity of genetic, environmental, and immunological drivers.6 Psoriasis patients can develop comorbidities including cardiovascular diseases, metabolic syndrome (ie, obesity, insulin resistance), type-2-diabetes, Crohn’s disease, psoriatic arthritis (PsA), depression, and psychiatric diseases.7–10 Several established clinical presentations of psoriasis are as guttate psoriasis, erythrodermic psoriasis, pustular psoriasis, and chronic plaque psoriasis vulgaris encountered for more than 90% of the patients. On the other hand, these types of psoriasis represent different features, unique molecular profiles, and responses to therapeutic agents; therefore, the same treatment protocols do not respond in every patient in the same manner. Hence, there is an urgent need to detect accurate biomarkers that will assist psoriasis diagnosis, prognosis, and therapy as well as developing personalized medicine strategies.
Theoretically, the logic underlying biomarker discovery is quite incomplex: If we had sufficient information about the underlying molecular pathophysiology, then it would possible to define disease driving pathways and target proteins and/or much better to take preventive precautions even before the disease arose.3 Since, psoriasis is a multifactorial disease whose manifestation is affected by genetic, epigenetic, environmental, and lifestyle factors, the knowledge coming from just one level omic data for psoriasis may only provide a snapshot. However, psoriasis mechanisms are needed to be handled in a multi-omics perspective within the systems biomedicine approach.
Over the last decade, technological and computational improvements have revolutionized our ability to reach high-throughput data on almost all molecular levels and biomarker discovery for various diseases have been comprehensively conducted via omics-driven methods. In the case of psoriasis, promising biomarkers have been established at the genome, transcriptome, proteome, epigenome, glycome, lipidome and metabolome levels.2,3,11–14 These developments have contributed to the understanding of the underlying molecular mechanisms of psoriasis physio-pathogenesis. Moreover, some of these proposed markers may distinguish disease features such as being diagnostics, prognostics, and therapeutic targets of psoriasis.
In this review, we recapitulate the hitherto findings in psoriasis biomarker discovery in different omics levels including transcriptomics, epigenomics, metabolomics, and glycomics. We will emphasize the limitations and current challenges of biomarker discovery for psoriasis and provide some future directions. We present a comprehensive review of what is known about biomarkers from different omics levels and their role in the pathogenesis of psoriasis.
The Workflow of Biomarker Discovery and Therapeutic Target Elucidation in Psoriasis
Candidate biomarkers of psoriasis could be elucidated utilizing emerging omics technologies including genomics, transcriptomics, proteomics, glycomics, lipidomics, and metabolomics. Several promising biomarkers of psoriasis have been established with these omics techniques, and more outstanding discoveries could be expected with the advancement of the sequencing platforms. Potential biomarkers have generally been validated through well-known techniques such as enzyme-linked immunosorbent assay (ELISA), immunohistochemistry (IHC), lectin array, Western blot (WB), and quantitative reverse transcription PCR (qRT-PCR) with a higher number of a cohort of subjects and/or samples, to obtain statistically significant and indicative results. The ones that passed from validation of the process will be candidate biomarkers and utilized in the clinical applications such as diagnosis, prognosis, recurrence, and therapy of psoriasis (Figure 1).
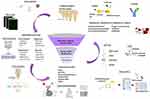 |
Figure 1 The workflow of biomarker discovery. |
Recent Insights from Different Omics Levels
Transcriptomic Signatures of Psoriasis
Analysis of the mRNA profiles of psoriatic and healthy skin has been performed in several studies and has resulted in many potential biomarkers for response to prognosis, diagnosis, and treatment (Table 1).
 |
Table 1 Transcriptomic Signatures of Psoriasis |
Interestingly, one of the early studies with microarray technology is on psoriasis. Oestreicher et al conducted the first transcriptome study on psoriasis employing the HuGeneFL panel including 159 genes and reported the differential expressions of GATA3, TXNA, and HBP17 in response to drugs, rhIL-11 and Cyclosporin A.15 In the following years, the transcriptomic responses to drug treatments were studied using genome-wide microarrays. For instance, Krueger et al evaluated the gene expression changes in a time-dependent manner using samples from patients treated with ixekizumab, an immunosuppressant, and reported IL-17 as a key “driver” cytokine in psoriasis that activates pathogenic inflammation.16 Again, to determine the molecular and cellular alterations in the skin after IL-17RA blockade with brodalumab, which is a recombinant human monoclonal antibody, skin biopsies were obtained at different time points from lesions. The microarray data indicated that the hindrance of IL-17 signaling in psoriatic patients provided a rapid molecular and histologic resolution of the inflammation associated interactions that characterize psoriasis.17
Transcriptome studies were not limited to investigating the drug effects of course. Identification of the differentially expressed genes (DEGs) between healthy and diseased samples has been considered as an indispensable step in biomarker discovery. Thousands of transcripts were been identified as DEGs in psoriatic lesional skin when compared to nonlesional (normal) skin samples in various studies.18–28 Among them, chemokines like CXCL16, MIP4, SYC19/21, SYCB9/10, SDF1, and TRIM22, implicated the presence of a mechanism involved in chronic disease activity through providing interactions to conduct T-cell wiring, activation, and dependent immune functions.23 Besides, several other genes associated with the inflammatory cytokines/chemokines or proteins responsive to cytokines, such as ACCP, CCL2, CCL22, CXCL5, DEFB4, ICAM1, IL15, IL1RN, IL36G, IL36RN PI3, S100A7A, S100A12, SERPINB4, TNF, TNFRSF1B, TNPO1, and VEGFA, were up-regulated in both serum and skin of psoriatic patients.19,20,22 Taking advantage of the ability to study defined cellular regions of the epidermis or dermis, Mitsui et al have carried out microarray analysis from laser micro-dissected psoriatic skins, and identified the previously reported DEGs that were driven from the bulk sets of tissues/extracts and also specific DEGs.21 They emphasized the roles of CCL19 and its receptor CCR7, which take place particularly within perivascular T cell and DC aggregates, and the importance of chemokine localization in psoriasis.21
With the development of next-generation sequencing technologies (NGS), the genome reprogramming under psoriasis was also studied using RNA-Seq technology in recent years. The RNA-seq analysis of PBMCs from pustular psoriasis patients revealed the function of leukocytes, particularly neutrophils, in disease mechanisms, and presented the significant down-regulation in expressions of S100A8, S100A9, S100A12, PLK1, and IRF7, and up-regulation in expressions of CIITA and NKTR in response to medical treatment.29
Although limited in number, meta-analyses of transcriptome datasets were also performed and resulted in proposals for a core transcriptome30 and a disease pathway.31 The core transcriptome, which was enriched mostly in IL-17A pathway, atherosclerosis signaling, and fatty acid metabolism, proposed several biomarker candidates for molecular diagnosis of psoriasis, and also molecular signatures to associate psoriasis and its systemic manifestations such as metabolic syndrome and cardiovascular diseases.30 From a comprehensive perspective on the molecular basis of psoriasis, a total of twelve transcriptome datasets were analyzed in an integrative framework and several candidate biomarkers and therapeutic targets and a JAK/STAT signaling pathway involving cytokines, interferon-stimulated genes, and antimicrobial peptides were proposed.31 More recently, a set of biomarker candidates was also proposed through a transcriptome based meta-analysis considering also the sex differences of patients.32 They suggested IRF9 and NMI in women, and SUB1 in men as potential biomarkers, and verified the biomarker potential of PI3, WIF1, and PC4 both at transcriptome level using qRT-PCR and protein level via ELISA.32 More recently, human beta defensin 1 (hBD-1) encoded by DEFB1 gene found significantly greater in psoriasis patients compared to controls and after narrow band ultraviolet B (nb-UVB) phototherapy hBD-1 levels compared the levels of healthy controls.33
Epigenomic Signatures of Psoriasis
Epigenome studies, focusing on histone modification, DNA methylation, and gene expression regulation through microRNA (miRNA), or long non-coding RNA (lncRNA), have revealed the potential of epigenetics in elucidating the mechanisms of different diseases including psoriasis and reported aberrations in epigenetic regulation for psoriasis (Table 2).
