Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
No Association Between FTO Gene Polymorphisms and Central Nervous System Tumor Susceptibility in Chinese Children
Authors Liao Y, Yuan L, Zhang Z, Lin A , Zhou J, Zhuo Z, Zhao J
Received 30 October 2020
Accepted for publication 14 December 2020
Published 19 January 2021 Volume 2021:14 Pages 109—115
DOI https://doi.org/10.2147/PGPM.S289345
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Yuxiang Liao,1 Li Yuan,2 Zhiping Zhang,1 Ao Lin,3 Jingying Zhou,4 Zhenjian Zhuo,3 Jie Zhao1
1Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha 410008, Changsha, People’s Republic of China; 2Department of Pathology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, People’s Republic of China; 3Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, People’s Republic of China; 4Department of Hematology, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou 325027, Zhejiang, People’s Republic of China
Correspondence: Jie Zhao
Department of Neurosurgery, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha 410008, Hunan, People’s Republic of China
Email [email protected]
Zhenjian Zhuo
Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, People’s Republic of China
Tel +86-13510841053
Email [email protected]
Background: Central nervous system (CNS) tumor is a malignancy commonly seen occurring in childhood, worldwide. Fat mass and obesity-associated (FTO) enzyme, initially identified as an obesity-related protein, also functions as a susceptibility gene for cancers. However, predisposing effect of FTO gene single nucleotide polymorphisms (SNPs) on CNS tumor risk remains unknown.
Methods: Herein, we genotyped 314 CNS tumor patients and 380 healthy controls samples from three hospitals to explore whether FTO gene SNPs impact CNS tumor risk. TaqMan SNP genotyping assay was applied for the genotyping. Odds ratios (ORs) and 95% confidence intervals (CIs), generated from multinomial logistic regression, were applied to determine the associations of SNPs (rs1477196 G>A, rs9939609 T>A, rs7206790 C>G, and rs8047395 A>G) in FTO gene with risk of CNS tumor.
Results: We failed to detect significant associations between FTO gene SNPs and CNS tumor risk, either in single-locus or combined analysis. A significantly increased ependymoma risk was found for carriers with 3– 4 risk genotypes in comparison to 0– 2 risk genotypes (adjusted OR=1.94, 95% CI=1.11– 3.37, P=0.020).
Conclusion: Our data indicated that FTO gene SNPs are unlikely to have large effects on CNS tumor risk but may have weaker effects.
Keywords: CNS tumor, risk, FTO, polymorphism, Chinese
Introduction
Central nervous system (CNS) tumors comprise about 20% of all malignancies occurring in childhood and adolescence.1 Gliomas account for the largest proportion of malignant brain tumors at 80%.2,3 Gliomas are heterogeneous primary brain tumors that originate from neuroglial stem or progenitor cells.4,5 Pediatric low-grade gliomas (LGGs), or WHO grade I or II gliomas, account for about 30% of all childhood CNS tumors. These subtype tumors rarely undergo malignant transformation with a favorable prognosis. However, for WHO grade III or IV high-grade gliomas (HGGs),6 the prognosis is poor at only 20% for 5-year survival rate.7 Of note, the development of new therapies such as immune therapies and target therapies has helped to improve the survival of CNS tumor patients.8
Exposure to high dose ionizing radiation is the most firmly established environmental risk factor for CNS tumor.9–11 Other factors such as cellular phones, tobacco uses, and allergies/atopic disease have been extensively investigated for CNS tumor risk.12,13 However, such relationships remain inconclusive. Heritable genetic variants are important factors associated with CNS tumor risk. Several genome-wide association studies of glioma patients have identified SNPs in RTEL1, PHLDB1, CCDC26, TP53, EGFR, and CDKN2B genes as CNS tumor risk-associated susceptibility polymorphisms.14–17 Candidate gene studies focusing on genes thought to be involved in gliomagenesis also identified multiple genetic variants associated with risk of CNS tumor.18,19 All the genetic variants identified so far only reveal a piece of the genetic landscape of CNS tumor.
