Back to Journals » Risk Management and Healthcare Policy » Volume 13
Nomogram for the Individualized Prediction of Survival Among Patients with H7N9 Infection
Authors Cheng Q , Sun Z , Zhao G , Xie L
Received 12 December 2019
Accepted for publication 6 March 2020
Published 20 March 2020 Volume 2020:13 Pages 255—269
DOI https://doi.org/10.2147/RMHP.S242168
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Marco Carotenuto
Qinglin Cheng,1,2,* Zhou Sun,1,* Gang Zhao,1,* Li Xie1,*
1Division of Infectious Diseases, Hangzhou Center for Disease Control and Prevention, Hangzhou 310021, People’s Republic of China; 2School of Public Health, Zhejiang Chinese Medical University, Hangzhou 310021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qinglin Cheng; Zhou Sun
Division of Infectious Diseases, Hangzhou Center for Disease Control and Prevention, 568 Mingshi Road, Hangzhou 310021, People’s Republic of China
Email [email protected]; [email protected]
Background: Until recently, almost all of these studies have identified multiple risk factors but did not offer practical instruments for routine use in predicting individualized survival in human H7N9 infection cases. The objective of this study is to create a practical instrument for use in predicting an individualized survival probability of H7N9 patients.
Methods: A matched case–control study (1:2 ratios) was performed in Zhejiang Province between 2013 and 2019. We reviewed specific factors and outcomes regarding patients with H7N9 virus infection (VI) to determine relationships and developed a nomogram to calculate individualized survival probability. This tool was used to predict each individual patient’s probability of survival based on results obtained from the multivariable Cox proportional hazard regression analysis.
Results: We examined 227 patients with H7N9 VI enrolled in our study. Stepwise selection was applied to the data, which resulted in a final model with 8 independent predictors [including initial PaO2/FiO2 ratio ≤ 300 mmHg, age ≥ 60 years, chronic diseases, poor hand hygiene, time from illness onset to the first medical visit, incubation period ≤ 5 days, peak C-reactive protein ≥ 120 mg/L], and initial bilateral lung infection. The concordance index of this nomogram was 0.802 [95% confidence interval (CI): 0.694– 0.901] and 0.793 (95% CI: 0.611– 0.952) for the training and validation sets, respectively, which indicates adequate discriminatory power. The calibration curves for the survival showed optimal agreement between nomogram prediction and actual observation in the training and validation sets, respectively.
Conclusion: We established and validated a novel nomogram that can accurately predict the survival probability of patients with H7N9 VI. This nomogram can serve an important role in counseling patients with H7N9 VI and guide treatment decisions.
Keywords: avian influenza virus, H7N9, survival, predictors, nomogram
Introduction
Human cases of avian influenza A (H7N9) virus infection (VI) had occurred annually during the spring and winter months in mainland China since 2013, reaching 1568 laboratory-confirmed cases and 615 deaths as of October 31, 2019.1 H7N9 virus infection was associated with severe and fatal respiratory disease in humans; most laboratory-confirmed human H7N9 infection cases were critically ill.2 The increasing number of H7N9 infection cases and concomitant high fatality risk have caused global concern regarding public health.3
Although no new cases of H7N9 VI were reported in China after April 5, 2019, H7N9 VI is the highest risk score of the 12 novel influenza A viruses assessed by the Influenza Risk Assessment Tool.4 Besides, the ongoing monitoring of clinicopathologic and molecular features of H7N9 VI recovered from infected humans, poultry and markets where live poultry or birds, continual epidemiological surveys and clinical severity estimates of H7N9 infection cases are critical to developing public health strategies for pandemic preparedness. However, available data that address this point are limited. It has been previously shown that clinicians who make diagnostic and treatment decisions based solely on their clinical judgment lack predictive accuracy regarding death risk in human H7N9 infection cases and are, therefore, unable to increase the survival probability in these cases.5–7 An accurate assessment of survival probability would help tailor treatment in human H7N9 infection cases. Unfortunately, the vast majority of published studies8,9 on the topic focus on risk factors of H7N9 infection and case fatalities.
Though previous studies have established several models based on predicting the outcome of H7N9 VI and showed certain application value,4,6-9 there is currently no model available for the prediction and assessment of survival in human H7N9 infection cases. To date, almost all of these studies have identified multiple risk factors but did not integrate them so as to comprehensively assess a patient’s specific survival probability in human H7N9 VI. It is now well established from a variety of studies that the nomogram model is an algorithm with integrating predictors and has been identified as a practical tool of preventive interventions.10 In addition, a nomogram can accurately predict and estimate the individualized risk of a disease and quantitatively demonstrate a personalized probability for predicting the incidence of disease outcome.11
To better evaluate the predictors associated with survival outcomes, based on a matched case−control study (1:2 ratios), we retrospectively investigated the sociodemographic, clinical and epidemiological data of 227 laboratory-confirmed cases of H7N9 VI in Zhejiang province, China, between March 1, 2013, and May 31, 2019. The objective of this study is to determine predictors for survival for H7N9 patients and create a practical instrument based on results obtained from a multivariable Cox proportional hazard model for use in predicting and evaluating the individualized survival probability in H7N9 patients.
