Back to Journals » OncoTargets and Therapy » Volume 10
Nuclear DDX3 expression predicts poor outcome in colorectal and breast cancer
Authors Heerma van Voss MR, Vesuna F, Bol GM, Meeldijk J, Raman A, Offerhaus GJ, Buerger H, Patel AH , van der Wall E, van Diest PJ, Raman V
Received 28 April 2017
Accepted for publication 10 June 2017
Published 17 July 2017 Volume 2017:10 Pages 3501—3513
DOI https://doi.org/10.2147/OTT.S140639
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Samir Farghaly
Marise R Heerma van Voss,1,2 Farhad Vesuna,2 Guus M Bol,1,2 Jan Meeldijk,1,3 Ana Raman,4 G Johan Offerhaus,1 Horst Buerger,5 Arvind H Patel,6 Elsken van der Wall,7,8 Paul J van Diest,1,8 Venu Raman1,2,8
1Department of Pathology, University Medical Center Utrecht, Utrecht, the Netherlands; 2Department of Radiology, Johns Hopkins University School of Medicine, Baltimore, MD, USA; 3Laboratory of Translational Immunology, University Medical Center Utrecht, Utrecht, the Netherlands; 4Department of Pharmacology, Johns Hopkins University School of Medicine, Baltimore, MD, USA; 5Institute of Pathology, Paderborn, Germany; 6Centre for Virus Research, MRC-University of Glasgow, Glasgow, UK; 7Cancer Center, University Medical Center Utrecht, Utrecht, the Netherlands; 8Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Purpose: DEAD box protein 3 (DDX3) is an RNA helicase with oncogenic properties that shuttles between the cytoplasm and nucleus. The majority of DDX3 is found in the cytoplasm, but a subset of tumors has distinct nuclear DDX3 localization of yet unknown biological significance. This study aimed to evaluate the significance of and mechanisms behind nuclear DDX3 expression in colorectal and breast cancer.
Methods: Expression of nuclear DDX3 and the nuclear exporter chromosome region maintenance 1 (CRM1) was evaluated by immunohistochemistry in 304 colorectal and 292 breast cancer patient samples. Correlations between the subcellular localization of DDX3 and CRM1 and the difference in overall survival between patients with and without nuclear DDX3 were studied. In addition, DDX3 mutants were created for in vitro evaluation of the mechanism behind nuclear retention of DDX3.
Results: DDX3 was present in the nucleus of 35% of colorectal and 48% of breast cancer patient samples and was particularly strong in the nucleolus. Nuclear DDX3 correlated with worse overall survival in both colorectal (hazard ratio [HR] 2.34, P<0.001) and breast cancer (HR 2.39, P=0.004) patients. Colorectal cancers with nuclear DDX3 expression more often had cytoplasmic expression of the nuclear exporter CRM1 (relative risk 1.67, P=0.04). In vitro analysis of DDX3 deletion mutants demonstrated that CRM1-mediated export was most dependent on the N-terminal nuclear export signal.
Conclusion: Overall, we conclude that nuclear DDX3 is partially CRM1-mediated and predicts worse survival in colorectal and breast cancer patients, putting it forward as a target for therapeutic intervention with DDX3 inhibitors under development in these cancer types.
Keywords: RNA helicase, DDX3X, nuclear export, CRM1
Introduction
DEAD box protein 3 (DDX3), also known as DDX3X, is a member of the DEAD box RNA helicase family of proteins. Like other family members, it has ATPase-dependent helicase activity, which allows for the unwinding of double-stranded RNA and restructuring of complex secondary RNA structures.1 DDX3 has thus been associated with several steps of endogenous RNA metabolism, such as splicing,2,3 nuclear mRNA export,4,5 RNA interference,6,7 ribosomal assembly and translation initiation.5,8 Recent studies have also indicated a role for DDX3 in neoplastic transformation. Functional studies have shown that DDX3 has antiapoptotic properties,9–11 and is necessary for cell cycle progression12,13 and invasion.14,15 The tumor-enhancing role of cytoplasmic DDX3 was corroborated by studies on DDX3 expression in patient cancer samples.6,13
DDX3 is known to shuttle between cytoplasm and nucleus,4 but in most human tissues and cell lines it is localized predominantly in the cytoplasm. We noticed that in a subgroup of colorectal and breast cancers, DDX3 can also be observed in the nucleus. It remains to be elucidated what exact role DDX3 plays in the nucleus of cancer cells and how the subcellular localization of DDX3 is regulated in these cells. It is known that DDX3 is exported out of the nucleus as part of mRNP complexes associated both with the tip-associated protein (TAP)-dependent bulk mRNA export pathway5,16 and the alternative chromosome region maintenance 1 (CRM1)-dependent pathway.4 However, the exact nature of the relation between DDX3 expression and expression of the nuclear exporter CRM1 requires further mechanistic exploration and validation in patient samples.