The fat mass and obesity-associated (FTO) gene was originally identified as an obesity susceptibility gene by genome-wide association studies (GWASs).20 Multiple single-nucleotide polymorphisms (SNPs) located in the intron 1 of FTO gene have been well recognized for their strong relationships with the risk of obesity.21–24 The contribution of FTO gene SNPs to cancer risk also has been demonstrated previously.25 For example, Kaklamani et al found that intron 1 (obesity-associated region) SNPs rs7206790, rs8047395, rs9939609, and rs1477196 in FTO gene were tightly associated with risk of breast cancer. It is highly possible that the obesity-associated SNPs could cause an increased risk to overweight and obese patients and lead to an increased cancer susceptibility.26 Notably, SNPs outside of intron 1 of FTO gene were also found to impact the susceptibility of cancer.27 Indeed, FTO has been reported as an oncogenic gene in various cancers. Of note, FTO was the first identified N6-methyladenosine (m6A) demethylase of eukaryotic mRNA.28 The aberrant m6A modifications mediated by FTO expression contributed to the development of cancers.29–31 m6A RNA methylation regulators are crucial participants in the malignant progression of CNS tumor. FTO may preferentially mediate demethylation of different methylation targets in CNS tumor.32 The strong relation between FTO gene as well as its SNPs and cancer motivated us to evaluate the association of FTO gene SNPs with CNS tumor risk. This study represents the first attempt to explore FTO gene SNPs in relation to CNS tumor risk in a Chinese population.
Materials and Methods
Study Subjects
In total, three hospitals located in Wenzhou, Guangzhou, and Changsha, participated in this project. All patients were histopathologically confirmed with CNS tumor without previous radiotherapy or chemotherapy, while cancer-free controls were enrolled from hospital visitors in the same hospital during the same period. The detailed criteria for diagnosing CNS tumor was based on the 2016 World Health Organization classification of tumors of the CNS.6 Controls were frequency-matched to the cases by age and gender. Controls with other tumors, neurological disease, congenital genetic disease, and infectious disease were excluded. None of the study subjects had a blood relationship. Written informed consent was provided from all enrolled patients’ guardians prior to study commencement. The research protocol for this study was reviewed and approved by the Institutional Review Boards of the participating hospitals (Xiangya Hospital, Guangzhou Women and Children’s Medical Center, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University). This study was conducted in accordance with the Declaration of Helsinki.
Genotyping
Potentially functional polymorphisms in FTO gene were screened from the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) and SNPinfo (http://snpinfo.niehs.nih.gov/snpfunc.htm). Detailed information on SNP selection and genotyping could be found previously.33–35 Four SNPs rs1477196 G>A, rs9939609 T>A, rs7206790 C>G, and rs8047395 A>G in FTO gene were screened out for analysis. Genomic DNA extraction from blood samples was accomplished through QIAamp DNA blood mini kit (QIAGEN Inc., Valencia, CA, USA). The DNA extraction process was carried out in accordance with the manufacturer’s instructions. To ensure genotyping accuracy, a blinded fashion was adopted without knowing the case-control status of the subject. Primers and fluorescently labeled probes were purchased from Applied Biosystems for detection of both the wild-type and variant alleles. Genotyping of SNPs was performed with a TaqMan SNP Genotyping Assay (Applied Biosystems, USA), with details in a previous study.33 The conditions of reactions were set as follow: preread stage at 60°C for 30 seconds, holding stage at 95°C for 10 minutes, repeated 45 cycles each of denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 1 minute. For quality control purpose, we also randomly selected 10% control samples to run a second genotype. A 100% rate was achieved for the second run data compared to the original data.
Statistical Analysis
Differences between case and control groups clinical variables were evaluated using Chi-square test or t-test, as appropriate. A goodness-of-fit χ2 test was applied to test whether the expected genotypes with the observed ones were in Hardy–Weinberg equilibrium (HWE) in the controls. The calculation of odds ratios (ORs) and 95% confidence intervals (95% CIs) were under multivariate logistic regression adjusted by sex and age, and further applied to determine the association between each SNP and CNS tumor risk. The P<0.05 indicated statistical significance. Statistical analysis was performed with SAS v10.0 (SAS Institute Inc., Cary, NC, USA).
Results
Population Characteristics
Frequency distributions of age (P=0.461) and gender (P=0.379) were similar between 314 patients and 380 controls, indicating adequate matching on these factors. The mean age for cases was 70.46 ± 48.39 months and for controls 64.19 ± 37.38 months. Cases were categorized as astrocytic tumors (214, 68.15%), ependymoma (61, 19.43%), neuronal and mixed neuronal-glial tumors (25, 7.96%), embryonal tumors (12, 3.82%), and not available (NA) (2, 0.64%). As to WHO stages, 151 (48.09%) CNS tumor cases were classified into stage I, 73 (23.23%) into stage II, 36 (11.46%) into stage III, 53 (16.88%) into stage IV, and 1 (0.32%) into NA (Table 1).
