Back to Journals » Infection and Drug Resistance » Volume 10
Occurrence of extended-spectrum and AmpC β-lactamases in multiple drug resistant Salmonella isolates from clinical samples in Lagos, Nigeria
Authors Akinyemi KO , Iwalokun BA , Oyefolu AOB, Fakorede CO
Received 1 October 2016
Accepted for publication 17 November 2016
Published 13 January 2017 Volume 2017:10 Pages 19—25
DOI https://doi.org/10.2147/IDR.S123646
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Video abstract presented by Professor Dr Akinyemi.
Views: 1989
KO Akinyemi,1 Bamidele Abiodun Iwalokun,2 Akeeb O Bola Oyefolu,1 CO Fakorede1
1Department of Microbiology, Lagos State University, Ojo, 2Molecular Biology and Biotechnology Division, Nigerian Institute of Medical Research, Yaba, Lagos, Nigeria
Purpose: Salmonella spp. are important foodborne pathogens exhibiting increasing resistance to antimicrobial drugs. Resistance to broad-spectrum β-lactams, mediated by extended-spectrum β-lactamase (ESBL) and AmpC β-lactamase enzymes is fast spreading and has had negative impacts on the clinical outcomes, particularly on third-generation cephalosporins. This study investigated the carriage of AmpC gene among multidrug-resistant Salmonella spp. from Lagos, Nigeria.
Methods: Forty Salmonella spp. from clinical samples (S. typhi = 13; S. typhimurium = 10; S. enteritidis = 8; S. choleraesuis = 5; S. paratyphi = 4) were subjected to in vitro susceptibility test by disk diffusion methods. Isolates that were resistant to cefoxitin and third-generation cephalosporins were screened for ESBL (Double Disk Synergy Test Method) and AmpC enzyme (AmpC disk test) production. Detection of AmpC fox gene was carried out by polymerase chain reaction.
Results: Thirty-two (80%) of the Salmonella isolates were cefoxitin resistant. Plasmid-mediated AmpC β-lactamase and ESBL enzymes were recorded in 10/40 (25%) and 16/40 (40%) of the Salmonella isolates, respectively. Specifically, 16/40 (40%) of the Salmonella isolates possessed 380 bp AmpC fox gene, with the highest occurrence found in S. typhi strains (43.8%) followed by S. typhimurium (25%). There was no AmpC fox gene detected in S. paratyphi strains. Interestingly, coproduction of enzymes occurred in some of the isolates, raising fears of resistance to a multitude of antibiotics in the treatment of bacterial infections.
Conclusion: Emergence of AmpC β-lactamase–producing Salmonella isolates in our environment was recorded for the first time, raising concern on increased antibiotic resistance among strains of Salmonella serovars in Lagos. Further genotypic study of the isolates could answer the questions on strain sources, clonal relatedness, and mechanism of spread.
Keywords: fox gene, plasmid, minimum inhibitory concentration, resistance, Salmonella
Introduction
Salmonella spp. are important pathogens that cause foodborne and waterborne diseases. The diseases associated with this pathogen in patients are generally treated using β-lactam antibiotics, particularly third- and fourth-generation cephalosporins. β-Lactamase production is an important mechanism of resistance to β-lactam antibiotics.1 The β-lactamase enzyme deactivates β-lactam antibiotics, thereby breaking and opening the common element in their molecular structure, that is, β-lactam. Some of these enzymes include extended-spectrum β-lactamase (ESBL), AmpC β-lactamase, and carbapenemase.2 In the last one decade, efforts to detect other β-lactamases, such as AmpC enzymes, in Klebsiella spp. and Escherichia coli isolates have been intensified, while those of Salmonella spp. seem to be scanty, particularly in developing countries. Fortunately, unlike the past two decades when the lack of standard guidelines for detecting AmpC-producing isolates was a problem, the situation is no longer the same as the expression of AmpC β-lactamases can be generated either by chromosomal or plasmid genes.3
The origin of plasmid-mediated AmpC β-lactamases was initially traceable to the chromosomes of several Enterobacteriaceae species and is rarely inducible.3 The transfer has resulted in plasmid-mediated AmpC β-lactamases in the isolates of E. coli, Klebsiella pneumoniae, Salmonella spp., Citrobacter freundii, Enterobacter aerogenes, Serratia spp., and Proteus mirabilis.4–6 There have been increasing trends in the number of infections caused by ESBL and AmpC-producing organisms. Several studies have reported that β-lactamases producing gram-negative bacteria are recovered from hospitalized patients after they receive treatment with β-lactam antibiotics such as cefoxitin and undergo a prolonged stay in hospital.4–7
Although plasmid-mediated AmpC β-lactamases had been discovered two decades ago, most clinical laboratories and physicians are still unaware of their clinical significance. The aftermath effect of this is that organisms producing these types of β-lactamases often go undetected, thus resulting in hospital outbreaks.3
In Nigeria, there is a paucity of information on the documentation of AmpC β-lactamases producing Salmonella spp. Resistance to broad-spectrum β-lactams, mediated by ESBL, is fast emerging and has had negative impacts on the clinical outcomes.5,8 The detection of AmpC- and ESBL-producing Salmonella strains is paramount, as it has both epidemiologic and therapeutic significance. Therefore, in view of the potential impact of β-lactamase–producing organisms on the clinical outcomes among infected patients in our environment, this study was undertaken to investigate the occurrence of ESBLs and AmpC enzymes among multidrug-resistant Salmonella isolates.