Apart from the biological relevance of understanding the nuclear role of DDX3 in cancer cells, identification of nuclear DDX3 as a prognostic and therapeutic biomarker could facilitate selection of patients who would benefit from adjuvant treatment, specifically with DDX3 inhibitors that are under development.12,17,18 In this study we, therefore, evaluated the correlation between nuclear DDX3 expression and survival in cohorts of breast and colorectal cancer patient samples. Because DDX3 is known to bind the nuclear exporter CRM1,4 we sought to determine whether nuclear retention of DDX3 can be explained by aberrant CRM1 expression. Finally, we carried out in vitro experiments to functionally validate mechanisms of nuclear DDX3 retention.
Materials and methods
Patient samples
Tissue microarrays (TMAs) with samples from 72 colorectal cancer patients from the Academic Medical Center, Amsterdam,19 292 colorectal cancers from Paderborn Germany13 and 315 breast cancers from the UMC Utrecht were used.20 All TMAs included multiple cores per patient. Missing cases were due to damaged or detached cores during cutting or staining, or due to cores not containing invasive carcinoma. Clinicopathological data were retrieved from the pathology report and patient files. The breast cancer cases in this cohort were classified into molecular subtypes as were described before.21,22 We used anonymous archival leftover pathology material. Therefore, no ethical approval or informed consent is required according to Dutch legislation,23 as this use of redundant tissue for research purposes is part of the standard treatment agreement with patients in our hospitals.24 The medical research ethics committee of the UMC Utrecht confirmed that official approval of this study is not required (reference number WAG/mb/16/029330).
Immunohistochemistry
Four μm thick sections were cut, mounted on SuperFrost slides (Menzel & Glaeser, Brunswick, Germany), deparaffinized in xylene and rehydrated in decreasing ethanol dilutions. Endogenous peroxidase activity was blocked with 1.5% hydrogen peroxide buffer for 15 minutes and was followed by antigen retrieval by boiling for 20 minutes in 10 mM citrate buffer (pH 6.0) for DDX3 and ethylenediaminetetraacetic acid buffer (pH 9.0) for CRM1. Slides were subsequently incubated in a humidified chamber for 1 hour with anti-DDX3 (1:1,000, pAb r647)25 or anti-CRM1 (1:500, ab84375; Abcam, Cambridge, UK). After washing with phosphate-buffered saline, slides were incubated with poly-HRP-anti-mouse/rabbit/rat IgG (Brightvision; Immunologic, Duiven, the Netherlands) as a secondary antibody for 30 minutes at room temperature. Peroxidase activity was developed with diaminobenzidine and hydrogen peroxide substrate solution for 10 minutes. The slides were lightly counterstained with hematoxylin and mounted. Appropriate positive and negative controls were used throughout.
Scoring was performed by consensus of two observers (P.v.D. and M.H.v.V. or G.B.). Although the intensity of nuclear DDX3 in cells was similar, the fraction of positive cells varied. Therefore, the percentage of DDX3-positive nuclei was scored. The optimal cutoff point was selected using the online tool cutoff finder, which helps to select a cutoff that maximizes the difference in survival between groups.26 Samples with ≥1% DDX3 staining were regarded positive. Almost all cells expressed cytoplasmic DDX3, but the intensity varied and was, therefore, scored semiquantitatively as absent (0), low (1), moderate (2) or strong (3). Cases with score 0–2 were classified as having low DDX3 expression and evaluated against cases with strong expression as before.13 Nuclear CRM1 was scored using a semiquantitative score that was previously described by Noske et al.27 The percentage of positive cells was scored as 0 (0%), 1 (<10%), 2 (10%–50%), 3 (51%–80%) or 4 (>80%). The intensity of positive nuclei was scored on the same scale as cytoplasmic DDX3. An overall score was calculated by multiplying the two scores. Cases with a score of 9 and higher were considered to have high nuclear CRM1. Cytoplasmic CRM1 was scored as absent or present.