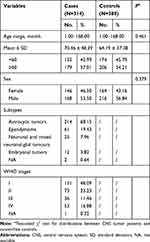 |
Table 1 Frequency Distribution of Selected Variables in CNS Tumor Patients and Cancer-Free Controls in Combined Subjects |
Effect of FTO Gene SNPs on CNS Tumor Risk
The SNPs of FTO gene and their relationships with CNS tumor risk are summarized in Table 2. As expected, all the four SNPs rs1477196 G>A, rs9939609 T>A, rs7206790 C>G, and rs8047395 A>G in FTO gene were in Hardy–Weinberg equilibrium (HWE), each with a value of 0.538, 0.835, 0.856, and 0.851, respectively. For the four single locus analysis, none of them could significantly predispose to CNS tumor risk, respectively. We then treated rs1477196 GG/GA, rs9939609 TA/AA, rs7206790 CG/GG, rs8047395 AA/AG genotypes as risk genotypes. Subjects with 1, 2, 3, and 4 risk genotypes still could not impact risk of CNS tumor. Similar negative results were obtained for 3–4 risk genotypes in comparison to 0–2 risk genotypes.
 |
Table 2 Association Between FTO Gene Polymorphisms and CNS Tumor Susceptibility in Chinese Children |
Stratification Analysis
The relationships between SNP rs7206790, risk genotypes and CNS tumor risk, respectively, were further determined by stratified analyses in age, gender, subtypes, and clinical stages subgroups (Table 3). Compared to CC genotype, rs7206790 CG/GG could not modify risk of CNS tumor, under any subgroup. Compared to 0–2 risk genotypes, carriers with 3–4 risk genotypes were more likely to develop ependymoma (adjusted OR=1.94, 95% CI=1.11–3.37, P=0.020).
 |
Table 3 Stratification Analysis of Risk Genotypes with CNS Tumor Susceptibility |
Discussion
Our study represents the first candidate gene-based association study to assess the relationship between FTO gene SNPs and CNS tumor risk in a Chinese population in three multicenter case-control studies. In general, no associations with risk of CNS tumor reached statistical significance in either individual or combined SNPs. We did observe that individuals with 3–4 risk genotypes were significantly associated with increased risk of CNS tumor.
FTO has been well documented to be involved in various types of cancers, such as acute myeloid leukemias (AMLs),36 breast cancer,37 and ovarian cancer.38 More recently, evidence regarding role of FTO on CNS tumor are also emerging. Cui et al39 demonstrated that inhibition of the RNA demethylase FTO could intensively suppress the development of glioblastoma stem (-like) cells (GSCs). Inhibition of FTO could enhances the anti-glioma effect of temozolomide by targeting MYC-miR-155/23a cluster-MXI1 feedback circuit.40 Tao et al41 found that protein level of FTO was significantly lower in gliomas than that in normal tissues. Further mechanism experiment revealed the glioma-suppressive role of FTO may through interacting with FOXO3a to promote its nuclear translocation and target gene expression.
Previous epidemiology studies have provided evidence regarding the associations of overweight/obesity related-FTO gene SNPs and risk of various types of cancers. For instance, SNPs rs7206790, rs9939609, rs8047395, and rs1477196, located in the intron 1 (obesity-associated region) in FTO gene, have been demonstrated to predispose to breast cancer. Notably, rs1477196 ranks the most significant relationship with risk of breast cancer.26 Gaudet et al42 genotyped 189 tagSNPs in FTO gene which were previously related to obesity by genome-wide association scans. However, none of the genotyped FTO gene SNPs could confer to risk of endometrial cancer. Sigurdson et al25 conducted a large independent papillary thyroid cancer (PTC) case-control study in Germany. They found that FTO gene rs8047395 is associated with PTC risk, using samples of 422 PTC cases and 752 controls recruited from three German clinical centers. Zeng et al43 genotyped 6 FTO gene SNPs (rs9939609, rs1477196, rs6499640, rs16953002, rs11075995, and rs1121980) using 537 breast cancer cases and 537 controls. Among them, FTO gene rs1477196 AA genotype was significantly related to decreased breast cancer risk compared to GG genotype, while rs16953002 AA genotype contributed to a higher breast cancer risk compared to GG genotype. Iles et al27 found that variants of FTO gene outside of intron 1 were associated with melanoma risk. To our knowledge, no studies have assessed the association of FTO gene SNPs and CNS tumor risk. Considering the implications of FTO gene SNPs to the risk of multiple cancers, it is of significance to explore the potential value of FTO gene SNPs to identify populations at high risk of CNS tumor for targeted prevention. In the current study, no significant correlations were detected between FTO gene SNPs (rs1477196 G>A, rs9939609 T>A, rs7206790 C>G, and rs8047395 A>G) and CNS tumor risk, either in single or combined locus analysis. In stratification analysis, carriers with 3–4 risk genotypes were more likely to develop ependymoma when compared to 0–2 risk genotypes. One possible explanation of the observation is a relatively small sample size. There may be other unknown environmental factors that also interact with the weak impact of these FTO SNPs and result in the observed effect.