Materials and methods
Bacterial isolates
A total of 40 nonreplicate stock cultures of clinical Salmonella isolates from the Microbiology Division of Lagos State University and Nigerian Institute of Medical Research, Lagos, were used. The institutional review boards of Lagos State University and Nigerian Institute of Medical Research approved the study. All patients who provided samples gave written informed consent for their specimens to be used in this research. The organisms were isolated from the patients attending public hospitals; Nigerian Institute of Medical Research clinics; Lagos State University Teaching Hospitals; Central Public Health Laboratory Services, Lagos; and infectious disease hospitals, Yaba, Lagos, between February 2014 and July 2015, who were diagnosed with pyrexia of unknown origin and/or gastroenteritis. The organisms were isolated from blood and stool specimens by standard procedures and the isolates were identified by standard methods as described by Cowan and Steel.9 Confirmation of Salmonella spp. was done with API 20E identification system (Bio-Mérieux, Saint Vulbas, France). Further identification of Salmonella spp.-based somatic (O) and flagella (H) antigen characteristics was done using polyvalent antisera (Wellcome Diagnostic, Temple Hill, Dartford, UK).
Antimicrobial susceptibility tests
All Salmonella isolates were investigated for their in vitro susceptibilities to 13 antibiotics by disk diffusion method as described by the Clinical and Laboratory Standard Institute (CLSI) guidelines.10 The disks (Oxoid, Basingstoke, Hampshire, UK) with the following were used for susceptibility testing: ampicillin (25 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), tetracycline (25 μg), nalidixic acid (30 μg), ciprofloxacin (20 μg), ofloxacin (20 μg), gentamicin (10 μg), cefotaxime (30 μg), augmentin (30 µg) (amoxicillin [20 µg]/clavulanic acid [10 µg] combination), ceftriaxone (30 µg), ceftazidime (30 µg), imipenem (30 µg), levofloxacin (10 µg), and azithromycin (15 µg). Agar dilution test was also performed to determine the minimum inhibitory concentration (MIC) of some of the antibiotics tested against plasmid-mediated AmpC-carrying isolates. Each of the disk diffusion or agar dilution tests carried out was performed in triplicate, and the mean values expressed as mm or µg/mL were computed. Interpretation of results was done in accordance with the CLSI guidelines.11,12 E. coli ATCC 25922 was used for quality control of the disk diffusion and agar dilution tests. Multidrug-resistant phenotype was defined as resistance to three or more classes of antibiotics.11
Detection of ESBL production
All Salmonella isolates that exhibited resistance to third-generation cephalosporin antibiotics were screened for ESBL production by Double Disk Synergy Test Method using CLSI interpretative standard guidelines.12 An isolate was considered ESBL positive if the diameter of zone of inhibition around the clavulanate or tazobactam containing disk was at least 4 mm greater than that of ceftazidime or cefotaxime alone.12
Detection of inducible AmpC production
Disk antagonism test
Salmonella isolates that fell within the susceptibility range to cefotaxime, ceftazidime, or ceftriaxone (third-generation cephalosporins) and cefoxitin in-line with CLSI were subjected to disk antagonism test for inducible AmpC detection method described by Sanders et al.13
Detection of plasmid-mediated AmpC production
Amp disk approximation test
Isolates that yielded a cefoxitin zone diameter less than 18 mm and that were resistant to third-generation cephalosporins were further screened for AmpC enzyme production using AmpC disk test method described by Black et al14 and Yang et al.15 Control strain E. coli ATCC 25922 was included.