 |
Table 2 Epigenomic Signatures of Psoriasis |
One decade ago, miRNAs were discovered as a unique branch of conserved small (18–24 nucleotides) non-coding RNA molecules, which are crucial regulators of gene expression. The initial skin-specific miRNA revealed was miR-203, which was significantly upregulated in psoriasis patients.34 Since then, several differentially expressed miRNAs were reported that are likely to alter the psoriasis pathogenesis including miR-21, miR-31, and miR-378 associated with angiogenesis; miR-135b, miR-205, and miR-203-AS associated with epidermal differentiation; miR-142-3p associated with inflammation; miR-99a and miR-146b playing role in the differentiation of keratinocytes.35–37 In addition, the expressions of miR-143 and miR-223 in PBMCs of psoriatic patients were associated with psoriasis disease severity index (PASI) and were suggested as early biomarker candidates for psoriasis treatment response.38 Considering the role of miR-146b in inhibition of proliferation of keratinocytes and miR-203 to reverse the IL-17 signaling pathway and reduce in JAK2/STAT3 signaling pathway activation, these miRNAs were suggested as potential therapeutic targes.36,39
LncRNAs, defined as non–protein‐coding RNAs that are in the length of 200 nucleotides or longer, are the most crowded and diverse group of non-coding RNA world40 and performed crucial functional roles in epigenetic regulation.41 Dysregulated lncRNAs could lead to aberrant keratinocyte differentiation in psoriasis.41 The first lncRNA study that was carried out with psoriasis reported that psoriasis susceptibility–related RNA gene induced by stress (PRINS) was overexpressed in the uninvolved epidermis of psoriatic patients and PRINS may involve in psoriasis pathogenesis via reducing the sensitivity to keratinocyte apoptosis.34 Tsoi et al carried out RNA sequencing from psoriatic skins and elucidated both previously annotated and novel skin-specific lncRNAs that were found co-expressed with genes associated with the immune processes and enriched in the epidermal differentiation complex.42 LncRNAs, namely, CYP4Z2P, HINT1, and TRHDE-AS1, were found differentially expressed in psoriasis lesional skin and were proposed in adjacent to known psoriasis susceptibility loci at CARD14, LCE3B/LCE3C, and IL23R.43 A gene co-expression study was also conducted via weighted gene co-expression network analysis (WGCNA) to determine differential differences between psoriasis patients and healthy controls concerning gene and lncRNA expressions and network analysis has resulted in previously unreported biological pathways such as olfactory receptor activity.44,45 A positive correlation between lncRNA-MSX2P1 and S100A7 expression and the association of these elements with miR‐6731‐5p led to the hypothesis that the complex of MSX2P1‐miR‐6731‐5p‐ S100A7 might be a novel candidate therapeutic target for the future treatment of psoriasis.46 More recently, the downregulation of lncRNA-AL162231.4-regulated CCL27 in psoriasis was presented via WGCNA and linked to the pathological development of psoriasis.47
DNA methylation and the enzymes that are associated with its regulation play critical roles in the various biological processes, including psoriasis.45 The first study of methylation analysis in psoriasis was carried out by Ruchusatsawat et al, who have shown the demethylation of SHP-1 promoter 2 in psoriatic patients when compared to normal skin.48 The hypermethylation of the ID4 promoter was associated with parakeratosis and cellular differentiation in psoriasis.48 Another study analyzed the promoter methylation status of p15 and p21 genes and the colony formation ability of high proliferative potential colony-forming cells (HPP-CFCs) and indicated higher transcription levels for p15 and p21 genes and lower methylation frequencies of these promoter regions were observed in psoriasis in comparison to normal volunteers. As a result, it was speculated that hypomethylations of these sites might lead to dysfunctional growth regulation pathways in high proliferative potential colony‐forming cells (HPP-CFCs) in psoriasis.49 Ngalamika et al analyzed the treg-specific demethylated region (TSDR) of FOXP3 in peripheral whole blood of patients with autoimmune diseases including psoriasis and systemic lupus erythematosus (SLE), and they revealed that SLE and psoriasis had significantly higher methylation levels.50 Furthermore, SELENBP1, PTPN22, and a couple of differentially methylated genes were suggested as potential targets for future clinical studies on psoriasis therapy.51 The effects of UV phototherapy on psoriasis and DNA methylation were also carried out and the altered DNA methylation was observed in psoriasis samples, plus reversion of abnormal methylation status was also shown following phototherapy in patients with clinical improvement.52 In another study, the methylation status was examined from psoriatic scales since it is less invasive than a punch biopsy to detect that parakeratotic cell-specific methylation profiles and it resulted in dramatically changed DNA methylation status within the psoriatic epidermis.53 Zhang et al developed a method termed methylated DNA immunoprecipitation sequencing (MeDIP-Seq) and demonstrated that DNA methylation profiles at the PDCD5 and TIMP2 loci were negatively correlated with the mRNA expression of each gene and might play roles in the pathogenesis of psoriasis.54
A restricted number of studies were conducted to identify histone modifications associated with the psoriasis pathogenesis. Ovejero‐Benito et al performed histone modification analysis associated with drug responses, and reported significantly reduced levels of acetylated histone H3 and H4 and increased levels of Histone H3 lysine K4 (H3K4) methylation.55 The hypoacetylation of H4 was also found as a signature in PBMCs of psoriasis vulgaris patients compared with normal controls and a negative correlation between the degree of histone H4 acetylation and disease activity in patients as measured by PASI were also reported.56
The up-to-date findings proposed that epigenetic alterations namely DNA methylation, histone modifications, and expression of miRNAs and lncRNAs might have crucial duties in psoriasis. On the other hand, their roles need to be further addressed via the employment of in vivo and in vitro studies through loss-of and gain-of-function methods. These insights will contribute a novel basis to construct diagnostic vehicles and treatment options for patients with psoriasis.