We attempted to apply strict measurements for the investigation of a relationship between FTO gene SNPs and CNS tumor risk; however, the following considerations are important when interpreting the results. First, due to relatively small sample size, our study is underpowered to detect all SNPs. Second, environmental factors such as cellphone use, occupation exposures, and medical history are important to consider for CNS tumor. These variables were absent from the current study, therefore, such variables remained unaccounted. Third, though subjects from all around China, all of them were Chinese descendant. Conclusions obtained from this single population limited the generalizability to other ethnicities. Meanwhile, the underlying mechanisms of how the FTO gene SNPs impacts CNS tumor risk remain to be further explored in the future.
Conclusions
In all, our multi-center study identified that FTO gene SNPs may not have enough impact on risk of CNS tumor in a population of Chinese children. Further functional characterization is warranted to provide a more accurate picture of the biological mechanisms underlying CNS tumor susceptibility.
Acknowledgments
This work was funded by grants from the National Natural Science Foundation of China (No: 81802346, 82002636).
Disclosure
The authors have no conflicts of interest to declare.
References
1. Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150. doi:10.1111/nan.12432
2. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16 Suppl 4:iv1–iv63. doi:10.1093/neuonc/nou223
3. Neglia JP, Robison LL, Stovall M, et al. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2006;98(21):1528–1537. doi:10.1093/jnci/djj411
4. Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi:10.1038/nrdp.2015.17
5. Udaka YT, Packer RJ. Pediatric brain tumors. Neurol Clin. 2018;36(3):533–556. doi:10.1016/j.ncl.2018.04.009
6. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
7. Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40(1):1–14. doi:10.1007/s10143-016-0709-8
8. Jiang T, Nam DH, Ram Z, et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2020. doi:10.1016/j.canlet.2020.10.050
9. Bondy ML, Scheurer ME, Malmer B, et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113(7 Suppl):1953–1968. doi:10.1002/cncr.23741
10. Vienne-Jumeau A, Tafani C, Ricard D. Environmental risk factors of primary brain tumors: A review. Rev Neurol (Paris). 2019;175(10):664–678. doi:10.1016/j.neurol.2019.08.004
11. Davis ME. Epidemiology and overview of gliomas. Semin Oncol Nurs. 2018;34(5):420–429. doi:10.1016/j.soncn.2018.10.001
12. Cordier D, Gerber M, Brand S. Effects of two types of exercise training on psychological well-being, sleep, quality of life and physical fitness in patients with high-grade glioma (WHO III and IV): study protocol for a randomized controlled trial. Cancer Commun (Lond). 2019;39(1):46. doi:10.1186/s40880-019-0390-8
13. Diwanji TP, Engelman A, Snider JW, Mohindra P. Epidemiology, diagnosis, and optimal management of glioma in adolescents and young adults. Adolesc Health Med Ther. 2017;8:99–113. doi:10.2147/AHMT.S53391
14. Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi:10.1038/ng.408
15. Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi:10.1038/ng.407
16. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. doi:10.1038/ng.3823
17. Kinnersley B, Labussiere M, Holroyd A, et al. Genome-wide association study identifies multiple susceptibility loci for glioma. Nat Commun. 2015;6:8559. doi:10.1038/ncomms9559
18. Yang T, Wen Y, Li J, et al. NRAS and KRAS polymorphisms are not associated with hepatoblastoma susceptibility in Chinese children. Exp Hematol Oncol. 2019;8:11. doi:10.1186/s40164-019-0135-z
19. Yang T, Wen Y, Li J, et al. Association of the TP53 rs1042522 C>G polymorphism and hepatoblastoma risk in Chinese children. J Cancer. 2019;10(15):3444–3449. doi:10.7150/jca.33063
20. Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi:10.1126/science.1141634