Statistics
Clinicopathological characteristics were compared between tumors with and without nuclear DDX3. Discrete variables were compared by χ2 or Fisher’s exact test and relative risks (RRs) were calculated with 95% CI. The distribution of continuous variables was assessed graphically. Transformation was performed where indicated. Student’s t-test and Mann–Whitney U-tests were calculated for normal and non-normal distributed variables, respectively. Survival in patients with and without nuclear DDX3 was compared by plotting Kaplan–Meier curves and log-rank tests. Multivariate analysis was performed by including all factors that were found predictive (P<0.1) in univariate analysis in a Cox-proportional hazard model. Stepwise backward selection based on Akaike Information Criterion was used to select the final model. Effect modifiers were identified by including multiplicative interaction terms into the model. P-values smaller than 0.05 were considered statistically significant. All statistical analyses were performed with R version 3.2.0.
Cloning and transfections
A pEGFP-C1 vector containing a parental DDX3 construct (GenBank Accession: AF061337) N-terminally fused to a 6xHis tag and EGFP sequence was used. An oligo containing three times the SV40 nuclear localization signal (NLS) sequence (5′-GATCCAAAAAAGAAGAGAAAGGTA-3′, AA: DPKKKRKV) with flanking restriction sites was annealed and subcloned into the parental vector to create an NLS-EGFP-6HIS-DDX3 and an EGFP-NLS-6HIS-DDX3 construct. The DDX3 deletion constructs were created by selective amplification of the parental DDX3 construct lacking the deleted area with Phusion High-Fidelity Polymerase (New England Biosciences, Ipswich, MA, USA), followed by Dpn1 digestion of the bacterial backbone, gel purification, phosphorylation and religation. All constructs were verified by sequencing before usage.
HeLa cells were chosen for DDX3 localization as before.4 The cell line was purchased from ATCC and STR-profiled in November 2015. 2×104 HeLa cells were plated in a 24-well plate. After 24 hours, the cells were transfected with 250 ng DDX3 construct, 50 ng H2B-mCherry construct and 1 μL TransIT-LT1 transfection reagent. After 12–16 hours, the cells were treated with 10 nM Leptomycin B (Sigma, St Louis, MO, USA) for 4 hours after which the number of DDX3-positive nuclei per high-power field was counted and the cells were imaged with a confocal Zeiss 780FCS system.
Results
Colorectal cancer patients with nuclear DDX3 have worse overall survival
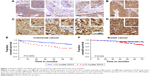
DDX3 expression could be scored in 304 out of 364 colorectal cancers, of which 34.9% had >1% nuclear DDX3. As shown in Figure 1, the percentage of cells with nuclear DDX3 varied. Some tumors expressed nuclear DDX3 in all cells, sometimes cells with nuclear DDX3 lay dispersed among negative cells. Heterogeneity between different areas of a tumor (different cores) was also observed. DDX3 was sometimes expressed in the nucleus of plasma cells and fibroblasts as well. Only DDX3 in the nuclei of cancer cells was considered for this analysis. As shown in Table 1, tumors with nuclear DDX3 more often had a larger tumor size (P=0.006). Some tumors expressed DDX3 both in the cytoplasm and the nucleus. The presence of nuclear DDX3 correlated negatively with cytoplasmic DDX3 (RR 0.62, P=0.005), which was seen in 125/304 (41%) of cases (Figure 1B).
Patients with nuclear DDX3 expression had a significantly decreased 5-year overall survival rate of 51.2%, as compared to 73.7% in those without nuclear DDX3 (hazard ratio [HR] 2.34, P=0.0005; Figure 1E). Other usual predictors of survival were tumor, node, metastasis (TNM) stage (P<0.001), differentiation grade (P=0.040) and tumor size (P=0.004). Nuclear DDX3 expression was retained in a Cox-proportional hazards model with TNM stage (HRadjusted 1.69, P=0.057; Table S1). No significant association between high cytoplasmic DDX3 and survival was observed in colorectal cancer patients (HR 0.69, P=0.160; Figure S1).
