Back to Journals » Therapeutics and Clinical Risk Management » Volume 16
Novel Deep Learning Technique Used in Management and Discharge of Hospitalized Patients with COVID-19 in China
Authors Meng Q , Liu W, Gao P , Zhang J, Sun A, Ding J, Liu H, Lei Z
Received 6 September 2020
Accepted for publication 23 November 2020
Published 8 December 2020 Volume 2020:16 Pages 1195—1201
DOI https://doi.org/10.2147/TCRM.S280726
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Garry Walsh
Qingcheng Meng,1 Wentao Liu,1 Pengrui Gao,1 Jiaqi Zhang,2 Anlan Sun,2 Jia Ding,2 Hao Liu,2 Ziqiao Lei3
1Department of Radiology, The Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, People’s Republic of China; 2Yizhun Medical AI Co. Ltd, Beijing, People’s Republic of China; 3Department of Radiology, The Wuhan Union Hospital, Wuhan, People’s Republic of China
Correspondence: Qingcheng Meng
Department of Radiology, The Affiliated Cancer Hospital of Zhengzhou University, No. 127 Dongming Road, Jinshui District, Zhengzhou 450008, People’s Republic of China
Tel/Fax +86-0371-65587152
Email [email protected]
Ziqiao Lei
Department of Radiology, The Wuhan Union Hospital, No. 1277 Jiefang Road, Jianghan District, Wuhan 430000, People’s Republic of China
Email [email protected]
Purpose: The low sensitivity and false-negative results of nucleic acid testing greatly affect its performance in diagnosing and discharging patients with coronavirus disease (COVID-19). Chest computed tomography (CT)-based evaluation of pneumonia may indicate a need for isolation. Therefore, this radiologic modality plays an important role in managing patients with suspected COVID-19. Meanwhile, deep learning (DL) technology has been successful in detecting various imaging features of chest CT. This study applied a novel DL technique to standardize the discharge criteria of COVID-19 patients with consecutive negative respiratory pathogen nucleic acid test results at a “square cabin” hospital.
Patients and Methods: DL was used to evaluate the chest CT scans of 270 hospitalized COVID-19 patients who had two consecutive negative nucleic acid tests (sampling interval > 1 day). The CT scans evaluated were obtained after the patients’ second negative test result. The standard criterion determined by DL for patient discharge was a total volume ratio of lesion to lung < 50%.
Results: The mean number of days between hospitalization and DL was 14.3 (± 2.4). The average intersection over union was 0.7894. Two hundred and thirteen (78.9%) patients exhibited pneumonia, of whom 54.0% (115/213) had mild interstitial fibrosis. Twenty-one, 33, and 4 cases exhibited vascular enlargement, pleural thickening, and mediastinal lymphadenopathy, respectively. Of the latter, 18.8% (40/213) had a total volume ratio of lesions to lung ≥ 50% according to our severity scale and were monitored continuously in the hospital. Three cases had a positive follow-up nucleic acid test during hospitalization. None of the 230 discharged cases later tested positive or exhibited pneumonia progression.
Conclusion: The novel DL enables the accurate management of hospitalized patients with COVID-19 and can help avoid cluster transmission or exacerbation in patients with false-negative acid test.
Keywords: X-ray, computed tomography, SARS-CoV-2, infectious disease, lung disease
Introduction
Coronavirus disease (COVID-19), caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a pandemic emergency worldwide1 at the beginning of 2020. Compared to the other members of the family Coronaviridae, which includes the well-known severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus, SARS-CoV-2 is transmitted more rapidly and may lead to severe respiratory distress syndrome or even death.2–5
Nucleic acid testing plays an indispensable role in the diagnosis and discharge of patients with COVID-19. However, the test’s low sensitivity and false-negative results have greatly affected its clinical utility. Consequently, the Diagnosis and Treatment Program (6th version) published by the National Health Commission of the People’s Republic of China used radiologic features to define the diagnostic and discharge criteria for COVID-19.6 An accurate chest computed tomography (CT)-based evaluation of viral pneumonia may indicate the need for isolation. Therefore, this radiologic modality plays an important role in the management of patients with suspected SARS-CoV-2 infection, especially when there are still no scientifically proven therapies for it.7 However, it is difficult to determine the suitability for discharge of a patient with COVID-19 based on non-standardized CT features in a clinical setting.
Recently, artificial intelligence (AI) using deep learning (DL) technology has demonstrated great success in the medical imaging field due to its remarkable feature-extracting capability.8,9 Attempts have also been made to detect various imaging features on chest CT.10 This study aimed to apply DL based on a three-dimensional deep convolutional neural network (3D-DCNN) framework to chest CT scans to standardize the discharge criteria of hospitalized COVID-19 patients with two consecutive negative nucleic acid tests.
