Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Neural stem cell-derived exosomes mediate viral entry
Authors Sims B, Gu L, Krendelchtchikov A, Matthews Q
Received 11 July 2014
Accepted for publication 3 September 2014
Published 21 October 2014 Volume 2014:9(1) Pages 4893—4897
DOI https://doi.org/10.2147/IJN.S70999
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Thomas Webster
Brian Sims,1,2,* Linlin Gu,3,* Alexandre Krendelchtchikov,3 Qiana L Matthews3,4
1Division of Neonatology, Department of Pediatrics, 2Department of Cell, Developmental, and Integrative Biology, 3Division of Infectious Diseases, Department of Medicine, 4Center for AIDS Research, University of Alabama at Birmingham, Birmingham, AL, USA
*These authors contributed equally to this work
Background: Viruses enter host cells through interactions of viral ligands with cellular receptors. Viruses can also enter cells in a receptor-independent fashion. Mechanisms regarding the receptor-independent viral entry into cells have not been fully elucidated. Exosomal trafficking between cells may offer a mechanism by which viruses can enter cells.
Methods: To investigate the role of exosomes on cellular viral entry, we employed neural stem cell-derived exosomes and adenovirus type 5 (Ad5) for the proof-of-principle study.
Results: Exosomes significantly enhanced Ad5 entry in Coxsackie virus and adenovirus receptor (CAR)-deficient cells, in which Ad5 only had very limited entry. The exosomes were shown to contain T-cell immunoglobulin mucin protein 4 (TIM-4), which binds phosphatidylserine. Treatment with anti-TIM-4 antibody significantly blocked the exosome-mediated Ad5 entry.
Conclusion: Neural stem cell-derived exosomes mediated significant cellular entry of Ad5 in a receptor-independent fashion. This mediation may be hampered by an antibody specifically targeting TIM-4 on exosomes. This set of results will benefit further elucidation of virus/exosome pathways, which would contribute to reducing natural viral infection by developing therapeutic agents or vaccines.
Keywords: neural stem cell-derived exosomes, adenovirus type 5, TIM-4, viral entry, phospholipids
Findings
Exosomes released from cells are nanostructures with diameters of 30–200 nm.1 These microvesicles are actively secreted into all body fluids, including blood, saliva, urine, and breast milk.2,3 Exosomes are routinely released at the plasma membranes of different cell types and are part of the intercellular communication network.1,3 Exosomes play important roles in adaptive immune responses to pathogens and tumors by transferring proteins, soluble factors, messenger RNAs, and microRNAs to the recipient cells.3,4 Notably, exosomes have been shown to transport retroviruses into macrophages5 and other immune cells.6 However, other particles such as prions can also be transported by exosomes.7 In this study, for the purpose of investigating the potential role of exosomes on virus entry, we prepared exosomes from neural stem cells and preliminarily evaluated the mediation of the exosomes in adenovirus type 5 (Ad5) entry into Coxsackie virus and adenovirus receptor (CAR)-deficient cells. CAR is a cellular receptor that specifically mediates the cellular entry of subgroup C adenoviruses (Ads), including Ad5.8
Generation of neural stem cell-derived exosomes
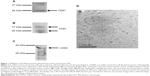
Exosomes have gained attention as effectors in the conditioned medium of stem cells. Neural stem cells have been shown to produce large amounts of exosomes.9 We sought to establish that we could collect a high concentration of neural stem cell-derived exosomes. Mouse neural stem cells were cultured in serum-free culture media for the purposes of extracting exosomes. In brief, culture medium from the cells was collected, filtered, and ultracentrifuged at 110,000× g to pellet exosomes.4 In order to confirm the successful purification of exosomes, Coomassie staining and Western blot analysis were applied on sample preparations to detect two key exosomal markers, cluster of differentiation (CD) 81 and CD63. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) was performed, followed by Coomassie staining. The Coomassie staining demonstrated the presence of CD81 proteins, with a major band of 26 kDa and a minor band at ~37 kDa (Figure 1A). Western blot analysis with an exosome-specific antibody, anti-CD81, further confirmed the CD81 protein bands at both 26 kDa and 37 kDa (Figure 1B). These multiple species of protein detected in both the Coomassie staining and Western blot analysis may indicate posttranslational modification (eg, glycosylation). In addition, SDS–PAGE and Western blot analysis were performed with an exosome-specific antibody, anti-CD63, further confirming the major, glycosylated CD63 protein band at 40 kDa (Figure 1C). In order to further verify the integrity of the exosomes, electron microscopy was performed on purified pelleted fractions. Figure 1D illustrates an electron micrograph of the mouse neural stem cell-derived exosomes. These validations ensured the integrity of exosomes for subsequent studies.