Nuclear DDX3 is associated with worse overall survival in breast cancer patients
To assess whether our finding that nuclear DDX3 correlates with survival was also applicable to other tumors, we analyzed a cohort of 315 consecutive breast cancer cases. DDX3 expression could be evaluated in 292 breast cancer patients, of which 141 (48.3%) had nuclear DDX3 (Figure 1C). As shown in Table 2, these patients more often exhibited ductal histology (P=0.006), higher grade (P=0.025), larger tumor size (P=0.046) and positive lymph nodes (P=0.003). In addition, a trend was observed for higher mitotic activity index (P=0.058). Nuclear localization of DDX3 did not correlate with cytoplasmic DDX3 expression in breast cancer patients (RR 0.91, P=0.493; Figure 1D). Cytoplasmic DDX3 expression in this breast cancer cohort will be discussed more elaborately in a separate report.
As seen in Figure 1F, similar to colon cancer patients, breast cancer patients with nuclear DDX3 had a worse 5-year survival rate (75%) than those without (90%; HR 2.39, P=0.004). Other variables that were significant predictors of poor survival were basal-like subtype (P=0.024), positive lymph node status (P=0.027), negative estrogen receptor (ER) status (P=0.019), negative progesterone receptor (PR) status (P=0.013) and age over 50 (P=0.017). Nuclear DDX3 remained a significant predictor (HRadjusted 2.63, P=0.010) in a multivariate Cox-proportional hazards model with basal-like subtype, lymph node status and age (Table S2).
The subcellular localization of the nuclear exporter CRM1 correlates with DDX3 localization
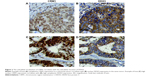
Because DDX3 is known to bind the nuclear exporter CRM1 we ascertained whether nuclear retention of DDX3 could be explained by aberrant CRM1 expression (Figure 2). CRM1 expression was mainly observed in the nucleus. High nuclear CRM1 expression was observed in 18% of colorectal cancer and 27% of breast cancer cases. Cytoplasmic expression was observed as well, more commonly in breast (36%) than colorectal cancer (8%). As shown in Figure 2A–B and Table 3, nuclear DDX3 was significantly associated with the presence of cytoplasmic CRM1 in colorectal cancers (RR 1.67, P=0.040). In breast cancer, no significant correlation between nuclear DDX3 and CRM1 expression was observed, suggesting that aberrant CRM1 expression could only partly explain nuclear DDX3 retention. However, a strong correlation between the intensity of cytoplasmic DDX3 and CRM1 expression (Figure 2C and D) was observed: nuclear CRM1 was higher in tumors with high cytoplasmic DDX3 in both colorectal cancer (RR 1.77, P<0.001) and breast cancer cases (RR 1.75, P=0.003). Contrary to what was observed in colorectal cancer, a significant correlation between cytoplasmic CRM1 and high cytoplasmic DDX3 was observed in breast cancer (RR 2.45, P<0.001).
Nuclear DDX3 localization is due to decreased CRM1-mediated export and increased import
To characterize the functional role of DDX3 in the nucleus, we made an attempt to mimic the nuclear DDX3 expression observed in patients in an in vitro setting. We generated a GFP-DDX3 fusion protein to study nucleocytoplasmic shuttling properties of DDX3 (Figure 3A). DDX3 is expressed primarily in the cytoplasm of HeLa cells (Figure 3B and C). Leptomycin B specifically inhibits CRM1 by covalent modification of Cys-529.28,29 When export of DDX3 by CRM1 was inhibited with leptomycin B, an increase in nuclear DDX3 was observed in 46% of cells, but the majority of DDX3 remained in the cytoplasm (Figure 3B and C).
Next we determined whether high nuclear DDX3 levels could potentially be explained by increased nuclear import of DDX3. We fused three tandem repeats of the SV40 NLS to the GFP-DDX3 construct (Figure 3A). Only a minor increase of nuclear DDX3 was observed in 20% of cells with the SV40 NLS, indicating that nuclear export of DDX3 is very efficient and not saturated. However, when export of the GFP-NLS-DDX3 construct was inhibited by leptomycin B, DDX3 localized strongly to the nucleus in 100% of cells, whereas cytoplasmic DDX3 was reduced to 25% of cells (Figure 3B and C). These data show that an increase in DDX3 import in combination with reduced CRM1-mediated export can result in high nuclear DDX3 expression. Experiments with an NLS-GFP-DDX3 construct yielded similar results. Unfortunately, it was not possible to create a DDX3 construct that localized to the nucleus spontaneously without inhibition of CRM1. We were, therefore, unable to study functional effects of nuclear DDX3 in an isolated fashion. However, the created constructs can be used for proof-of-principle studies on regulation of the subcellular DDX3 localization.