Patients and Methods
Patients
A cohort of 270 patients (102 men and 168 women; age range, 18–65 years; mean age ± standard deviation, 51.9 ± 15.6 years) at a Wuhan square cabin hospital confirmed to test positive on SARS-CoV-2 nucleic acid testing was retrospectively identified from February 16 to March 7, 2020. All respiratory secretions for testing were collected using oropharyngeal swabs, and testing was performed at the laboratory of Union Hospital in Wuhan, China. The study was conducted in compliance with the ethical standards of the Helsinki Declaration and approved by the Wuhan Union Hospital and Affiliated Tumor Hospital of Zhengzhou University Medical Ethics Committee. All patients provided written informed consent.
Patients who met the following criteria were included in the analysis: 1) aged 18–65 years, 2) had normal body temperature for >3 days, 3) experienced significant alleviation of respiratory symptoms, and 4) had two consecutive negative respiratory pathogen nucleic acid test results (sampling interval ≥1 day). The exclusion criteria consisted of: 1) COVID-19 accompanied by moderate or severe liver and kidney function impairment; 2) pregnancy concurrent with COVID-19; and 3) psychiatric symptoms or tumors. After applying these criteria, the CT images evaluated by DL as requiring hospitalization and clinical data of the eligible patients were analyzed.
CT Technique
All patients underwent low-dose CT (LDCT) scanning using a 40-slice multidetector CT scanner (uCT 550, Shanghai United Imaging Healthcare Co., Ltd., Shanghai, China) with the following parameters: tube voltage, 120 kV; tube current, 30 mAs; field of view, 400 mm × 400 mm to 500 mm × 500 mm; matrix, 512 × 512; rotation time, 0.6 s; section width, 2.0 mm; reconstruction interval, 2.0 mm; and scan duration, 3–10s.
Unenhanced spiral image acquisitions were obtained from the thoracic inlet to the lung base with breath-holding. The “Dose Report” function was used to record the dose parameters during spiral CT, and a standard algorithm was used to reconstruct images. All studies were reviewed on a PACS workstation at a window level of −500 to −700 Hounsfield units (HU) and window width of 1400 HU. Multiplanar images were obtained using the multiplanar reformatting (MPR) technique on the workstation.
DL Framework Based on 3D-DCNN Algorithms
In this work, we used a 3D-DCNN network named as U-net [13] as shown in Figure 1, because of its U-shape architecture. U-net is a fully convolutional neural network, suitable for image segmentation. In this study, three-dimensional convolution operations were applied to replace the traditional two-dimensional operations in order to obtain the information between different CT image layers. Compared to traditional detection network in the pneumonia detection, the advantages of U-net were displayed by the following two reasons. One reason was that the lesion of pneumonia varied the size from center meters to the whole lung during the whole illness. Traditional best performance detection network for example Faster reversal convolutional neural network (R-CNN) [11–12] could not efficiently detect the lesion with so large size variation, because the box regression would be very hard for it. Another reason was that the pneumonia volume was an important clinical index to evaluate the stage of illness and prognostis. So, the precisely pixel-level segmentation was necessary, and the U-net as the best and most popular segmentation architecture [13] became our first choice. Theoretically, the structure of U-net could keep multi scale’s feature information when extracting the high-level context features by network downsampling and gave accurate classification results by merging multi-layer context information which was obtained during the up-sampling process, shown in Figure 1. The reconstruction capability was strengthened by reconstructing each scale’s feature from the deep layer aggregation. In this way, there was less information loss when multi-layer features were merged. And the loss function of deep supervision performed the supervision learning to each reconstruction result at each scale, which accelerated the convergent of the model.
Outcomes and Measures
The degree of pneumonia was determined by the total volume ratio of lesions to lung (total volume ratio=volume of lesion1 + 2 + 3 + … /the volume of lung). The standard set for discharge was a volume ratio of lesions to lung <50%. The LDCT data of all patients were interpreted independently by three thoracic radiologists with 7, 10, and 15 years of experience. All observations were assessed for major and ancillary features, after which the lung lesions were evaluated using the 4-grade pneumonia scale.
Statistical Analysis
Statistical analysis was performed using SPSS 24.0 (SPSS Inc, Chicago, IL, USA). Measurement data are reported as means ± standard deviations (SDs). Continuous data are presented as frequencies and percentages.
Results
Patient and Lesion Characteristics
Table 1 demonstrates that of 270 patients enrolled in this study, 57 (21.1%) had normal lung features, and 213 (78.9%) had pneumonia. Additionally, 100 (46.9%) and 15 (7.0%) of the 213 cases had mild and moderate interstitial fibrosis. Regarding the locations of lesions in the lungs, the distribution was mainly in the peripheral zone in 136 cases (63.8%), diffuse in 75 cases (35.2%), and the central zone in 2 cases (0.9%). Sixteen (7.5%), 31 (14.6%), and 166 cases presented with lesions in 1, 2, or ≥3 lung lobes, respectively. We identified 21 cases with vascular enlargement on CT, 33 with pleural thickening, 4 with nodule, and 4 with mediastinal lymphadenopathy.