Materials and Methods
Sample Size Calculation
To calculate the sample size, we used the following formula:12
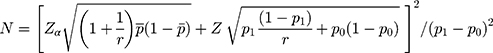
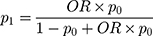
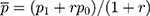
where 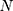 = sample size; α = alpha (expected significant level, two-tailed test); β = 1– power (expected power, two-tailed test); Z statistic (Z) —Z statistic for confidence level; r — number of control subjects matched to each case subject;
= sample size; α = alpha (expected significant level, two-tailed test); β = 1– power (expected power, two-tailed test); Z statistic (Z) —Z statistic for confidence level; r — number of control subjects matched to each case subject;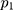 — probability of exposure in the case group;
— probability of exposure in the case group;  probability of exposure in the control group (
probability of exposure in the control group (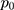 can be estimated as the population prevalence of exposure); OR = odds ratio (odds ratio of exposures between cases and controls; OR can be estimated as the population OR of exposure).
can be estimated as the population prevalence of exposure); OR = odds ratio (odds ratio of exposures between cases and controls; OR can be estimated as the population OR of exposure).
In this study, the investigators present their results with 95% confidence interval (CI), 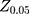 =1.96 (α= 0.05),
=1.96 (α= 0.05), 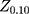 =1.64 (β= 0.10), r = 2,
=1.64 (β= 0.10), r = 2, 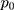 = 35.3%, and OR= 2.5.13 In addition, considering the loss of follow-up, participants’ rejection rate, and sampling error, the final sample size was determined to be 200 in the training set. The validation set was selected in this study using the ‘all comers’ design.14
= 35.3%, and OR= 2.5.13 In addition, considering the loss of follow-up, participants’ rejection rate, and sampling error, the final sample size was determined to be 200 in the training set. The validation set was selected in this study using the ‘all comers’ design.14
Workflow
The workflow of the study is summarized in Figure 1. Two separate datasets were used to develop and validate the prediction tool based on the survival of H7N9 patients. Data from Institution I (i.e., the First Affiliated Hospital, Zhejiang University School of Medicine; n =171) were used as a training data set to derive the survival in H7N9 patients, while data from Institution II (i.e., other designated hospitals including Shaoxing People’s Hospital, Ningbo Second Hospital and the First Hospital of Jiaxing that provided H7N9 VI treatment in Zhejiang province; n = 56) were used as an independent dataset to validate the prediction tool. After the quantitative survival features were extracted from human H7N9 infection cases, univariate analysis, including a Pearson Chi-square test, independent samples t test and univariable Cox proportional hazard regression model, and a multivariate Cox proportional hazard regression model were applied to select optimal independent predictors to build a practical instrument for predicting and evaluating the survival in patients with H7N9 VI.
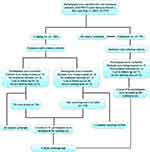 |
Figure 1 Workflow in this study. Patients being treated were defined as patients with an antiviral drug therapy during the course of study. |
Study Design and Settings
A matched case−control study (1:2 ratios) was conducted in Zhejiang Province. All H7N9 patients with laboratory-confirmed VI who were enrolled in this retrospective study between March 1, 2013, and May 31, 2019, constituted the case−control study.
For the present study, the H7N9 cases were selected from all of the H7N9 VI designated hospitals in Zhejiang Province. In this observational case−control study, the subjects were classified into “died” (i.e., the case group) and “survived” groups (such as the control group) according to the ultimate outcome. The cases were selected for this study using the “all comers” principle,14 as long as they met inclusion and exclusion criteria, and the controls were selected by using a random sampling method from the same H7N9 VI designated hospitals. The subjects were ultimately selected if they (a) had a history of H7N9 VI treatment; (b) had a definite treatment outcome; and (c) could be followed up. The subjects were excluded if (a) H7N9 patients were being treated; (b) no treatment outcome could be obtained; (c) subjects who lost during the follow-up visit and (d) the missing data were severe (Figure 1).
The starting date of H7N9 VI was defined as the starting time of the observation study, while we defined a patient’s observation ending date as the end date of the study, which was the data of incident death or survival. Incident death or survival for all years was collected between March 1, 2013, and May 31, 2019.