DDX3 localizes to the nucleolus
High expression levels of DDX3 in the nucleus allowed for closer examination of the subnuclear expression pattern of DDX3. As can be observed in Figure 3C, DDX3 is expressed diffusely within the nucleus, but is often most intense in nucleoli. Nucleolar DDX3 was seen in 2%–6% of cells after addition of either an NLS or leptomycin B. When import was increased and export was inhibited simultaneously, 30% of cells had clear localization to the nucleolus. This expression pattern resembled that observed in patient samples, although DDX3 expression also was strong at the nuclear membrane in those (Figure 1A and C).
Influence of CRM1 inhibition on subcellular localization of DDX3 mutants
CRM1 is known to export cargo from the nucleus by binding to a leucine-rich nuclear export signal,30 which is conserved in the first 21 amino acids of the Ded1/DDX3 subfamily of DEAD box RNA helicases.31 However, binding studies indicated that CRM1 binding occurs between amino acid 260 and 517 of DDX3.4 We evaluated whether deletion of either the N-terminal nuclear export signal (NES) domain or the 260–517 region of DDX3 results in increased retention of DDX3 in the nucleus, and whether these proteins were still responsive to CRM1 inhibition. As shown in Figure 3, deletion of the first 21 amino acids containing the NES resulted in 46% of cells having DDX3 in the nucleus. No further increase was observed after leptomycin B treatment and the expression pattern was comparable to that of wild-type DDX3 after CRM1 inhibition, indicating that deletion of the N-terminal NES largely abrogates nuclear export by CRM1.
Deletion of amino acids 260–517 resulted in a speckled DDX3 nuclear expression pattern in almost all cells, but the intensity of DDX3 remained strongest in the cytoplasm. When CRM1 was inhibited this pattern shifted to all cells having intense nuclear DDX3 expression, whereas only 7% of cells showed cytoplasmic DDX3 (Figure 3C). This indicates that although the 260–517 region has influence on the subcellular localization of DDX3, DDX3 localization is still responsive to CRM1 inhibition after deletion of this region.
Discussion
In this study we evaluated the relationship between nuclear DDX3 expression and survival in breast and colorectal cancer. We found the presence of nuclear DDX3 to be an independent predictor of worse survival in both colorectal and breast cancer. Mechanistically, in colorectal cancer nuclear DDX3 retention could in part be explained by dysregulation of the nuclear exporter CRM1, but no correlation between nuclear DDX3 and altered CRM1 expression could be observed in breast cancer. We functionally validated this finding in vitro by showing that inhibition of CRM1 with leptomycin B resulted in an increase in nuclear DDX3 levels. Analysis of the subcellular localization of DDX3 deletion mutants before and after CRM1 inhibition indicated that the N-terminal NES sequence of DDX3 is most important for this interaction. A much stronger increase in nuclear DDX3 retention was observed after addition of an NLS in combination with CRM1 inhibition, suggesting that nuclear DDX3 localization can also be regulated through nuclear import, by unknown mechanisms. Interestingly, we found DDX3 to strongly localize to the nucleolus in vitro, which resembled the expression pattern in patient samples.
DDX3 is an actively investigated molecule in cancer biology, but previous studies have focused on its cytoplasmic expression pattern in cancer cells.32 Understanding the role of nuclear DDX3 expression in tumors is relevant given DDX3’s function in RNA processing and its known nucleocytoplasmic shuttling capacities. DDX3 has been found to promote oncogenesis and DDX3 inhibitors are being developed for colorectal13 and breast cancers17 among other malignancies.12,18,33,34 In addition, DDX3 is essential for the nuclear export of the human immunodeficiency virus 1 (HIV-1) protein Rev and is, therefore, also a potential therapeutic target in the treatment of HIV infections.35,36 Understanding the cellular mechanisms behind increased nuclear retention of DDX3 may facilitate the development of therapies specifically targeting the export function of DDX3.37 In addition, nuclear DDX3 could serve as a prognostic and potentially therapeutic biomarker for selecting cancer patients that may benefit from treatment with DDX3 inhibitors.