Metabolomic Signatures of Psoriasis
Metabolomics is a fresh-sprouted “omics” field that covers the systematic elucidation of the metabolites in a biological system. Since, the metabolome demonstrates the end products of proteomics or cellular processes, dysregulated metabolic pathways associated with the psoriasis pathogenesis might be evaluated with metabolomics to gain insights for diagnostic and therapeutic developments. Although the metabolite aberrations directly drive the disease process, they represent a molecular snapshot of the cellular processes that are affected during the disease pathogenesis. The metabolic pattern of uninvolved skin, psoriatic skin, and corticosteroid treated psoriatic skin were investigated via several chromatographies, spectrometries, and NMR (nuclear magnetic resonance) technologies, and altered levels of several metabolites were detected (Table 3).
 |
Table 3 Metabolomic Signatures of Psoriasis |
The increased arachidonic acid and 12-HETE (Hydroxy-eicosatetraenoic acid) in psoriatic skin samples and the role of lipoxygenase pathway products in the pathogenesis of psoriasis were known for almost four decades.57 Armstrong et al analyzed the serum metabolite levels of psoriasis and healthy controls by using GC-TOF-MS (gas chromatography/time-of-flight mass spectrometry) and they revealed that higher serum levels of α-ketoglutaric acid and glucuronic acid and lower levels of asparagine and glutamine.58 These findings pointed out psoriasis pathogenesis associated with altered joint destruction processes and immune-mediated pathways. In another study, the plasma metabolite levels from individuals with mild or severe psoriasis as well as healthy controls were measured by LC–MS (liquid chromatography mass spectrometry) and findings revealed the lower levels of threonine, ornithine, and methionine and these levels turn in normal ranges after etanercept treatment59 which indicated plasma levels of amino acids might be employed for assessing PASI and investigation of response to treatment. Similarly, the lower levels of the myo-inositol and glucose, and higher levels of choline and taurine were detected in psoriatic patients; however, patients treated with corticosteroids showed high levels of glucose, myo-inositol, GPC and glycine whereas choline was reduced.60 Another serum metabolite analysis reported the higher levels of homocysteine and ADMA (asymmetrical dimethyl arginine) in psoriasis patients using UPLC (ultra performance liquid chromatography) and HPLC (high performance liquid chromatography) and suggested that these metabolites could be an indicator of the disease severity.61
Psoriatic skin metabolome was also investigated through biocompatible hydrogel micropatch probes and subsequent direct mass spectrometry scanning which were less invasive and patient-friendly.62 Findings unveiled higher levels of choline, citrulline, glutamic acid, and lower levels of urocanic acid, lactic acid, and phenylalanine. Also, polar metabolites such as choline and glutamic acid positively or urocanic acid and citrulline negatively correlated with the plaque severity scores.62 The metabolic perturbations of the glycolysis pathway and amino acid metabolic activity were established in psoriatic patients serum through GC-MS mediated metabolome analysis and higher levels of amino acids including asparagine, aspartic acid, isoleucine, phenylalanine, ornithine, and proline; higher levels of lactic acid and urea; and lower levels of crotonic acid, azelaic acid, ethanolamine, and cholesterol were reported. Aberrations in these pathways might be sourced from the elevated requirement for protein biosynthesis and keratinocyte hyperproliferation, as well.63 Insulin, C-peptide levels and homeostasis model assessment-insulin resistance (HOMA-IR) score was found significantly higher in patients with psoriasis than in controls.63 Moreover, endothelial dysfunction demonstrated by flow-mediated dilation (FMD) and FMD scores were reported decreased in patients with psoriasis compared with healthy controls.63
Although urine is the most easy to collect and non-invase sample undoubtedly, the metabolome studies from urine samples of psoriasis patients were very rare. One of these studies reported lower citrate, alanine, methylsuccinate, trigonelline, carnitine, and hippurate levels from urine of psoriasis patients compared to healthy controls.64 Setkowicz et al reported that psoriasis urine samples had higher urinary tetranor-12(S)-HETE (hydroxyeicosatetraenoic acid) and lower 12(S)-HETE, the ratio of these metabolites were proposed as diagnostic marker of psoriasis.65
These studies have unveiled novel contributions to disease pathogenesis and proposed new molecular signatures that might be employed as a marker of psoriasis disease. Also, these findings might aid to reveal the pathogenesis of psoriasis and assisted in researches for the discovery of early diagnosis and therapeutic interventions.
Glycomic Signatures of Psoriasis
Glycosylation is a crucial post-translational modification of proteins and more than half of proteins in humans are glycoproteins.66 Glycans are the molecules which covalently adherent to proteins and perform essential roles in determining the protein structures and functions.66 If we consider the biological system or whole organism as a switchboard, glycans could be thought of as “on” and “off” switches in modulating glycoproteins.67 Unfortunately, glycans are not as valued as they deserved.68 In this section, we are emphasizing the glycome studies that proposed clinical glycomic biomarkers in psoriasis. The new milestones in the quantitative glycomics provide more glycans or glycosylation alterations that might be revolutionized into better potential biomarkers showed greater diagnostic sensitivity and specificity than existing markers. Recent studies presented various glycomic signatures that might be considered as an activity marker, a diagnostic marker, an inflammatory marker, a pathogenesis marker, a risk assessment marker, or a therapeutic target in psoriasis (Table 4).