Molecular detection of plasmid-mediated AmpC β-lactamases
Detection of AmpC fox genes
This was done by polymerase chain reaction (PCR) using the primers designed by Pérez-Pérez and Hanson,16 with a modification in the amplification condition after PCR optimization in our laboratory. The primers were synthesized and supplied by Fermentas (Carlsbad, Canada). Each PCR is of 20 μL volume, comprising 1× PCR buffer (pH 8.3), 1.5 mM of MgCl2, 200 nM each of the deoxynucleotide triphosphates, and 40 pmol each of the forward primer 5′-AAC ATG GGG TAT CAG GGA GAT G-3′ and the reverse primer 5′-CAA AGC GCG TAA CCG GAT TGG-3′. The reaction was carried out in a Techne™ thermal cycler (TC-312; Thermo Fisher Scientific, Waltham, MA, USA) with a program that consisted of an initial denaturation at 94°C for 5 min, followed by 25 cycles of DNA denaturation at 94°C for 30 s, primer annealing at 58°C for 30 s, and primer extension at 72°C for 60 s. This was completed by a final extension step at 72°C for 7 min. Aliquots of PCR products (10 µL each) were separately loaded into wells created on 2% agarose gel prestained with ethidium bromide at 0.5 µg/mL and electrophoresed at 70 V for 1 h. This was followed by visualization on a UV transillumator. A 100 bp DNA ladder from Promega (Mannheim, Germany) was used as a marker. A fox AmpC carrying E. coli ATCC 8739 was used for quality control.
Statistical methods
Data were expressed as numbers and percentages. Comparison was done using chi-square or Fischer’s exact test for percentages and Student’s t-test for mean values. P values <0.05 were considered to be significant. Data were analyzed using the statistical package for social sciences SPSS version 15.
Results
In this study, five distinct species of Salmonella were isolated from fecal samples, while only Salmonella typhi (n=7) was recovered from blood (Figure 1). All ESBL producers showed enhanced inhibition of one or more extended-spectrum β-lactams by the disk potentiation method and exhibited a 5 mm or greater increase in the zone diameter of ceftazidime plus inhibitor with respect to ceftazidime alone. ESBL enzymes were detected in 16/40 (40%) of the Salmonella isolates. S. typhi recorded the highest number of ESBL-positive serovars (6), followed by Salmonella enteritidis (4) in this study (Table 1; Figure 2).
  | Figure 1 Distribution of Salmonella isolates in the clinical samples studied. |
  | Table 1 ESBL, AmpC production and AmpC fox gene detection and coexistence of two enzymes among the Salmonella isolates Abbreviation: ESBL, extended-spectrum β-lactamase. |
  | Figure 2 Extended-spectrum β-lactamase production by Salmonella enteritidis and Salmonella typhi in plates (A) and (B), respectively. |
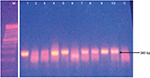
The AmpC-producing isolates demonstrated cefoxitin zone diameters <18 mm. The results showed that 32/40 (80%) of the Salmonella isolates were cefoxitin resistant. None of the isolates were positive for inducible AmpC β-lactamases. Phenotypically, plasmid-mediated AmpC β-lactamase was recorded in 10/40 (25%) Salmonella isolates. AmpC maker fox gene was used to detect plasmid-mediated AmpC β-lactamase in Salmonella isolates by molecular method, and 16/40 (40%) of the Salmonella isolates were positive (Table 1). Consequently, 10/40 (25%) of the AmpC β-lactamase producers screened phenotypically were positive, compared to 16/40 (40%) AmpC fox genes detected by PCR molecular method. All the plasmid-mediated AmpC β-lactamase Salmonella isolates possessed 380 bp AmpC fox genes (Figure 3), with the highest occurrence of fox gene found in S. typhi strains (43.8%) followed by Salmonella typhimurium (25%). There was no AmpC fox gene detected in S. paratyphi strains (Table 1). In this study, coexistence of ESBL and AmpC β-lactamases (fox gene) occurred in 3/40 (7.5%) of the isolates, mainly in two strains of S. typhi and one strain of S. enteritidis (Table 1).