Data Collection
We collected the medical records of all patients with H7N9 VI in Zhejiang Province from a surveillance database of infectious diseases managed by China’s Center for Disease Control and Prevention (CDC) and the provincial CDC. Each confirmed patient with H7N9 VI was asked to be reported to CDC within 1 day by a national monitoring system of infectious diseases. Staff members from the local CDC were assigned to collect data using a standardized questionnaire according to field investigations. We extracted data regarding patients’ clinical, epidemiological, and sociodemographic features, as well as time-to-event dates and prognoses. The way of all data collection has been detailedly described in the previous study.13 All data were entered in duplicate into an electronic database. We retrospectively studied 227 laboratory-confirmed cases of H7N9 VI in Zhejiang Province from March 1, 2013, to May 31, 2019 (no new cases of H7N9 VI were reported in Zhejiang province after December 31, 2017). We excluded 52 human H7N9 infection patients according to inclusion and exclusion criteria. Patients in the training set (n =171) and the external validation set (n =56) were analyzed, respectively (Figure 1).
Variables and Definitions
Definitions of case and exposure and laboratory examination for the A (H7N9) virus have been detailedly described elsewhere.13 The main outcome variable was measured as incident death or survival. Table 1 shows the definition of variables in this study.
 |
Table 1 Definitions of This Study |
Statistical Analysis
Univariate analysis was performed to determine factors (including patients’ clinical, epidemiological, and demographic features, as well as time-to-event dates and prognoses) associated with the survival of patients with H7N9 VI in the training dataset and the validation dataset. Researchers divide the subjects into two groups based on their death or survival from the infection. A Pearson Chi-square test was used for categorical variables and an independent sample t-test for continuous variables based on baseline characteristics of the subjects (including training and validation sets). The laboratory findings and the questionnaire variables were analyzed using a hazard ratio (HR) generated by univariable Cox proportional hazard regression.
Subsequently, independent predictors associated with the survival of H7N9 VI were evaluated using HR generated by a multivariate Cox regression model. All variables with P value <0.10 were included in the multivariate Cox regression analysis using a backward stepwise method based on the minimum statistics of the Akaike information criterion. Variables with P value less than 0.10 were considered statistically significant in the multivariate Cox regression model and were included in the final predictive model. A nomogram was developed based on all the patient variables in the training set. This model was used to predict each individual patient’s survival probability.
Based on the results of multivariate Cox proportional hazard regression analysis in the training set, a nomogram was developed and validated. Nomogram validation included two components. First, internal validation of the clinic nomogram was performed using a concordance index (C-index) by subjecting the nomogram of bootstrapping with 200 resamples.15 Next, for external validation, we predicted the survival of H7N9 patients using data from the other 56 individuals of the validation set. Calibration of the nomogram for 1-, 2- and 3-month probability of survival in H7N9 patients was performed by comparing the predicted probability of survival with the observed probability of survival after bias correction.
All statistical analyses were conducted with R software (version i 386 3.6.1; www.R-project.org, 2019). The multivariable Cox proportional hazard regression model was created using the R software’s “survival” package, while the nomogram and calibration curves were plotted using the “rms” package.
Results
Patient Characteristics
A flow diagram summarizing the identified eligible patients and the study participants is shown in Figure 1. The baseline characteristics of the study population are listed in Table 2. We retrospectively studied 227 patients with H7N9 VI in Zhejiang province from 2013 to 2019. Patients in the training set (n=171) and the external validation set (n= 56) were analyzed, respectively. The mean age was 58.06 ± 14.05 and 57.17 ± 11.61, and the ratio of males to females was 2.42 to 1 and 3.03 to 1 in the training and validation set, respectively.
 |
Table 2 Baseline Characteristics of Patients with H7N9 Virus Infection in the Study Population |
Notably, 128 confirmed cases (56.4%) were aggregated in Hangzhou city in this study population. Most of the cases were due to poor hand hygiene [PHH; 70.0% (159 of 227 cases)], chronic diseases [CD; 66.1% (150 of 227 cases)], and low education level [i.e., primary school or illiterate; 59.5% (135 of 227 cases)] in the study population. The average age of H7N9 patients who died was older than that of patients for the survival group in the study population (P < 0.05). Additionally, almost all cases [99.6% (226 of 227 cases)] in the study population had experienced direct contact with poultry or birds or visited a market where live poultry or birds were sold within 14 days preceding illness onset. The case-fatality rate was 32.8% (56 of 171 cases) and 30.4% (17 of 56 cases) for the training and validation sets, respectively.