 |
Table 4 Glycomic Signatures of Psoriasis |
The cathepsin D and zinc‐α2‐glycoprotein (CatD and ZAG), two catalytic enzymes linked with apoptosis and desquamation were found strongly upregulated by IFN-γ in the normal keratinocytes, whereas, found absent from the psoriatic plaque through immunohistochemistry and RT-PCR analysis.69 The corneodesmosin (CDSN) is a late differentiation epidermal glycoprotein that has roles in keratinocyte adhesion and a significant increase in CDSN expression reported in lesional psoriasis patients.70 CD134 is a cell surface glycoprotein, belongs to the TNF receptor family and its expression occurred in activated T-cells. CD134L is CD134 ligand which identified as a cell surface molecule. The expression of CD134 and CD134L were examined immunohistochemically, and Matsumura et al found that the inflammatory state may upregulate CD134L expression on vascular cells.71 The overexpression of membrane proteins including MMP-2 (matrix metalloproteinase 2) and TIMP-2 (Tissue inhibitor of metalloproteinases 2) were reported in the cytoplasm of suprabasal keratinocytes of psoriatic patients and Fleischmajer et al proposed that they might organize the proteolysis of keratinocyte adhesion molecules and have parts in transepidermal migration of these adhesion molecules.72 The chondroitin sulphate (CS) is a member of glycosaminoglycan family and four novel anti-CS antibodies including IO3H10, IO3D9, IO3H12, and IO4C2 were identified and proposed as useful tools for psoriasis.73 The adhesion molecules including sE-selectin, sP-selectin, sICAM-1, and pro-inflammatory cytokine IL8 were found over-expressed in psoriatic serum and these immunological parameters showed a significant decrease in expression after Goeckerman’s therapy.74 Elevated serum levels of calcium-binding S100A8 and S100A9 were associated with disease activity and abnormal keratinocyte differentiation in psoriasis.75 The CD163 as the most useful marker of macrophages and its expression was found upregulated and associated with the pathogenic inflammation in psoriasis via releasing key inflammatory products.76 Galectin-3 is a member of the galectin family of β-galactoside-binding lectins and distributed to all over the organs. Galectin-3 was found under-expressed in psoriatic keratinocytes and suggested as a therapeutic target for the treatment of psoriasis.77 GlycA is an emerging and novel integrated glycosylation marker associated with inflammation and has the potential to predict cardiovascular events.78 Since psoriasis sometimes manifests with cardiovascular comorbidities, Joshi et al searched for the association of GlycA with psoriasis and they found that GlycA related to psoriasis severity and subclinical cardiovascular events.79 Also, psoriasis treatment decreased GlycA and these findings provide the potential utility of GlycA in risk assessment of in subclinical cardiovascular events in psoriasis.
Haptoglobin, an acute-phase glycoprotein, is produced mostly in the liver to attached free hemoglobin for degradation and iron recycling and three glycoforms (Lk, Ly, and Lx) of haptoglobin were elucidated only in psoriatic skin and might be employed as markers of the psoriasis pathology.13 Human endothelial cell-specific molecule-1 (endocan), a soluble dermatan sulfate proteoglycan, secreted by endothelial cells and strongly associated with the development of blood vessels and signature of vascular pathogenesis.80 In psoriasis patients’ serum samples, endocan levels correlated with PASI and endocan levels suggested as risk assessment marker of cardiovascular events and treatment targets.81 Vascular endothelial growth factor (VEGF), a 40–45 kDa dimeric glycoprotein, is mostly associated with angiogenesis and serum levels of VEGF were found elevated in psoriatic patients before treatment.82 The significant reduction of VEGF levels and significant correlation with PASI were observed after treatment with acitretin and psoralen plus ultraviolet-A (PUVA) which proposed a crucial role of VEGF in the pathogenesis of psoriasis and represented as a therapeutic target for psoriasis.82
The expression of CEACAMs (Carcinoembryonic antigen-related cell adhesion molecules), mammalian immunoglobulin-related glycoproteins, were also studied immunohistochemically in the skin of patients with psoriasis and the over-expressions of CEACAM1, CEA and CEACAM6 were observed. These CEACAMs were associated with epidermal cell de-differentiation in the diseased skin of psoriasis vulgaris and suggested as pathogenesis marker.83 Podoplanin, a transmembrane glycoprotein, is a well-distinguished lymphatic endothelial marker and reported as over-expressed independently by TGF-β1 and IFN-γ in keratinocytes via TGF- β receptor-Smad pathway and JAK-STAT pathway. It was also reported that podoplanin strongly related to the wound healing process and pathogenesis of hyperproliferation of psoriatic epidermis.84
Identification of clinically essential glycan-based biomarkers with the possible aberration of glycosylation might contribute to understanding the disease pathogenesis and diagnosis and treatment of psoriasis.
Limitations
Accumulating evidence showed that there have been many studies conducted to investigate psoriasis disease mechanism and decode molecular signatures. Although too many studies proposed different types of biomarkers, there are limited validation data to corroborate the employment of potential biomarkers in clinical practice for diagnosis, evaluation of disease activity, or the prognosis of psoriasis disease.85
Researchers have faced with some drawbacks while investigating novel and clinically usable biomarkers in different omics levels. One of them is the obvious dissociation of omics data and immunohistochemistry (IHC) data. Researchers are confused if the IHC marker used in pathology do not be utilized as significant in the omics results, and vice versa.86 That controversy is originated from the qualitative and quantitative nature of these methods. Although an IHC marker generally not succeeded to be an omics validated biomarker, an omics biomarker might be a validated functional IHC marker.86
Another important drawback is heterogeneity phenomenon most scientists encountered within two ways; 1) disease heterogeneity occurs between individuals that have the same disease, 2) specimen heterogeneity represents multiple phenotypes in an individual such as FFPE tissue sample of a patient with a brain tumor could have healthy and tumor tissues at the same time. Disease heterogeneity is stemming from ethnicity, the way lives the life, microbiota, nutrition, and genetic predisposition. So many drugs and compounds sometimes fail to treat some patients because of disease heterogeneity. Specimen heterogeneity could be encountered in almost every high throughput omics technology, as well. For example, in gene-expression microarrays, a simple asynchronous pattern of expression could take place where a couple of samples in the analysis have both normal and aberrant expression values for the same genes.86 Therefore, in an omics biomarker discovery context, the challenge is how to logically and effectively mine heterogeneous data. Furthermore, irrational choice of cut-offs including fold change, and strong reliability on biostatistics might diminish by narrowing diverse patterns of expressions and their multiple profiles, consequently, observe unreal interpretations of specimens’ relatedness.86 These problems could be defeated by systematic analysis of heterogeneity of the data. There are some tools and approaches to analyze the heterogeneity of data such as MDP molecular degree of perturbation (MDP) which can distinguish potentially problematic subject data from transcriptome dataset and quantify the perturbation score of healthy and diseased samples, as well.87 DeconRNASeq (deconvolution on mRNA-seq) is another pipeline that provides deconvolution of heterogeneous tissues based on mRNA-Seq data.88
In metabolomics studies, the most challenging issue is to confirm the identity of a biomarker since metabolites vary widely in electrical charge, concentration, and molecular weight.2
In epigenomic research, current limitations can be summarized as diverse assays, large gene lists, few comparative data, and controversies in published papers. Therefore, the major requirement in epigenetic diagnostics today is the rational validation of reported candidate biomarkers.89 In addition, there are a couple of bottlenecks of usage of lncRNAs as biomarkers. The plasma levels of lncRNAs are generally low on contrary to short non‐coding RNAs, such as miRNAs. Hence, a great deal of lncRNAs is not tractable in plasma by most used methods, such as microarrays or quantitative PCR.41 Glycomics is an emerging field of omics compared with other omics technologies. A major pitfall is stemming from analyzing tools that used for glycan identification. Lectin based assays are the mostly used method in glycan identification studies. Besides, Western blot, northern blot, types of mass spectrometry, and types of chromatography techniques are the main technologies employed in glycan analysis. In lectin-based assays, two crucial limitations that required to be overcome i) sensitivity (the weak affinity for their target glycan) and ii) presence and availability of lectins with uncommon/less-studied glycan structures.90
Individual drawbacks of each technology needed to be explored and studies on using a set of markers for a disease condition would help to eliminate variations between individuals.