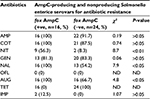
Sixteen of 40 (50%) of the cefoxitin-resistant isolates were AmpC β-lactamase producers and 50% of non-AmpC Salmonella isolates were also recorded. In all, 60% (24/40) of the Salmonella isolates were non-AmpC β-lactamase producers. On comparison of the resistance ability of fox gene-positive and -negative isolates to other antibiotics tested, data presented in Table 2 show significant association (P<0.05) between fox AmpC carriage and resistance to nitrofurantoin, nalidixic acid, and augmentin (amoxicillin/clavulanic acid). The mean MIC values of antibiotics used against fox AmpC and ESBL Salmonella isolates varied among the serotypes. For example, an average MIC value of 0.19 µg/mL was obtained for levofloxacin against fox AmpC Salmonella choleraesuis producer similar to S. enteritidis (Table 3). There was a general increase in the average MIC values recorded for most of the antibiotics evaluated against Salmonella serotypes with dual carriage of fox AmpC and ESBL, compared with fox AmpC carriage alone. For example, there was an increase in the mean MIC values for ceftazidine and cotrimoxazole from 7.1 and 9.7 µg/mL against fox AmpC S. typhi single carriage to 19.4 and 11.4 µg/mL against dual carriers of fox AmpC and ESBL S. typhi, respectively (Table 3).
Discussion
The prevalence of ESBL among Salmonella serotypes in this study was 16/40 (40%). This result is on the high side when compared with earlier studies conducted in Nigeria and elsewhere, particularly among the bacterial genera of the family Enterobacteriaceae. For example, in two separate studies conducted in Kano State, Nigeria by Yusha’u et al17 and Yusuf and Haruna,5 ESBL prevalence of 9.3% and 15.8%, respectively, was recorded in gram-negative bacteria. This is an indication of the increasing trend of ESBL in the clinical settings in our environment. In our previous report, ESBL production due to blaCTX-M-I gene cluster was detected in 45.8% of Salmonella isolates.18 This finding further affirmed the circulation of ESBL-producing Salmonella spp. in our locality.
In this study, none of the isolates was positive for inducible AmpC β-lactamases, thus ruling out the presence of chromosomal-mediated AmpC β-lactamase resistance in the Salmonella isolates evaluated. Consequently, 10/40 (25%) of the AmpC β-lactamase producers screened phenotypically were positive, compared to 16/40 (40%) AmpC fox genes detected by PCR molecular method. The observed difference in the number of AmpC β-lactamase producers by the molecular and phenotypic methods used may be attributed to the false-negative and false-positive results associated with the phenotypic method. It has been documented that possible misleading results arising from the phenotypic tests for AmpC are one of the biggest challenges associated with its specificity and sensitivity, which may impact negatively on the accuracy of surveillance and hospital infection control measures.19
Plasmid-mediated AmpC β-lactamases were first reported in the late 1980s, but till date, many infectious disease personnel are still unaware of their clinical significance due to lack of surveillance or inaccurate laboratory testing. The number of infections caused by AmpC-producing pathogens in the family Enterobacteriaceae is increasing in several countries like Italy,20 Indian,4 Egypt,6 and Japan.21 All the plasmid-encoded AmpC β-lactamase Salmonella isolates detected in this study possessed 380 bp AmpC fox genes.
The overall results of 40% AmpC β-lactamase producers detected by fox group genes are on the high side. However, scanty data on the prevalence of plasmid-mediated AmpC β-lactamase-producing Salmonella spp. in this part of the world and elsewhere had made objective comparison of these results very difficult. Nevertheless, we recorded the highest occurrence (43.8%) of AmpC fox genes in S. typhi strains followed by S typhimurium (25%). Our finding is similar to that of the study carried out by El-Hady and Adel6 who reported 30.4% AmpC β-lactamase producers detected by fox group genes, 56.5% by cit group genes (including CMY-2), and 73.9% by mox group genes (including CYM-1) in the clinical isolates of Enterobacteriaceae. There was no AmpC fox gene detected in S. paratyphi strains (Table 2). Interestingly, we observed varying capability of Salmonella serotypes to produce AmpC and express fox genes. For example, AmpC fox gene in S. typhi was found among three ESBL positives, but not expressed. In S. typhimurium, two of the ESBL-positive isolates also carried AmpC fox gene, but did not express detectable level by the AmpC disk test. These observations may be due to other possible AmpC types and/or mechanisms mediating cefoxitin resistance in Salmonella isolates. In S. choleraesuis, fox AmpC gene carriage was not detected by PCR. Cefoxitin resistance found in ESBL-positive isolates (n=2) could have been mediated by other mechanisms, including other AmpC β-lactamases not analyzed in this study.