Predictors’ Selection
Tables 2 and 3 summarize the results of univariate analyses of the association between an individual covariate and the survival from influenza A (H7N9). Seventeen of the 43 tested covariates were significantly associated with the survival from influenza A (H7N9) in the training sets (P < 0.05). The significant covariates were (a) demographic characteristics, including age, age ≥60 years, being retired, CD, and PHH, (b) time characteristics of the diagnosis and treatment, including the amount of time from illness onset to the first medical visit (TIOFMV), time from illness onset to treatment outcome, time treated in an outpatient clinic, and time treated in a hospital, and (c) clinical characteristics, including initial PaO2/FiO2 ratio (IPFR) ≤300 mmHg, incubation period (IP), the IP of ≤5 days, peak C-reactive protein (CRP) ≥120 mg/L, increased initial neutrophil percentage (INP), initial bilateral lung infection (IBLI), peak CRP, and INP. The remaining 26 covariates, including gender, onset time, incidence area, type of residence, education level, surgery, smoking, drinking, direct contact with poultry or birds or having visited a market where live poultry or birds were sold within 14 days preceding illness onset, time from illness onset to hospitalization, time from illness onset to oseltamivir treatment, time from illness onset to admission to the intensive care unit, time from illness onset to laboratory confirmation, time from illness onset reported to the CDC, first symptom count, body temperature peak, initial white blood cell count (IWBCC), increased IWBCC, decreased IWBCC, decreased initial lymphocyte percentage (ILP), decreased initial hemoglobin value, decreased initial blood platelet count, ILP, initial hemoglobin value, and initial blood platelet count, were not associated with the survival from influenza A (H7N9) (P > 0.05).
 |
Table 3 Univariable Cox Proportional Hazard Regression Analysis of Predictors for the Survival with Influenza A (H7N9) in Zhejiang Province, China |
To further explore the possible predictors of survival from influenza A (H7N9), we performed a multivariate Cox proportional hazard regression analysis. The analysis showed that age ≥60 years, CD, PHH, TIOFMV, IP ≤ 5 days, IPFR ≤300 mmHg, peak CRP ≥ 120 mg/L, and IBLI were significantly linked to the survival in H7N9 patients. The models also indicated that of all of the independent variables, IPFR ≤300 mmHg was the strongest impact factor for the survival from influenza A (H7N9) (HR = 5.90, 95% CI: 2.99–11.64, P <0.001) (Table 4). These findings were used to create a practical clinic nomogram for predicting the survival probability from influenza A (H7N9) (Figure 2).
 |
Table 4 Multivariable Cox Proportional Hazard Regression Model for the Survival on Patients with H7N9 Virus Infection in Zhejiang Province, China |
Construction of the Nomogram
A nomogram was developed to assess the survival using significant factors from the 171 patients’ data in the training set. With 8 variables of training set such as IPFR ≤300 mmHg, IBLI, peak CRP ≥ 120 mg/L, TIOFMV, IP ≤ 5 days, CD, PHH, and age ≥60 years in Table 4, it was possible to create a nomogram to predict the probability of survival from influenza A (H7N9) (Figure 2). The top row of the nomogram corresponds to the general score. For each variable listed on the left, there is a corresponding row on the right indicating possible descriptors. After characterizing the patient for each variable, a perpendicular line toward the first row should be drawn to identify the value. This action should be performed for all 8 variables, followed by tallying the final score. This final score should be identified in a total point row and then a perpendicular line is drawn that corresponds to the probability of survival from influenza A (H7N9).
Calibration and Validation of the Nomogram
After internal validation using the bootstrap technique, the C-index of this nomogram was 0.802 (95% CI: 0.694–0.901) and 0.793 (95% CI: 0.611–0.952) for the training and validation sets, respectively, which indicated adequate discriminatory power. The calibration plots were also performed separately using the training and external validation sets. As shown in Figure 3, the calibration plot shows that the predicted 1-, 2-, and 3-month probabilities of survival from influenza A (H7N9) correspond closely with the actual 1-, 2-, and 3-month probabilities of survival from influenza A (H7N9) estimated in the training set. Figure 4 illustrates that the nomogram appears well calibrated, and there is a strong correlation between predicted and observed outcomes across the spectrum of predictions in the external validation set.
Predicting an Individual Patient’s Risk
To make it easier to interpret our results, we represented the final reduced model with a nomogram that can be used to calculate a prognostic score and estimate the survival from influenza A (H7N9) for an individual patient (Figure 2). The nomogram produced the following mathematical predictive model for the presence of survival from influenza A (H7N9) in the training set, with h (t, x) denoting the probability of survival in H7N9 patients:16
h (t, x) = h0 (t) exp (β1x1 + … + βixi+ … + βkxk) = h0 (t) exp [0.634×(age ≥ 60 years) +1.071×CD+1.431×PHH+0.258×TIOFMV+1.356×(IP ≤ 5 days) +1.774×(IPFR ≤300 mmHg)+1.482×(peak CRP ≥ 120 mg⁄L)+0.879×IBLI]
where h (t, x) is the hazard at time t after a defined starting point for an individual with variables x = (x1 … xi … xk) is being predicted by h0 (t), the so-called underlying hazard at time t, and the predictor variables x1 to xk (recorded at time zero), each variable xi being multiplied by a corresponding regression coefficient βi. Here, exp stands for the exponential function, e.g., exp (βx) = e βx, and the underlying hazard h0 (t) is the hazard at time t of an individual whose xi’s are all zero.