Future Directions and Concluding Remarks
The omics revolution provides researchers to investigate molecular, cellular, and functional candidates that make it possible to enlighten the psoriasis disease pathogenesis. A great deal of biomarkers has been investigated, and, some of them offer hope but further replication and validation are required. Novel and promising biomarkers are anticipated their discovery. New advances in both omics technology and dermatology might enable us to employ personalized medicine approaches through emerging “omics” techniques to the personalized treatment of psoriasis. Elucidation of the different levels of omics data is crucial to understanding the systemic manifestations of psoriasis. The substantial amount of data could be generated from different omics levels including transcriptome, proteome, metabolome, lipidome, glycome, epigenome, and integrome whose complex and holistic nature allows researchers to investigate deeper with this lens and connects the dots between each level. Hence, each technique will converge to conclude, integrate, and overlap for biomarker discovery and validation at different levels and platforms that eventually promise crucial findings into the pathophysiology of psoriasis.
The integration of multi-omics data offers information on biomolecules and how they interplay from different levels to understand the complex biology systematically and holistically.91 Integrative approaches aid in assessing the flow of information from one omics level to the other and thus aid in filling the gap from genotype to phenotype. Multi-omics data boosts the uncovering of a molecular response at the pathway level. The handling of biological phenomenon holistically, have the ability to scan individual treatment responses in a multi-layered manner, enhance prognostics and predictive accuracy of disease phenotypes, and therefore can eventually aid in better treatment and prevention.91,92 Foulkes et al provided a study with valuable insights that represent the integration of the transcriptome and proteome data from blood, lesional/non-lesional psoriatic skin and serum samples. They also found association between clinical response and TNF-regulated genes in blood and skin, by so, they catch psoriasis signatures that indicative of patient response to biologic therapies.93
The major issue in biomarker discovery is the validation of the proposed biomarkers, lack of both quality control and statistical analysis, so, standards for establishing the validation criteria for biomarker selection are urgently required. Another issue is the heterogeneity of high- throughput data which must be addressed with specific approaches to eliminate them. Also, individual variation of biomarkers should be investigated, and research should be constructed to use a set of biomarkers that would aid to overcome variations between individuals.94
Despite the all mentioned bottlenecks, the urgent emergence, availability and the convergence of omic technologies, and advanced bioinformatics approaches will provide new avenues to the discovery of novel and specific psoriasis biomarkers with significant diagnostic and/or therapeutic values.2 Longitudinal studies in larger cohorts of patients will provide to corroborate the rationale for biomarker use in clinical practice including studying for comorbidities, diagnostics, and potential drug targets, as well.85 A promising field, drug repositioning, might be a promising option that provides the new uses for approved or investigational drugs that are outside the scope of the original medical indication for psoriasis treatment.95 Furthermore, there is a study that has perfect fit the vision of personalized medicine, monitoring the status of the patient skin by using artificial intelligence and cloud-based deep learning through a mobile application that can perform image analysis evaluation of treatment response.3,96 These insightful studies will continue to comprehend broader visions of psoriasis.
As a final remark, cutting-edge, systemic, and holistic disease understanding which connects the gaps between diagnostics and treatment options will emerge decoding the pathomechanisms behind psoriasis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Griffiths CE, Barker JN. Pathogenesis and clinical features of psoriasis. The Lancet. 2007;370:263–271. doi:10.1016/S0140-6736(07)61128-3
2. Jiang S, Hinchliffe TE, Wu T. Biomarkers of an autoimmune skin disease—psoriasis. Genom Proteom Bioinf. 2015;13:224–233. doi:10.1016/j.gpb.2015.04.002
3. Litman T. Personalized medicine—concepts, technologies, and applications in inflammatory skin diseases. Apmis. 2019;127:386–424.
4. Michalek IM, Loring B, John SM. A systematic review of worldwide epidemiology of psoriasis. J Eur Acad Dermatol Venereol. 2017;31:205–212. doi:10.1111/jdv.13854
5. Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi:10.1056/NEJMra0804595
6. Villanova F, Di Meglio P, Nestle FO. Biomarkers in psoriasis and psoriatic arthritis. Ann Rheum Dis. 2013;72:ii104–10. doi:10.1136/annrheumdis-2012-203037
7. Gottlieb AB, Chao C, Dann F. Psoriasis comorbidities. J Dermatolog Treat. 2008;19:5–21. doi:10.1080/09546630701364768
8. Wolf N, Quaranta M, Prescott NJ, et al. Psoriasis is associated with pleiotropic susceptibility loci identified in type II diabetes and Crohn disease. J Med Genet. 2008;45:114–116. doi:10.1136/jmg.2007.053595
9. Grozdev I, Korman N, Tsankov N. Psoriasis as a systemic disease. Clin Dermatol. 2014;32:343–350. doi:10.1016/j.clindermatol.2013.11.001
10. Singh S, Taylor C, Kornmehl H, et al. Psoriasis and suicidality: a systematic review and meta-analysis. J Am Acad Dermatol. 2017;77:425–440. doi:10.1016/j.jaad.2017.05.019
11. Capon F. The genetic basis of psoriasis. Int J Mol Sci. 2017;18:2526. doi:10.3390/ijms18122526
12. Pollock RA, Abji F, Gladman DD. Epigenetics of psoriatic disease: a systematic review and critical appraisal. J Autoimmun. 2017;78:29–38. doi:10.1016/j.jaut.2016.12.002
13. Maresca B, Cigliano L, Spagnuolo MS, et al. Differences between the glycosylation patterns of haptoglobin isolated from skin scales and plasma of psoriatic patients. PLoS One. 2012;7:12. doi:10.1371/journal.pone.0052040
14. Łuczaj W, Wroński A, Domingues P, et al. Lipidomic analysis reveals specific differences between fibroblast and keratinocyte ceramide profile of patients with psoriasis vulgaris. Molecules. 2020;25:630. doi:10.3390/molecules25030630
15. Oestreicher JL, Walters IB, Kikuchi T, et al. Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. Pharmacogenomics J. 2001;1:272–287. doi:10.1038/sj.tpj.6500067
16. Krueger JG, Fretzin S, Suárez-Fariñas M, et al. IL-17A is essential for cell activation and inflammatory gene circuits in subjects with psoriasis. J Allergy Clin Immunol. 2012;130:145–154. doi:10.1016/j.jaci.2012.04.024
17. Blauvelt A, Chiricozzi A. The immunologic role of IL-17 in psoriasis and psoriatic arthritis pathogenesis. Clin Rev Allergy Immunol. 2018;55:379–390. doi:10.1007/s12016-018-8702-3
18. Gao Y, Yi X, Ding Y. Combined transcriptomic analysis revealed AKR1B10 played an important role in psoriasis through the dysregulated lipid pathway and overproliferation of keratinocyte. Biomed Res Int. 2017;2017.