To the best of our knowledge, this is the first report to describe the detection of plasmid-mediated AmpC Salmonella spp. in Lagos and, thus, calls for serious concerns.
Coexistence of ESBL and AmpC β-lactamases (fox gene) was detected in three (9.1%) isolates, particularly in S. typhi and S. enteritidis strains (Table 1). The dual carriage of the fox AmpC gene and the phenotypic expression of ESBL observed in S. enteritidis and S. typhi isolates resulted in enhanced ceftazidime resistance (>2-fold increase), which was statistically significant (P<0.05) when compared with fox AmpC gene carriage alone. Reduced, but not significant (P>0.05) susceptibility to levofloxacin, imipenem, cotrimoxazole, and gentamicin was also observed among these isolates (P>0.05; Table 3). The coexistence of bacteria in the family Enterobacteriaceae has been documented. For instance, Yusuf and Haruna5 reported a prevalence of 5.8% in E. coli. Similar observations were observed in Klebsiella spp. (11.1%) and Proteus spp. Sinha et al22 recorded 8% coexistence among E. coli. Also, coexistence of ESBL blaSHV28 and the AmpC blaCMY-2 in K. pneumoniae isolated from animals has been documented in Italy.20 Therefore, the observed coproduction of ESBL and AmpC β-lactamases in our isolates suggests that ESBL coexistence with AmpC works synergistically and increases the MIC (Table 3). It also suggests the possible spread of plasmid-mediated multiple drug resistance (MDR) isolates to aminoglycosides, trimethoprin–sulfamethoxazole, and fluoroquinolones.
The inability to use other AmpC makers such as mox group genes, cit group genes, and ebc group genes to accommodate other most common types of plasmid-mediated AmpC β-lactamases is one of the limitations of this study.
Conclusion
This study revealed the occurrence of plasmid-mediated AmpC β-lactamases producing MDR Salmonella isolates in our environment for the first time. It also showed the coexistence of ESBL and AmpC β-lactamases (fox gene) in some strains of Salmonella isolates. A need to screen the ESBL potential of MDR Salmonella isolates for plasmid-mediated AmpC β-lactamases, along with performing antibiogram is advocated before making therapeutic decisions in patient management in our hospitals. This is essential to prevent treatment failure and stem mortality. Further investigations are required on other families of AmpC β-lactamase–encoded genes, to study the mechanisms of cefoxitin resistance among Salmonella isolates, and to sequence fox AmpC genes detected for source attribution. Therefore, application of molecular typing techniques such as multilocus sequencing techniques and pulse field gel electrophoresis on plasmid-mediated AmpC β-lactamases and ESBL-producing strains of Salmonella enterica serovars is envisaged. This is essential in order to assess the levels of homogeneity and heterogeneity and for better understanding of the mechanisms of antibiotic resistance and spread.
Acknowledgments
We are grateful to the staff of Department of Microbiology and both the managements of Lagos State University and Nigerian Institute of Medical Research for providing consumables and technical support, respectively. The abstract for this study was presented at the Interscience Conference on Antimicrobial Agents and Chemotherapy in San Diego, California, USA September 17–21, 2015 and was published online at icaac.posterview.com. The presentation of this study at ICAAC/ICC 2015 in conjunction with ASM Program was sponsored by Bill & Melinda Gates Foundation ICAAC travel grant award for scientists from low-income countries.
Disclosure
The authors report no conflicts of interest in this work.