21. Yang J, Loos RJ, Powell JE, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490(7419):267–272. doi:10.1038/nature11401
22. Ningombam SS, Chhungi V, Newmei MK, et al. Differential distribution and association of FTO rs9939609 gene polymorphism with obesity: A cross-sectional study among two tribal populations of India with East-Asian ancestry. Gene. 2018;647:198–204. doi:10.1016/j.gene.2018.01.009
23. Peters U, North KE, Sethupathy P, et al. A systematic mapping approach of 16q12.2/FTO and BMI in more than 20,000 African Americans narrows in on the underlying functional variation: results from the Population Architecture using Genomics and Epidemiology (PAGE) study. PLoS Genet. 2013;9(1):e1003171. doi:10.1371/journal.pgen.1003171
24. Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi:10.1371/journal.pgen.0030115
25. Sigurdson AJ, Brenner AV, Roach JA, et al. Selected single-nucleotide polymorphisms in FOXE1, SERPINA5, FTO, EVPL, TICAM1 and SCARB1 are associated with papillary and follicular thyroid cancer risk: replication study in a German population. Carcinogenesis. 2016;37(7):677–684. doi:10.1093/carcin/bgw047
26. Kaklamani V, Yi N, Sadim M, et al. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC Med Genet. 2011;12:52. doi:10.1186/1471-2350-12-52
27. Iles MM, Law MH, Stacey SN, et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet. 2013;45(4):428–32, 432e1. doi:10.1038/ng.2571
28. Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi:10.1038/nchembio.687
29. Zhou C, Zhang Z, Zhu X, et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine. 2020;59:102955. doi:10.1016/j.ebiom.2020.102955
30. Jin H, Ying X, Que B, et al. N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207. doi:10.1016/j.ebiom.2019.07.068
31. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. doi:10.1186/s12943-019-1033-z
32. Chai RC, Wu F, Wang QX, et al. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY). 2019;11(4):1204–1225. doi:10.18632/aging.101829
33. Zhuo ZJ, Liu W, Zhang J, et al. Functional polymorphisms at ERCC1/XPF genes confer neuroblastoma risk in Chinese children. EBioMedicine. 2018;30:113–119. doi:10.1016/j.ebiom.2018.03.003
34. Zhuo ZJ, Hua RX, Chen Z, et al. WTAP gene variants confer hepatoblastoma susceptibility: a seven-center case-control study. Mol Ther Oncolytics. 2020;18:118–125. doi:10.1016/j.omto.2020.06.007
35. Zhuo Z, Zhou C, Fang Y, et al. Correlation between the genetic variants of base excision repair (BER) pathway genes and neuroblastoma susceptibility in eastern Chinese children. Cancer Commun (Lond). 2020;40(11):641–646. doi:10.1002/cac2.12088
36. Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141. doi:10.1016/j.ccell.2016.11.017
37. Xu Y, Ye S, Zhang N, et al. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). 2020;40(10):484–500. doi:10.1002/cac2.12075
38. Huang H, Wang Y, Kandpal M, et al. FTO-dependent N (6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking cAMP signaling. Cancer Res. 2020;80(16):3200–3214. doi:10.1158/0008-5472.CAN-19-4044
39. Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. doi:10.1016/j.celrep.2017.02.059
40. Xiao L, Li X, Mu Z, et al. FTO inhibition enhances the antitumor effect of temozolomide by targeting MYC-miR-155/23a cluster-MXI1 feedback circuit in glioma. Cancer Res. 2020;80(18):3945–3958. doi:10.1158/0008-5472.CAN-20-0132
41. Tao B, Huang X, Shi J, et al. FTO interacts with FOXO3a to enhance its transcriptional activity and inhibits aggression in gliomas. Signal Transduct Target Ther. 2020;5(1):130. doi:10.1038/s41392-020-00234-3
42. Gaudet MM, Yang HP, Bosquet JG, et al. No association between FTO or HHEX and endometrial cancer risk. Cancer Epidemiol Biomarkers Prev. 2010;19(8):2106–2109. doi:10.1158/1055-9965.EPI-10-0515
43. Zeng X, Ban Z, Cao J, et al. Association of FTO mutations with risk and survival of breast cancer in a Chinese population. Dis Markers. 2015;2015:101032. doi:10.1155/2015/101032
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