19. Gudjonsson JE, Ding J, Johnston A, et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 2010;130:1829–1840. doi:10.1038/jid.2010.36
20. Keermann M, Kõks S, Reimann E, et al. Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC Genomics. 2015;16:322. doi:10.1186/s12864-015-1508-2
21. Mitsui H, Suárez-Fariñas M, Belkin DA, et al. Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Invest Dermatol. 2012;132:1615–1626. doi:10.1038/jid.2012.33
22. Suárez-Farinas M, Li K, Fuentes-Duculan J, et al. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012;132:2552–2564. doi:10.1038/jid.2012.184
23. Zhou X, Krueger JG, Kao MC, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi:10.1152/physiolgenomics.00157.2002
24. Kulski JK, Kenworthy W, Bellgard M, et al. Gene expression profiling of Japanese psoriatic skin reveals an increased activity in molecular stress and immune response signals. J Mol Med. 2005;83:964–975. doi:10.1007/s00109-005-0721-x
25. Reischl J, Schwenke S, Beekman JM, et al. Increased expression of Wnt5a in psoriatic plaques. J Invest Dermatol. 2007;127:163–169. doi:10.1038/sj.jid.5700488
26. Yao Y, Richman L, Morehouse C, et al. Type I interferon: potential therapeutic target for psoriasis? PLoS One. 2008;3:7. doi:10.1371/journal.pone.0002737
27. Jabbari A, Suárez-Fariñas M, Dewell S, et al. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol. 2012;132:246. doi:10.1038/jid.2011.267
28. Bigler J, Rand HA, Kerkof K, et al. Cross-study homogeneity of psoriasis gene expression in skin across a large expression range. PLoS One. 2013;8:1. doi:10.1371/journal.pone.0052242
29. Wang L, Yu X, Wu C, et al. RNA sequencing-based longitudinal transcriptomic profiling gives novel insights into the disease mechanism of generalized pustular psoriasis. BMC Med Genomics. 2018;11:52. doi:10.1186/s12920-018-0369-3
30. Tian S, Krueger JG, Li K, et al. Meta-analysis derived (MAD) transcriptome of psoriasis defines the “core” pathogenesis of disease. PLoS One. 2012;7:9. doi:10.1371/journal.pone.0044274
31. Sevimoglu T, Arga KY. Computational systems biology of psoriasis: are we ready for the age of omics and systems biomarkers? OMICS. 2015;19:669–687. doi:10.1089/omi.2015.0096
32. Sevimoglu T, Turanli B, Bereketoglu C, et al. Systems biomarkers in psoriasis: integrative evaluation of computational and experimental data at transcript and protein levels. Gene. 2018;647:157–163. doi:10.1016/j.gene.2018.01.033
33. Uzuncakmak TK, Karadag AS, Ozkanli S, et al. Alteration of tissue expression of human beta defensin-1 and human beta defensin-2 in psoriasis vulgaris following phototherapy. Biotechnic Histochem. 2020;95:243–248. doi:10.1080/10520295.2019.1673901
34. Sonkoly E, Bata-Csorgo Z, Pivarcsi A, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi:10.1074/jbc.M501704200
35. Joyce CE, Zhou X, Xia J, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. 2011;20:4025–4040. doi:10.1093/hmg/ddr331
36. Hou RX, Liu RF, Zhao XC, et al. Increased miR-155-5p expression in dermal mesenchymal stem cells of psoriatic patients: comparing the microRNA expression profile by microarray. Genet Mol Res. 2016;15:3. doi:10.4238/gmr.15038631
37. Lerman G, Avivi C, Mardoukh C, et al. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS One. 2011;6:6. doi:10.1371/journal.pone.0020916
38. Løvendorf MB, Zibert JR, Gyldenløve M, et al. MicroRNA-223 and miR-143 are important systemic biomarkers for disease activity in psoriasis. J Dermatol Sci. 2014;75:133–139. doi:10.1016/j.jdermsci.2014.05.005
39. Xu Y, Ji Y, Lan X, et al. miR-203 contributes to IL-17-induced VEGF secretion by targeting SOCS3 in keratinocytes. Mol Med Rep. 2017;16:8989–8996. doi:10.3892/mmr.2017.7759
40. Quinn JJ, Zhang QC, Georgiev P, et al. Rapid evolutionary turnover underlies conserved lncRNA–genome interactions. Genes Dev. 2016;30(2):191–207. doi:10.1101/gad.272187.115
41. Tang L, Liang Y, Xie H, et al. Long non‐coding RNAs in cutaneous biology and proliferative skin diseases: advances and perspectives. Cell Proliferat. 2020;53:e12698. doi:10.1111/cpr.12698
42. Tsoi LC, Iyer MK, Stuart PE, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi:10.1186/s13059-014-0570-4
43. Gupta R, Ahn R, Lai K, et al. Landscape of long noncoding RNAs in psoriatic and healthy skin. J Invest Dermatol. 2016;136:603–609. doi:10.1016/j.jid.2015.12.009
44. Ahn R, Gupta R, Lai K, Chopra N, Arron ST, Liao W. Network analysis of psoriasis reveals biological pathways and roles for coding and long non-coding RNAs. BMC Genomics. 2016;17(1):841. doi:10.1186/s12864-016-3188-y
45. Capell BC, Seykora JT. Loss of methylation modification marks the presence of psoriasis. J Invest Derm. 2020;140:1127–1128. doi:10.1016/j.jid.2020.01.011
46. Qiao M, Li R, Zhao X, et al. Up-regulated lncRNA-MSX2P1 promotes the growth of IL-22-stimulated keratinocytes by inhibiting miR-6731-5p and activating S100A7. Exp Cell Res. 2018;363:243–254. doi:10.1016/j.yexcr.2018.01.014
47. Li H, Yang C, Zhang J, et al. Identification of potential key mRNAs and LncRNAs for psoriasis by bioinformatic analysis using weighted gene co-expression network analysis. Mol Genet Genomics. 2020;3:1–9.