This is the first study to describe prognostic value of DDX3 expression specifically in the nucleus. Several other studies have reported a correlation between cytoplasmic DDX3 and survival in breast and colorectal cancers.32 A study by Su et al found no significant difference in breast cancer patients with high and low cytoplasmic DDX3 expression38 and reported a correlation between low cytoplasmic DDX3 expression and worse survival in colorectal cancer patients. In our larger colorectal cancer cohort, patients with high cytoplasmic DDX3 also did slightly better (HR 0.69), but this was not statistically significant. Potential explanations for the observed differences are the use of different cutoffs for positivity and different antibodies. We previously found cytoplasmic DDX3 to be associated with nuclear beta-catenin expression in patient samples and to promote oncogenic Wnt signaling. Although most colorectal cancers are driven by genetic alterations in the Wnt-signaling pathway, only a subset of cancers shows strong nuclear beta-catenin expression. Interestingly, this subgroup of colorectal cancer patients has a relatively favorable prognosis,39 explaining how cytoplasmic DDX3 can be driving Wnt signaling, without being associated with worse overall survival. As DDX3 spontaneously localizes to the nucleus only in a very small percentage of cells in vitro and we were unable to create a stable cell line overexpressing full-length DDX3 in the nucleus, it is very hard to decipher the exact role of DDX3 in the nucleus on oncogenesis. However, the fact that the presence of DDX3 in the nucleus was associated with worse survival in two different cancer types indicates that cancers may benefit from high nuclear DDX3 levels as well.
When trying to understand a protein’s nuclear function, it is useful to know its location in the nucleus. High DDX3 expression was specifically seen in the nucleolus. This was previously observed after HIV-1 Tat and Rev overexpression40,41 and DDX3 was also identified in nucleolar extracts by proteomics.42 The nucleolus is the structural-functional domain of the cell in which ribosomal biogenesis occurs. Prominent nucleoli have been recognized as a cytological hallmark of cancer as early as the 19th century,43 but recently received renewed attention as evidence accumulates that several onco- and tumor suppressor genes are directly involved in the regulation of ribosome production to meet the altered metabolic needs of cancer cells.44 Interestingly, DDX3 is known to play a role in ribosomal assembly and translation initiation in the cytoplasm.45 Our observation that DDX3 localizes specifically to the nucleolus, and that this feature corresponds with worse prognosis, indicates that DDX3 may also play a role in pre-ribosomal assembly in the nucleolus. The presence of high nuclear DDX3 could reflect increased protein synthesis demands in cancers. A recent study identified essential genes in hematological malignancies and found this group to be enriched in RNA processing genes including DDX3X. Many of these essential genes localized to the nucleolus.46 Interestingly, the nucleolus is also increasingly recognized as a target for cancer therapy.47 Further research is required to fully understand and characterize the function of DDX3 in this subcellular compartment.
With regard to the mechanism behind nuclear DDX3 retention, we observed a correlation between cytoplasmic expression of CRM1 and nuclear DDX3 expression in colorectal cancer. This suggests that dysregulation of CRM1 is one of the mechanisms of nuclear DDX3 expression. The binding site of DDX3 to CRM1 is a matter of debate. DDX3 has a classical N-terminal NES sequence that is conserved and required for CRM1 binding in the Saccharomyces cerevisiae homolog Ded1p48,49 and the Xenopus laevis homolog An3.31 However, Yedavalli et al observed binding between CRM1 and amino acids 260–517 of DDX3.4 Our analysis of DDX3 deletion mutants showed that deletion of both areas resulted in an increase in nuclear DDX3, but only the construct that lacked the NES lost its sensitivity to CRM1 inhibition, indicating that this is the essential domain for CRM1-mediated export. A similar conclusion was recently made by Fröhlich et al, who found the N-terminal to be essential for DDX3 transportation out of the nucleus into cytoplasmic unspliced HIV-1 mRNA ribonucleoprotein complexes.50 It is possible that the 260–517 region of DDX3 is necessary for binding other exporters of DDX3 like TAP.5
As cytoplasmic CRM1 expression was infrequent in colorectal cancer and did not correlate with nuclear DDX3 in breast cancer, dysregulation of CRM1 is not likely the sole mechanism behind nuclear DDX3. When, in addition to nuclear export inhibition, we stimulated nuclear import by addition of an NLS, we observed a complete shift of all DDX3 in the cell to the nucleus, showing that increased import can also contribute to increased nuclear DDX3 levels. However, the mechanism behind nuclear import of DDX3 remains unknown. DDX3 has a classical NLS sequence at amino acid 212,51 but it is also possible that DDX3 enters the nucleus as part of a complex. Future research on this topic is warranted.