The predicted probabilities associated with each factor are mapped into points on a scale from 0 to 100. The presence or the level of each predictive factor is associated with a point system, allowing summing up the points for 8 independent predictors. The total points accumulated by the various covariates correspond to the predicted probability of survival. For example, for an individual with the characteristics of 60 years of age and older, CD, TIOFMV= 14 days, IP ≤ 5 days, IPFR ≤300 mmHg, peak CRP ≥ 120 mg/L, and IBLI (see Table 5).
 |
Table 5 Predicting an Individual H7N9 Patient’s Survival |
Discussion
H7N9 VI, as we all know, usually presents as a very severe disease. Although the H7N9 VI can be treated, the mortality of H7N9 cases is very high.7 In reviewing the literature, no data was found on validated prognostic models for the survival of H7N9 infection cases. Previous studies17,18 demonstrated that a central component of preventive strategies is the identification of individuals at risk for the development of the disease. Accurate prediction and estimation on the prognosis of the disease are essential for patient counseling and treatment decision-making.19 Admittedly, patients are very interested to inquire about the chance of recovery and the probability of survival according to data from their individual case. Unfortunately, far too little attention has been paid to developing a nomogram so as to comprehensively estimate an H7N9 patient’s specific probability of survival in clinical practice.
In this study, we performed a matched case−control study to explore the predictors of survival of H7N9 cases. Our study has identified 8 independent predictors of survival obtained from primary patient data, including sociodemographic data, clinical findings, laboratory data, and epidemiology. Based on the results of the present study, we established and validated a robust nomogram for providing simple and precise personalized prediction of the survival of H7N9 cases (Figure 2). Our findings may provide more reliable evidence in developing prevention and control strategies of H7N9 VI. We anticipate that these results will be useful in reducing the mortality of H7N9 VI and the design of clinical interventions for improving the life quality of patients with H7N9 VI.
The current study is important because it offers a few important features. First, this is the first nomogram for predicting the survival of H7N9 patients that has collected enough risk factors to allow reliable prediction and evaluation analyses. Second, in the validation analysis of nomogram, whether internal (i.e., the C-index of this nomogram was 0.802 and 0.793 for the training and validation sets, respectively) or external, the comprehensive model produced both sufficient precision and satisfied uniformity in predicting the survival of H7N9 VI. Third, H7N9 infection cases were enrolled in this study using the “all comers” principle, regardless of etiology or demographics. Thus, the sample is more likely to be representative of the general population in Zhejiang Province.
Another important aspect is that the applying of nomogram is very simple, convenient, and economy-friendly (i.e., an accurate assessment is made by just using eight dominant predictors of survival among H7N9 patients). The nomogram model is comparatively easy to use, which can assess the combination of various predictors, and help clinicians accurately predict the prognosis of an individual case. The clinic nomogram may demonstrate better individualized identification than a general regressive forecasting model.4,6-9 Although previous reports7 have suggested that a logistic regression model is used to evaluate the death risk of H7N9 VI, it may not accurately reflect the effect of variables on prognostication. In fact, the survival analysis needs to consider the impact of time effect.20 As such, we specifically consider assessing the survival probability by using a semi-parametric model (i.e., Cox proportional hazard model) to maximize the Wald χ2 statistic.
Interestingly, it could be inferred that a significant association exists between a shorter incubation period and a greater risk of death among human H7N9 cases. Generally, when the incubation period or the duration of premonitory symptoms are shorter in respiratory diseases, the symptoms are more severe and the mortality rate is higher,21,22 this rule of thumb is supported by the present study’s findings. This association may be attributed to the avian influenza virus’ growth characteristics, receptor usage, and host response leading to more severe cytopathic effects.23
Likewise, PHH is significantly associated with the survival in patients with H7N9 VI. Research has indicated that a high viral load is associated with illness severity and prognosis,24 which might have originated from poor hand hygiene and self-protection measures after contact with poultry or other animals.13,25 To improve the survival of H7N9 VI, it is vital that appropriate hand hygiene practices are implemented by the susceptible population after direct contact with poultry or birds. Furthermore, it can be inferred that PHH is associated with a nonlinear effect on the survival of H7N9 patients (Figure 2). Although worse survival among H7N9 patients with PHH has been previously noted,25 our study better defines the nonlinear relation of PHH to outcome.