48. Ruchusatsawat K, Wongpiyabovorn J, Protjaroen P, et al. Parakeratosis in skin is associated with loss of inhibitor of differentiation 4 via promoter methylation. Hum Pathol. 2011;42:1878–1887. doi:10.1016/j.humpath.2011.02.005
49. Zhang K, Zhang R, Li X, et al. Promoter methylation status of p15 and p21 genes in HPP-CFCs of bone marrow of patients with psoriasis. Eur J Dermatol. 2009;19:141–146. doi:10.1684/ejd.2008.0618
50. Ngalamika O, Liang G, Zhao M, et al. Peripheral whole blood FOXP3 TSDR methylation: a potential marker in severity assessment of autoimmune diseases and chronic infections. Immunol Rev. 2015;44:126–136.
51. Chandra A, Ray A, Senapati S, et al. Genetic and epigenetic basis of psoriasis pathogenesis. Mol Immunol. 2015;64:313–323. doi:10.1016/j.molimm.2014.12.014
52. Gu X, Boldrup L, Coates PJ, et al. Epigenetic regulation of OAS2 shows disease-specific DNA methylation profiles at individual CpG sites. Sci Rep. 2016;6:32579. doi:10.1038/srep32579
53. Nobeyama Y, Umezawa Y, Nakagawa H. Less-invasive analysis of DNA methylation using psoriatic scales. J Dermatol Sci. 2011;83:70–73. doi:10.1016/j.jdermsci.2016.03.013
54. Zhang P, Zhao M, Liang G, et al. Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J Autoimmun. 2013;41:17–24. doi:10.1016/j.jaut.2013.01.001
55. Ovejero‐Benito MC, Reolid A, Sánchez‐Jiménez P, et al. Histone modifications associated with biological drug response in moderate‐to‐severe psoriasis. Exp Dermatol. 2018;27:1361–1371. doi:10.1111/exd.13790
56. Zhang P, Su Y, Zhao M, Huang W, Lu Q. Abnormal histone modifications in PBMCs from patients with psoriasis vulgaris. Eur J Dermatol. 2011;21:552–557. doi:10.1684/ejd.2011.1383
57. Barr RM, Wong E, Mallet AI, et al. The analysis of arachidonic acid metabolites in normal, uninvolved and lesional psoriatic skin. Prostaglandins. 1984;28:57–65. doi:10.1016/0090-6980(84)90113-8
58. Armstrong AW, Wu J, Johnson MA, et al. Metabolomics in psoriatic disease: pilot study reveals metabolite differences in psoriasis and psoriatic arthritis. F1000Research. 2014;3.
59. Kamleh MA, Snowden SG, Grapov D, et al. LC–MS metabolomics of psoriasis patients reveals disease severity-dependent increases in circulating amino acids that are ameliorated by anti-TNFα treatment. J Prot Res. 2015;14:557–566. doi:10.1021/pr500782g
60. Sitter B, Johnsson MK, Halgunset J, et al. Metabolic changes in psoriatic skin under topical corticosteroid treatment. BMC Dermatol. 2013;13:8. doi:10.1186/1471-5945-13-8
61. Ö B, Altınyazar HC, Baran H, Ünlü A. Serum homocysteine, asymmetric dimethyl arginine (ADMA) and other arginine–NO pathway metabolite levels in patients with psoriasis. Arch Dermatol Res. 2015;307:439–444. doi:10.1007/s00403-015-1553-3
62. Dutkiewicz EP, Hsieh KT, Wang YS, et al. Hydrogel micropatch and mass spectrometry–assisted screening for psoriasis-related skin metabolites. Clin Chem. 2016;62:1120–1128. doi:10.1373/clinchem.2016.256396
63. Kang H, Li X, Zhou Q, et al. Exploration of candidate biomarkers for human psoriasis based on gas chromatography‐mass spectrometry serum metabolomics. Br J Dermatol. 2017;176:713–722. doi:10.1111/bjd.15008
64. Alonso A, Julià A, Vinaixa M, et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016;14:133. doi:10.1186/s12916-016-0681-8
65. Setkowicz M, Mastalerz L, Gielicz A, et al. Lack of association of ALOX 12 and ALOX 15B polymorphisms with psoriasis despite altered urinary excretion of 12 (S)‐hydroxyeicosatetraenoic acid. Br J Dermatol. 2015;172:337–344. doi:10.1111/bjd.13225
66. Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem. 2017;409:395–410. doi:10.1007/s00216-016-9880-6
67. Walt D, Aoki-Kinoshita KF, Bendiak B, et al. Transforming Glycoscience: A Roadmap for the Future. Washington: National Academies Press; 2012.
68. Özdemir V, Arga KY, Aziz RK, et al. Digging deeper into precision/personalized medicine: cracking the sugar code, the third alphabet of life, and sociomateriality of the cell. OMICS. 2020;24:62–80. doi:10.1089/omi.2019.0220
69. Chen S, Arany I, Apisarnthanarax N, et al. Response of keratinocytes from normal and psoriatic epidermis to interferon-γ differs in the expression of zinc-α2-glycoprotein and cathepsin D. FASEB J. 2000;14:565–571. doi:10.1096/fasebj.14.3.565
70. Allen M, Ishida-Yamamoto A, McGrath J, et al. Corneodesmosin expression in psoriasis vulgaris differs from normal skin and other inflammatory skin disorders. Lab Invest. 2001;81:969–976. doi:10.1038/labinvest.3780309
71. Matsumura Y, Hori T, Nishigori C, et al. Expression of CD134 and CD134 ligand in lesional and nonlesional psoriatic skin. Arch Dermatol Res. 2003;294:563–566. doi:10.1007/s00403-002-0363-6
72. Fleischmajer R, Kuroda K, Hazan R, et al. Basement membrane alterations in psoriasis are accompanied by epidermal overexpression of MMP-2 and its inhibitor TIMP-2. J Invest Dermatol. 2000;115:771–777. doi:10.1046/j.1523-1747.2000.00138.x
73. Smetsers TF, van de Westerlo EM, Gerdy B, et al. Human single-chain antibodies reactive with native chondroitin sulfate detect chondroitin sulfate alterations in melanoma and psoriasis. J Invest Dermatol. 2004;122:707–716. doi:10.1111/j.0022-202X.2004.22316.x
74. Borská L, Fiala Z, Krejsek J, et al. Selected immunological changes in patients with Goeckerman’s therapy TNF-alpha, sE-selectin, sP-selectin, sICAM-1 and IL-8. Physiol Res. 2001;55:6.