Conclusion
Nuclear DDX3 expression predicts worse survival in breast and colorectal cancer. Mechanistically, this can be partly explained by altered expression of the nuclear exporter CRM1.
Acknowledgments
This work was financially supported by the Utrecht University Alexandre Suerman Stipend, the Dutch Cancer Foundation (UU2013-5851), the Saal van Zwanenberg foundation and the JoKolk Scholarship Foundation (Marise R Heerma van Voss), the Medical Research Council, UK grant MC_UU_12014/2 (Arvind H Patel), and the Flight Attendant Medical Research Institute (Venu Raman). We would like to thank Ashley Irving and Yehudit Bergman for their help with cloning the DDX3 constructs, Natalie ter Hoeve and Petra van der Groep for their help with setting up immunohistochemistry protocols, Cathy Moelans, Liudmila Kodach and Folkert Morsink for generating tissue microarrays, Kai Kammers for his critical evaluation of statistical methods and Nikolas Stathonikos for his help with digital imaging.
Disclosure
Venu Raman, Guus M Bol and Paul J van Diest have received a patent for the use of DDX3 as a cancer biomarker (US9322831). Venu Raman and Paul J van Diest are on the advisory board of Natsar Pharmaceuticals. The authors report no other conflicts of interest in this work.
References
Leitao AL, Costa MC, Enguita FJ. Unzippers, resolvers and sensors: a structural and functional biochemistry tale of RNA helicases. Int J Mol Sci. 2015;16(2):2269–2293. | ||
Deckert J, Hartmuth K, Boehringer D, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26(14):5528–5543. | ||
Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13(1):116–128. | ||
Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119(3):381–392. | ||
Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19(9):3847–3858. | ||
Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci U S A. 2011;108(29):12007–12012. | ||
Kasim V, Wu S, Taira K, Miyagishi M. Determination of the role of DDX3 a factor involved in mammalian RNAi pathway using an shRNA-expression library. PLoS One. 2013;8(3):e59445. | ||
Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36(14):4708–4718. | ||
Li Y, Wang H, Wang Z, et al. Inducible resistance of tumor cells to tumor necrosis factor-related apoptosis-inducing ligand receptor 2-mediated apoptosis by generation of a blockade at the death domain function. Cancer Res. 2006;66(17):8520–8528. | ||
Sun M, Song L, Li Y, Zhou T, Jope RS. Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ. 2008;15(12):1887–1900. | ||
Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441(1):119–129. | ||
Bol GM, Vesuna F, Xie M, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med. 2015;7(5):648–669. | ||
Heerma van Voss MR, Vesuna F, Trumpi K, et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6(29):28312–28326. | ||
Botlagunta M, Vesuna F, Mironchik Y, et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27(28):3912–3922. | ||
Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2015;34(21):2790–2800. | ||
Topisirovic I, Siddiqui N, Lapointe VL, et al. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28(8):1087–1098. | ||
Xie M, Vesuna F, Botlagunta M, et al. NZ51, a ring-expanded nucleoside analog, inhibits motility and viability of breast cancer cells by targeting the RNA helicase DDX3. Oncotarget. 2015;6(30):29901–29913. | ||
Wilky BA, Kim C, McCarty G, et al. RNA helicase DDX3: a novel therapeutic target in Ewing sarcoma. Oncogene. 2016;35(20):2574–2583. | ||
Kodach LL, Wiercinska E, de Miranda NF, et al. The bone morphogenetic protein pathway is inactivated in the majority of sporadic colorectal cancers. Gastroenterology. 2008;134(5):1332–1341. | ||
Moelans CB, de Weger RA, van Blokland MT, et al. HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cell Oncol. 2009;31(1):1–10. | ||
Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295(21):2492–2502. | ||
Vermeulen JF, van de Ven RA, Ercan C, et al. Nuclear Kaiso expression is associated with high grade and triple-negative invasive breast cancer. PLoS One. 2012;7(5):e37864. | ||
Wet medisch-wetenschappelijk onderzoek met mensen, WMO [The Medical Research Involving Human Subjects Act]. In: Burgerlijk Wetboek. Dutch government; the Netherlands; 1998. Dutch. | ||