Most notably, IPFR is a used clinical predictor of H7N9 VI’s outcome in the present study, though its diagnostic utility is disputed in clinical work.26 In accordance with the present result, previous studies27,28 have demonstrated that PFR is a very essential indicator to assess the acute lung injury or acute respiratory distress syndrome of a patient. As known, the oxygen transport is diffusion-limited from the alveolus to the pulmonary capillary in patients with pulmonary diseases (i.e., pulmonary fibrosis).29 Several reports26–29 have shown that the PFR responds to an increase of diversion at a constant oxygen saturation by indicating poorer pulmonary function. Thus, it is reasonable to assume that the PFR may be used in clinical settings as an indicator of predicting pulmonary function status of patients with H7N9 VI. In the model presented here, the survival of H7N9 VI is associated with IPFR less than or equal to 300 mmHg (see Table 4). This finding, while preliminary, suggests that IPFR would seem an adequate measure of pulmonary dysfunction in patients with H7N9 VI.
In addition to the predictors for above, several other predictors are selected based on the multivariate Cox regression analysis, including peak CRP ≥ 120 mg/L, 60 years of age and older, CD, TIOFMV, and IBLI. All these predictors have a strong effect on the survival and are therefore included in the individualized nomogram model. Data in the present study also quantitate the authentic effect that these predictors have on survival. In brief, by aggregating independent predictors into a clinic nomogram model, we can comprehensively predict an H7N9 patient’s personalized probability of survival (Figure 2).
Most importantly, the best way to interpret and apply these findings is not in terms of how the individual factors contribute to risk but how these parameters can be modified or improved to potentially increase the survival of H7N9 patients.30 Since the death mechanism of H7N9 patients is still unclear, our findings and algorithm should be used to modify identified patient risk factors in an effort to minimize mortality.
In terms of our findings, identifying 8 predictors of survival may have an impact on the treatment, healthcare and surveillance of H7N9 cases. Our study confirms that decreased IPFR is defined as a strong predictor of survival of patients with H7N9 VI. An implication of this is the possibility that the uppermost treatment strategy is to improve pulmonary function for H7N9 cases. We also find that the peak CRP level may be considered a particularly important factor when evaluating the probability associated with survival in H7N9 cases. According to a study conducted by Gao and colleagues,31 CRP is involved in the deterioration observed in severe influenza infection and may be a candidate molecule for immunotherapy treatment. A previous study31 indicated that CRP treatment can improve the prognosis of patients with H7N9 VI. It is well known that therapeutic measures are elective, so we should select a treatment measure that can improve CRP. This clinic tool may be able to help physicians to solve such problems. Moreover, this nomogram can provide information in guiding clinicians’ decision-making regarding the most effective intervention strategies for H7N9 patients. For example, according to this algorithm, an H7N9 patient is found to be a high risk for death. This finding has an important implication for developing the strategy of early intervention in the high-risk death population of H7N9 patients. In addition, we find that the cost of applying this nomogram is mainly involved in developing a practice tool of nomogram (i.e., an evaluation scale) and incorporating the nomogram into the treatment information system of hospital. To the government department, it should be able to afford the cost and expense of applying this tool. Overall, this nomogram may display the advanced public health concept of predictive, preventive, and personalized medicine.32 This tool deserves to be further explored in future researches of clinic and public health. Considering these advantages of nomogram, we suggest that the government should guide, support and foster the development of this tool. Additionally, a treatment scheme of H7N9 cases including this nomogram should be formulated by the government.
Our study does have some limitations. First, our nomogram is limited by the retrospective nature of the data, which could suffer from recall bias and failure to incorporate some recognized prognostic parameters (e.g., the frequency or intensity of exposure and bacterial coinfection) and some important molecular factors (e.g., pro-inflammatory cytokine and angiotensin II). Second, further efforts regarding prospective data collection and patient follow-up, wider geographic recruitment, and the incorporation of additional factors are encouraged to improve this tool. Third, further studies with large sample populations are necessary to confirm our model. Despite these limitations, as we know, there are limited numbers of published data on the survival of H7N9 patients. Therefore, this study could contribute information about the novel concept of predictive, preventive, and personalized medicine for the survival of H7N9 patients.
Conclusion
So far, little has been done to enhance the clinician’s ability to properly counsel patients about individual risk prior to initiating treatment for H7N9 infection. In the matched case–control study, we constructed and validated a novel nomogram based on the status of 60 years of age and older, CD, PHH, TIOFMV, IP ≤ 5 days, IPFR ≤300 mmHg, peak CRP ≥120 mg/L, and IBLI, which accurately predict the probability of survival among patients with H7N9 VI.