75. Benoit S, Toksoy A, Ahlmann M, et al. Elevated serum levels of calcium‐binding S100 proteins A8 and A9 reflect disease activity and abnormal differentiation of keratinocytes in psoriasis. Br J Dermatol. 2006;155:62–66. doi:10.1111/j.1365-2133.2006.07198.x
76. Fuentes-Duculan J, Suárez-Fariñas M, Zaba LC, et al. A subpopulation of CD163-positive macrophages is classically activated in psoriasis. J Invest Dermatol. 2010;130:2412–2422. doi:10.1038/jid.2010.165
77. Shi ZR, Tan GZ, Cao CX, et al. Decrease of galectin-3 in keratinocytes: a potential diagnostic marker and a critical contributor to the pathogenesis of psoriasis. J Autoimmun. 2018;89:30–40. doi:10.1016/j.jaut.2017.11.002
78. Riggs KA, Joshi PH, Khera A, et al. Impaired HDL metabolism links GlycA, A novel inflammatory marker, with incident cardiovascular events. J Clin Med. 2019;8(12):2137. doi:10.3390/jcm8122137
79. Joshi AA, Lerman JB, Aberra TM, et al. GlycA is a novel biomarker of inflammation and subclinical cardiovascular disease in psoriasis. Circ Res. 2016;119:1242–1253. doi:10.1161/CIRCRESAHA.116.309637
80. Turan T, Akyuz AR, Aykan AC, et al. Plasma endocan levels in patients with isolated coronary artery ectasia. Angiology. 2016;67:932–936. doi:10.1177/0003319716637789
81. Balta I, Balta S, Demirkol S, et al. Elevated serum levels of endocan in patients with psoriasis vulgaris: correlations with cardiovascular risk and activity of disease. Br J Dermatol. 2013;169:1066–1070. doi:10.1111/bjd.12525
82. Nofal A, Al-Makhzangy I, Attwa E, et al. Vascular endothelial growth factor in psoriasis: an indicator of disease severity and control. J Eur Acad Dermatol Venereol. 2009;23:803. doi:10.1111/j.1468-3083.2009.03181.x
83. Fujisawa A, Egawa K, Honda Y, et al. CEA (carcinoembryonic antigen) and CEACAM6 (CEA-related cell adhesion molecul 6) are expressed in psoriasis vulgaris. Open Dermatol J. 2013;7:1. doi:10.2174/1874372220130822005
84. Honma M, Minami-Hori M, Takahashi H, Iizuka H. Podoplanin expression in wound and hyperproliferative psoriatic epidermis: regulation by TGF-β and STAT-3 activating cytokines, IFN-γ, IL-6, and IL-22. J Dermatol Sci. 2012;65:134–140. doi:10.1016/j.jdermsci.2011.11.011
85. Molteni S, Reali E. Biomarkers in the pathogenesis, diagnosis, and treatment of psoriasis. Psoriasis (Auckl). 2012;2:55.
86. Abu-Asab MS, Chaouchi M, Alesci S, et al. Biomarkers in the age of omics: time for a systems biology approach. OMICS. 2011;15:105–112. doi:10.1089/omi.2010.0023
87. Aquime Gonçalves AN, Lever M, Russo P, et al. Assessing the impact of sample heterogeneity on transcriptome analysis of human diseases using MDP webtool. Front Genet. 2019;10:971. doi:10.3389/fgene.2019.00971
88. Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–1085. doi:10.1093/bioinformatics/btt090
89. Lorincz AT. The promise and the problems of epigenetic biomarkers in cancer. Expert Opin Med Diagn. 2011;5:375–379. doi:10.1517/17530059.2011.590129
90. Haab BB. Using lectins in biomarker research: addressing the limitations of sensitivity and availability. Proteom Clin Appl. 2012;6:346–350. doi:10.1002/prca.201200014
91. Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief in Bioinform. 2018;19:1370–1381.
92. Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:1–5. doi:10.1186/s13059-017-1215-1
93. Foulkes AC, Watson DS, Carr DF, et al. A framework for multi-omic prediction of treatment response to biologic therapy for psoriasis. J Invest Dermatol. 2019;139:100–107. doi:10.1016/j.jid.2018.04.041
94. Rashmi R, Rao KS, Basavaraj KH. A comprehensive review of biomarkers in psoriasis. Clin Exp Dermatol. 2009;34:658–663. doi:10.1111/j.1365-2230.2009.03410.x
95. Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi:10.1038/nrd.2018.168
96. Greis C, Meier Zürcher C, Djamei V, et al. Unmet digital health service needs in dermatology patients. J Dermatolog Treat. 2018;29:643–647. doi:10.1080/09546634.2018.1441488
97. Ruchusatsawat K, Wongpiyabovorn J, Shuangshoti S, et al. SHP-1 promoter 2 methylation in normal epithelial tissues and demethylation in psoriasis. J Mol Med. 2006;84:175–182. doi:10.1007/s00109-005-0020-6
98. Chen M, Chen ZQ, Cui PG, et al. The methylation pattern of p16INK4a gene promoter in psoriatic epidermis and its clinical significance. Br J Dermatol. 2008;158:987–993. doi:10.1111/j.1365-2133.2008.08505.x
99. Hermann H, Runnel T, Aab A, et al. miR-146b probably assists miRNA-146a in the suppression of keratinocyte proliferation and inflammatory responses in psoriasis. J Invest Dermatol. 2017;13:1945–1954. doi:10.1016/j.jid.2017.05.012
100. Wei T, Folkersen L, Biskup E, et al. Ubiquitin‐specific peptidase 2 as a potential link between micro RNA‐125b and psoriasis. Br J Dermatol. 2017;176:723–731. doi:10.1111/bjd.14916
101. Wang R, Zhao Z, Zheng L, et al. MicroRNA-520a suppresses the proliferation and mitosis of HaCaT cells by inactivating protein kinase B. Exp Ther Med. 2017;14:6207–6212. doi:10.3892/etm.2017.5323
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