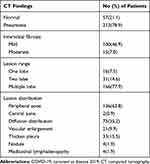 |
Table 1 Initial CT Findings of 270 Patients with Coronavirus Disease 2019 (COVID-19) |
Performance Metrics of DL
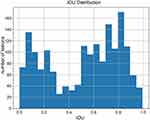
The mean number of days between hospitalization and the execution of the DL technique was 14.3 (± 2.4) days. We used 110 CT scans to evaluate the performance of DL and obtained a dice value of 0.63. The evaluation was performed by using intersection over union (IOU): 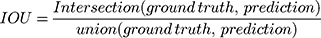 . The average IOU over every lesion was:
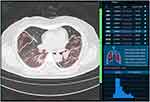
. The average IOU over every lesion was: 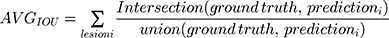 . A higher IOU value meant the prediction was more consistent with the label given by the radiologist (ground truth). The IOU of every prediction is shown in Figure 2. The mean IOU was 0.7894, although there were two peaks in the distribution, which meant most predictions might have a high or very low IOU value. This may be due to some COVID-19 lesions consisting of ground-grass opacities, making it difficult for both the radiologist and AI system to discriminate the margins. Some of the AI system’s predictions proposed in this paper are shown in Figure 3. The detected lesions and their areas affected with pneumonia labeled by AI were similar to those labeled by radiologists, which indicated that AI also performed well for discrete pneumonia lesions.
. A higher IOU value meant the prediction was more consistent with the label given by the radiologist (ground truth). The IOU of every prediction is shown in Figure 2. The mean IOU was 0.7894, although there were two peaks in the distribution, which meant most predictions might have a high or very low IOU value. This may be due to some COVID-19 lesions consisting of ground-grass opacities, making it difficult for both the radiologist and AI system to discriminate the margins. Some of the AI system’s predictions proposed in this paper are shown in Figure 3. The detected lesions and their areas affected with pneumonia labeled by AI were similar to those labeled by radiologists, which indicated that AI also performed well for discrete pneumonia lesions.
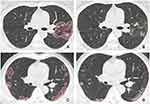 |
Figure 3 Predict results of the AI system. (A, C): predicted by AI system, (B, D): labeled by radiologist. |
Of the 1720 pneumonia lesions in our study, these lesions’ volumes varied from tens to tens of thousands of mm3, with the volume distribution seen in Figure 4. The volume of these lesions from 500 mm3 to 4000 mm3 accounted for 27.9%, 4000 mm3 to 8000 mm3 for 26.2%, and 8000 mm3 to 12,000 mm3 for 14.0%. Therefore, more than half of the lesions were sized between 500 and 8000 mm3, and >80% of the lesions were <12,000 mm3. Using our DL-determined discharge standard of total volume ratio <50%, 40 of 270 cases (14.8%) remained hospitalized for continuous observation, and 230 (85.2%) were discharged.
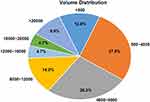 |
Figure 4 Volume distribution for all lesions. |
During the 14-day isolation period, 3 of the 40 hospitalized cases again tested positive on nucleic acid testing, one of which is shown in Figure 5, and the ratio of the total volume of lesions to lung volume, which was >95%. The 3D display and density of lesions are also shown in Figure 5. None of the 230 discharged cases had a subsequent positive nucleic acid test or manifested progression of pneumonia.
Discussion
In this study, we designed and evaluated a 3D DL model to provide standardized discharge criteria for common COVID-19 patients. We found that most discharged patients with common COVID-19 were managed well with favorable outcomes. The three patients who later tested positive on follow-up nucleic acid testing after previously testing negative were hospitalized based on our criteria and thus, avoided cluster transmission or disease exacerbation due to prompt treatment.