In conclusion, this nomogram can serve an important role in counseling patients with H7N9 VI and guide treatment decisions. Considering the high mortality of H7N9 patients and high medical cost of H7N9 patients' care, the early identifying of at-risk individuals and early intervening before the onset of illness can reduce the high mortality and economic burden attributed to this disease.
Abbreviations
VI, virus infection; CDC, Center for Disease Control and Prevention; OR, odds ratio; CI, confidence interval; C-index, concordance index; PHH, poor hand hygiene; CD, chronic diseases; TIOFMV, time from illness onset to the first medical visit; IP, incubation period; CRP, C-reactive protein; IPFR, initial PaO2/FiO2 ratio; INP, initial neutrophil percentage; IBLI, initial bilateral lung infection; IWBCC, initial white blood cell count; ILP, initial lymphocyte percentage; IVH, initial value of hemoglobin; IBPC, initial blood platelet count.
Ethics and Consent Statement
We obtained ethical approval to carry out this study from the Institutional Review Board (IRB) of Hangzhou Center for Disease Control and Prevention, Hangzhou, China. The medical directors of the selected hospitals acknowledged this IRB and delivered their permission to start data collection. All patients or their relatives provided written informed consent for this research, which was carried out in compliance with the Helsinki Declaration. All individual information was strictly kept confidential and anonymous in the manuscript.
Acknowledgment
We extend our gratitude to the individuals vital to executing this study. In particular, we thank the First Affiliated Hospital, School of Medicine, Zhejiang University and other designated hospitals of avian influenza A (H7N9) VI treatment in Zhejiang province for supplying clinical data and Hangzhou CDC’s field investigators for supplying epidemiological data.
Author Contributions
QLC and ZS developed the study concept and supervised data collection. All authors were involved in the study design. QLC, GZ and LX analyzed the data with input from all authors. QLC drafted the initial manuscript. ZS and GZ provided input on early drafts. All authors contributed to data analysis and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work is supported by the Medical Science and Technology Project of Zhejiang Province (grant number: 2020PY064) and the Health Science and Technology Project of Hangzhou Municipality (grant number: OO20190783).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Department of Health of the Government of the Hong Kong Special Administrative Region. Avian Influenza Report. Available from: https://www.chp.gov.hk/files/pdf/2019_avian_influenza_report_vol15_wk44_chi.pdf.
2. Sha J, Chen X, Ren Y, et al. Differences in the epidemiology and virology of mild, severe and fatal human infections with avian influenza A (H7N9) virus. Arch Virol. 2016;161(5):1239–1259. doi:10.1007/s00705-016-2781-3
3. Qiu W, Chu C, Mao A, Wu J. The impacts on health, society, and economy of SARS and H7N9 outbreaks in China: a case comparison study. J Environ Public Health. 2018;2018:2710185. doi:10.1155/2018/2710185
4. Burke SA, Trock SC. Use of influenza risk assessment tool for prepandemic preparedness. Emerg Infect Dis. 2018;24(3):471–477. doi:10.3201/eid2403.171852
5. Sheshadri A, Shah DP, Godoy M, et al. Progression of the radiologic severity index predicts mortality in patients with parainfluenza virus-associated lower respiratory infections. PLoS One. 2018;13(5):e0197418. doi:10.1371/journal.pone.0197418
6. Gustafson L, Jones R, Dufour-zavala L, et al. Expert elicitation provides a rapid alternative to formal case-control study of an H7N9 avian influenza outbreak in the United States. Avian Dis. 2018;62(2):201–209. doi:10.1637/11801-011818-Reg.1
7. Yang Y, Li X, Birkhead GS, Zheng Z, Lu JH. Clinical indices and mortality of hospitalized avian influenza A (H7N9) patients in Guangdong, China. Chin Med J (Engl). 2019;132(3):302–310. doi:10.1097/CM9.0000000000000043
8. Yang Y, Wong G, Yang L, et al. Comparison between human infections caused by highly and low pathogenic H7N9 avian influenza viruses in wave five: clinical and virological findings. J Infect. 2019;78(3):241–248. doi:10.1016/j.jinf.2019.01.005
9. Subbarao K. Avian influenza H7N9 viruses: a rare second warning. Cell Res. 2018;28(1):1–2. doi:10.1038/cr.2017.154
10. Kattan MW. Comparison of Cox regression with other methods for determining prediction models and nomograms. J Urol. 2003;170(6):S6–S10. doi:10.1097/01.ju.0000094764.56269.2d
11. Thurtle D, Rossi SH, Berry B, Pharoah P, Gnanapragasam VJ. Models predicting survival to guide treatment decision-making in newly diagnosed primary non-metastatic prostate cancer: a systematic review. BMJ Open. 2019;9(6):e029149. doi:10.1136/bmjopen-2019-029149
12. Kasiulevičius V, Šapoka V, Filipavičiūtė R. Sample size calculation in epidemiological studies. Gerontologija. 2006;7(4):225–231.