References
Adak GK, Long SM, O’Brien SJ. Trends in indigenous foodborne disease and dealths, England and Wales, 1992 to 2000. Gut. 2002;51(6);832–841. | ||
Bush K, Jacoby G, Medeiros A. A functional classification scheme for b-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother.1995;39(6):1211–1233. | ||
Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type lactamases. Antimicrob. Agents Chemother. 2002:46(1):1–11. | ||
Mohamudha Parveen R, Harish BN, Parija SC. AmpC beta-lactamases among gram negative clinical isolates from a tertiary hospital, south India. Braz J Microbiol. 2010;41(3):596–602. | ||
Yusuf I, Haruna M. Detection of AmpC and ESBL producers among enterobacteriaceae in a tertiary health care in Kano, Nigeria. Int J S Technol. 2013;3:220–225. | ||
El-Hady SA, Adel LA. Occurrence and detection of AmpC b-lactamases among Enterobacteriaceae isolates from patients at Ain Shams University Hospital. Egypt J Med Hum Genet. 2015;16:239–244. | ||
Manchanda V, Singh NP, Shamweel A, Eideh HK, Thukral SS. Molecular epidemiology of clinical isolates of AmpC producing Klebsiella pneumoniae. Indian J Med Microbiol. 2006;24(3):177–181. | ||
Akinyemi KO, Iwalokun BA, Agboyinu JA, Ogunyemi O, Fassure AK. Emergence of third generation cephalosporin resistance and typing by random amplified polymorphic DNA (RAPD) among clinical Salmonella isolates from Lagos, Nigeria. Br Microbiol Res J. 2014;4(6):668–678. | ||
Barrow GI, Feltham RKA, editors. Cowan and Steel’s Manual for the Identification of Medical Bacteria. 3rd ed. Cambridge, UK: Cambridge University Press; 1993:331. | ||
National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing: Ninth Informational Supplement. version 19. Wayne, PA, USA: National Committee for Clinical Laboratory Standards, 1999;1–112 (NCCLS document no. M-100-S9). | ||
National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing: fifth informational supplement: M100-S5. National Committee for Clinical Laboratory Standards, 1994. Wayne, Pa. | ||
Clinical and Laboratory Standards Institute. Performanc Standards for Antimicrobial Susceptibility Testing. Nineteenth Informational Supplement. CLSI document M100-S21. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2011. | ||
Sanders CC, Sanders WE, Goering HV. In vitro antagonism of _beta-lactam antibiotics by cefoxitin. Antimicrob Agents Chemother. 1982;21(6):968–975. | ||
Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid- mediated AmpC b-lactamases in Enterobacteriaceae lacking chromosomal AmpC β-lactamases. J Clin Microbiol. 2005;43(7):3110–3113. | ||
Yang J, Wang M, Cheng A, Pan K, Li C, Deng S. A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection. World J Gastroenterol. 2008;14(18):2872–2876. | ||
Pérez-Pérez FJ, Hanson ND. Detection of plasmid mediated AmpC b-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. | ||
Yusha’u M, Olonitola SO, Aliyu BS. Prevalence of Extended–Spectrum Beta lactamases (ESBLs) Among members of the Enterobacteriaceae isolates obtained from Mohammed Abdullahi Wase Specialist Hospital, Kano, Nigeria. Int J Pure Appl Sci. 2007;1(3):42–48. | ||
Akinyemi KO, Iwalokun BA, Alafe OO, Mudashiru SA, Fakorede C. blaCTX-M-1group extended spectrum beta lactamase-producing Salmonella typhi from hospitalized patients in Lagos, Nigeria. Infect Drug Resist. 2015;8:99–106. | ||
Handa D, Pandey A, Asthana A, Rawat A, Handa S, Thakuria B. Evaluation of phenotypic tests for the detection of AmpC betalactamase in clinical isolates of Escherichia coli. Indian J Pathol Microbiol. 2013;56(2):135–138. | ||
Donati V, Feltrin F, Hendriksen RS, et al. Extended spectrum-beta-lactamases, AmpC beta-lactamases and plasmid mediated quinolone resistancein klebsiella spp. from companion animals in Italy PLoS One. 2014;9(3):1–7. | ||
Yamasaki K, Komatsu M, Abe N, et al. Laboratory surveillance for prospective plasmid-mediated AmpC beta-lactamases in the Kinki region of Japan. J Clin Microbiol 2010;48(9):3267–3273. | ||
Sinha P, Rajni S, Suman R, Raman S, Smita S, Deepali P. Prevalence of extended spectrum beta lactamase and AmpC beta lactamase producers among Escherichia coli isolates in a tertiary care hospital in Jaipur. Indian J Pathol Microbiol. 2008;51(3):367–369. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.