van Diest PJ. No consent should be needed for using leftover body material for scientific purposes. For. BMJ. 2002;325(7365):648–651. | ||
Angus AG, Dalrymple D, Boulant S, et al. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol. 2010;91(Pt 1):122–132. | ||
Budczies J, Klauschen F, Sinn BV, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. | ||
Noske A, Weichert W, Niesporek S, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer. 2008;112(8):1733–1743. | ||
Kudo N, Matsumori N, Taoka H, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A. 1999;96(16):9112–9117. | ||
Ishizawa J, Kojima K, Hail N Jr, Tabe Y, Andreeff M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther. 2015;153:25–35. | ||
Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. | ||
Askjaer P, Rosendahl R, Kjems J. Nuclear export of the DEAD box An3 protein by CRM1 is coupled to An3 helicase activity. J Biol Chem. 2000;275(16):11561–11568. | ||
Bol GM, Xie M, Raman V. DDX3, a potential target for cancer treatment. Mol Cancer. 2015;14(1):188. | ||
Samal SK, Routray S, Veeramachaneni GK, Dash R, Botlagunta M. Ketorolac salt is a newly discovered DDX3 inhibitor to treat oral cancer. Sci Rep. 2015;5:9982. | ||
Koshio J, Kagamu H, Nozaki K, et al. DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked is an immunogenic target of cancer stem cells. Cancer Immunol Immunother. 2013;62(10):1619–1628. | ||
Yedavalli VS, Zhang N, Cai H, et al. Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication. J Med Chem. 2008;51(16):5043–5051. | ||
Radi M, Falchi F, Garbelli A, et al. Discovery of the first small molecule inhibitor of human DDX3 specifically designed to target the RNA binding site: towards the next generation HIV-1 inhibitors. Bioorg Med Chem Lett. 2012;22(5):2094–2098. | ||
Mahboobi SH, Javanpour AA, Mofrad MR. The interaction of RNA helicase DDX3 with HIV-1 Rev-CRM1-RanGTP complex during the HIV replication cycle. PLoS One. 2015;10(2):e0112969. | ||
Su CY, Lin TC, Lin YF, et al. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer. Oncotarget. 2015;6(21):18602–18612. | ||
Chung GG, Provost E, Kielhorn EP, Charette LA, Smith BL, Rimm DL. Tissue microarray analysis of beta-catenin in colorectal cancer shows nuclear phospho-beta-catenin is associated with a better prognosis. Clin Cancer Res. 2001;7(12):4013–4020. | ||
Jarboui MA, Bidoia C, Woods E, et al. Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus. PLoS One. 2012;7(11):e48702. | ||
Yasuda-Inoue M, Kuroki M, Ariumi Y. Distinct DDX DEAD-box RNA helicases cooperate to modulate the HIV-1 Rev function. Biochem Biophy Res Commun. 2013;434(4):803–808. | ||
Andersen JS, Lyon CE, Fox AH, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12(1):1–11. | ||
Derenzini M, Montanaro L, Trere D. What the nucleolus says to a tumour pathologist. Histopathology. 2009;54(6):753–762. | ||
Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3(3):179–192. | ||
Geissler R, Golbik RP, Behrens SE. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res. 2012;40(11):4998–5011. | ||
Wang T, Birsoy K, Hughes NW, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–1101. | ||
Hein N, Hannan KM, George AJ, Sanij E, Hannan RD. The nucleolus: an emerging target for cancer therapy. Trends Mol Med. 2013;19(11):643–654. | ||
Senissar M, Le Saux A, Belgareh-Touze N, Adam C, Banroques J, Tanner NK. The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res. 2014;42(15):10005–10022. | ||
Hauk G, Bowman GD. Formation of a trimeric Xpo1-Ran[GTP]-Ded1 exportin complex modulates ATPase and helicase activities of Ded1. PLoS One. 2015;10(6):e0131690. | ||
Fröhlich A, Rojas-Araya B, Pereira-Montecinos C, et al. DEAD-box RNA helicase DDX3 connects CRM1-dependent nuclear export and translation of the HIV-1 unspliced mRNA through its N-terminal domain. Biochim Biophy Acta. 2016;1859(5):719–730. | ||
Nassar H, Qureshi H, Volkanadsay N, Visscher D. Clinicopathologic analysis of solid papillary carcinoma of the breast and associated invasive carcinomas. Am J Surg Pathol. 2006;30(4):501–507. |
Supplementary materials
  | Figure S1 Kaplan–Meier plot depicting overall survival in high and low DDX3-expressing colorectal cancer samples. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.