13. Cheng QL, Ding H, Sun Z, et al. Retrospective study of risk factors for mortality in human avian influenza A (H7N9) cases in Zhejiang Province, China, March 2013 to June 2014. Int J Infect Dis. 2015;39(10):95–101. doi:10.1016/j.ijid.2015.09.008
14. Franzone A, Heg D, Räber L, et al. External validity of the “all-comers” design: insights from the BIOSCIENCE trial. Clin Res Cardiol. 2016;105(9):744–754. doi:10.1007/s00392-016-0983-z
15. Kulesa A, Krzywinski M, Blainey P, Altman N. Sampling distributions and the bootstrap. Nat Methods. 2015;12(6):477–478. doi:10.1038/nmeth.3414
16. Christensen E. Multivariate survival analysis using Cox’s regression model. Hepatology. 1987;7(6):1346–1358. doi:10.1002/(ISSN)1527-3350
17. Evans JP, Skrzynia C, Burke W. The complexities of predictive genetic testing. BMJ. 2001;322(7293):1052–1056. doi:10.1136/bmj.322.7293.1052
18. Golubnitschaja O, Costigliola V. General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European association for predictive, preventive and personalised medicine. EPMA J. 2012;3(1):14. doi:10.1186/1878-5085-3-14
19. Callegaro D, Miceli R, Bonvalot S, et al. Development and external validation of two nomograms to predict overall survival and occurrence of distant metastases in adults after surgical resection of localised soft-tissue sarcomas of the extremities: a retrospective analysis. Lancet Oncol. 2016;17(5):671–680. doi:10.1016/S1470-2045(16)00010-3
20. Moore DF. Applied Survival Analysis Using R. Switzerland: Springer; 2016.
21. Virlogeux V, Fang VJ, Wu JT, et al. Brief report: incubation period duration and severity of clinical disease following severe acute respiratory syndrome coronavirus infection. Epidemiol Camb Mass. 2015;26(5):666–669. doi:10.1097/EDE.0000000000000339
22. Virlogeux V, Park M, Wu JT, Cowling BJ. Association between severity of MERS-CoV infection and incubation period. Emerg Infect Dis. 2016;22(3):526–528. doi:10.3201/eid2203.151437
23. Zhang F, Bi Y, Wang J, et al. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect. 2017;75(1):71–75. doi:10.1016/j.jinf.2017.04.001
24. de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203. doi:10.1038/nm1477
25. Liu M-D, Chan T-C, Wan C-H, et al. Changing risk awareness and personal protection measures for low to high pathogenic avian influenza in live-poultry markets in Taiwan, 2007 to 2012. BMC Infect Dis. 2015;15(1):241. doi:10.1186/s12879-015-0987-8
26. Narayanan A, Batra P, Faridi MMA, Harit D. PaO2/FiO2 ratio as predictor of mortality in neonates with meconium aspiration syndrome. Am J Perinatol. 2019;36(6):609–614. doi:10.1055/s-0038-1672171
27. Feiner JR, Weiskopf RB. Evaluating pulmonary function: an assessment of PaO2/FIO2. Crit Care Med. 2017;45(1):e40–e48. doi:10.1097/CCM.0000000000002017
28. Guo L, Wang W, Zhao N, et al. Mechanical ventilation strategies for intensive care unit patients without acute lung injury or acute respiratory distress syndrome: a systematic review and network meta-analysis. Crit Care. 2016;20(1):226. doi:10.1186/s13054-016-1396-0
29. Datta M, Via LE, Chen W, et al. Mathematical model of oxygen transport in tuberculosis granulomas. Ann Biomed Eng. 2016;44(4):863–872. doi:10.1007/s10439-015-1415-3
30. Hernandez-alejandro R, Bertens KA, Pineda-solis K, Croome KP. Can we improve the morbidity and mortality associated with the associating liver partition with portal vein ligation for staged hepatectomy (ALPPS) procedure in the management of colorectal liver metastases? Surgery. 2015;157(2):194–201. doi:10.1016/j.surg.2014.08.041
31. Gao R, Wang L, Bai T, Zhang Y, Bo H, Shu Y. C-reactive protein mediating immunopathological lesions: a potential treatment option for severe influenza a disease. EBioMedicine. 2017;22:133–142. doi:10.1016/j.ebiom.2017.07.010
32. Trovato GM. Sustainable medical research by effective and comprehensive medical skills: overcoming the frontiers by predictive, preventive and personalized medicine. EPMA J. 2014;5(1):14. doi:10.1186/1878-5085-5-14
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