Previous histological examinations of biopsy specimens collected from the lungs of COVID-19 patients showed diffuse alveolar damage with edema, cellular proteinaceous exudates, focal reactive pneumocytic hyperplasia with patchy inflammatory cellular infiltration, and multinucleated giant cells.14,15 On CT, these lesions might appear as areas of ground-glass opacities or consolidation.16 Previous studies had successfully applied DL techniques to detect pneumonia in pediatric chest radiographs and chest CT.10,17 In this study, a 3D DL framework was proposed to evaluate the severity of disease in COVID-19 patients. Using the U-net network, this framework was able to extract 3D global representative features. These extracted features helped integrate the 3D spacing information and clarify the distinction between the different lesional textures. Image segmentation yielded a dice value of 0.63, which suggested that the voxel prediction accuracy exceeded 85% in each image dimension. The discharge criteria of common COVID-19 patients were managed well by DL, and three cases with positive follow-up tests after false-negative tests in our study were also hospitalized again, which avoided the cluster transmission or exacerbation. Therefore, the management and discharge of COVID-19 patients must involve both nucleic acid testing and timely performed and accurately read chest CT.7,18
COVID-19 pneumonia manifests with chest CT imaging abnormalities, even in asymptomatic patients. It evolves rapidly from focal unilateral to diffuse bilateral ground-glass opacities that progress to or co-exist with consolidations in 1–3 weeks. In our study, most patients with pneumonia (75.2%) met the criteria for the total volume ratio of lesions to lung <50%. Moreover, 21 patients with pneumonia (9.1%) also presented with vascular enlargement, in contrast to 82.4% reported by Yan Li et al10 We attribute this inter-study difference to our study population’s disease status, wherein a negative nucleic acid test indicated they were in the recovery phase of COVID-19. We speculate that vascular dilatation may be predictive of patients’ short-term prognosis with COVID-19 and suggest a large-scale validation study of this deduction.
Our study is not without limitations. First, the square cabin hospital is a temporary hospital. Accordingly, the populations of medical workers and patients have a rapid turnover rate. Therefore, the changes in CT of COVID-19 pneumonia could not be evaluated by DL due to the lack of baseline chest CT data. Second, further studies are necessary to evaluate the value of density variation of pneumonia detected by DL, which might help the clinician manage the COVID-19.
Conclusion
In conclusion, our study found that DL enables the accurate management and discharge of COVID-19 patients quantitatively. Further medical isolation is needed for COVID-19 patients with a total volume of lesions to lung ≥ 50%, which can help avoid cluster transmission or exacerbation. Therefore, the model presented by the “square cabin” hospital enabled optimal management for COVID-19.
Abbreviations
COVID-19, coronavirus disease 2019; DL, deep learning; R-CNN, reversal convolutional neural network; CT, computed tomography; IOU, intersection over union.
Acknowledgments
We would like to thank Editage for English language editing.
Author Contributions
All authors made a significant contribution to the work reported, whether the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Perlman S. Another decade, another coronavirus. N Engl J Med. 2020;382(8):760–762. doi:10.1056/NEJMe2001126.
2. Richman DD, Whitley RJ, Hayden FG, eds. Clinical Virology, 4th Edn. Washington: ASM Press; 2016.
3. Zhu N, Zhang D, Wang W, et al. China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi:10.1056/NEJMoa2001017.
4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5.
5. Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi:10.1001/jama.2020.1585
6. National Health Commission of the People’s Republic of China website. Diagnosis and treatment of novel coronavirus infection (trial version6); 2020. Avialble from: www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2.shtml.
7. Li Y, Xia XL. Coronavirus Disease 2019 (COVID-19): role of Chest CT in Diagnosis and Management. AJR Am J Roentgenol. 2020;214(6):1280–1286. doi:10.2214/AJR.20.22954.
8. Kong B, Wang X, Bai J, et al. Learning tree-structured representation for 3D coronary artery segmentation. Comput Med Imaging Graph. 2020;80:101688. doi:10.1016/j.compmedimag.2019.101688.
9. Ye H, Gao F, Yin Y, et al. Precise diagnosis of intracranial hemorrhage and subtypes using a threedimensional joint convolutional and recurrent neural network. Eur Radiol. 2019;29:6191–6201. doi:10.1007/s00330-019-06163-2
10. Li Z, Zhong Z, Li Y, et al. From community-acquired pneumonia to COVID-19: a deep learning-based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur Radiol. 2020:1–10. doi:10.1007/s00330-020-07042-x.
11. Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans Pattern Anal Mach Intell. 2017;39(6):1137–1149. doi:10.1109/TPAMI.2016.2577031.
12. Ding J, Li A, Hu Z, Wang L. Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham; 2017:
13. Ronneberger O, Philipp F, Thomas B. U-Net: Convolutional Networks for Biomedical Image Segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham; 2015.
14. Tian S, Hu W, Niu L, Liu H, Xu H, Xiao S-Y. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID-19) Pneumonia in Two Patients With Lung Cancer. J Thorac Oncol. 2020;15(5):700–704. doi:10.1016/j.jtho.2020.02.010.
15. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi:10.1016/S2213-2600(20)30076-X.
16. Hansell DM, Bankier AA, MacMahon H, et al. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246(3):697–722. doi:10.1148/radiol.2462070712.
17. Rajaraman S, Candemir S, Xue Z, et al. A Novel Stacked Generalization of Models for Improved TB Detection in Chest Radiographs. Conf Proc IEEE Eng Med Biol Soc. 2018:718–721. doi:10.1109/EMBC.2018.8512337
18. Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425–434. doi:10.1016/S1473-3099(20)30086-4.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.