Mediation of exosomes on Ad transfer
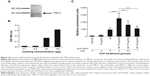
Previous studies have shown that exosomes and other microvesicles increase the ability of viruses such as the human immunodeficiency virus (HIV) to enter the cells.10 Ad has a similar size as HIV. Therefore, as proof of principle, receptor-independent Ad5 entry via neural stem cell-derived exosomes was assessed. Using Ad5 (with a luciferase reporter) and exosomes, we validated exosome-mediated entry of Ad5 into a mouse B cell line, A20, which has deficient levels of CAR.11 Luciferase activity was evaluated in different cell treatment groups (cell-only, exosomes, Ad, and Ad + exosomes groups) to illustrate the Ad5 entry level under different conditions. In brief, in the treatment group of Ad + exosomes, exosomes at three concentrations (0.1 μg, 1.0 μg, and 10 μg) were incubated with Ad5 (100 viral particles per cell) separately for 1 hour at 37°C before being applied to cells. Cells (1×106 per group) were then treated (cell-only, exosomes, Ad, and Ad + exosomes groups) for 24 hours. Cell-only, exosomes, and Ad groups were used as assay controls. Each group was washed to eliminate virus/exosome residues in the assay medium, and the cells were subsequently lysed. Lysates were subjected to luciferase analysis. In order for Ad to express the luciferase gene, the Ad/exosome complex would have to be taken up into the cells, followed by transcription and translation of the Ad genome and luciferase gene/protein. The data demonstrated that exosomes mediated the transfer of Ad5 to A20 cells, and there was significance at a quantity of 10 μg of exosomes (Figure 2). These data clearly suggested a significant quantity of receptor-independent transfer of virus into the B cell line via the mediation of exosomes.
Blockage of exosome-mediated Ad entry by interfering with T-cell immunoglobulin mucin protein-4 interactions
In order to elucidate the mechanisms that facilitate exosome interactions, we focused on T-cell immunoglobulin mucin protein 4 (TIM-4)-mediated exosome interactions. It is well known that TIM-4 acts as a phosphatidylserine (PS) receptor. Exosomes derived from various cell types have been shown to contain PS-binding proteins.12 We evaluated the presence of TIM-4 on neural stem cell-derived exosomes by Western blot (Figure 3A) and enzyme-linked immunosorbent assay (ELISA) analyses (Figure 3B). Purified exosomes were obtained as described in the section “Generation of neural stem cell-derived exosomes” (Figure 1). Exosomes were subjected to SDS–PAGE and Western blot analysis, using anti-TIM-4 antibody and donkey anti-goat secondary antibody conjugated with horseradish peroxidase. The data showed a protein band that resolved at ~64 kDa, which is likely to be glycosylated TIM-4 associated in exosomal preparations (Figure 3A). As further confirmation of the association of TIM-4 with exosomes, we performed an ELISA. Samples containing 0.1 μg, 0.5 μg, 50 μg, and 100 μg of exosomes were bound to an ELISA plate. The plate was then washed, blocked, and treated with anti-TIM-4 antibody. Subsequently, the plate was incubated with donkey anti-goat secondary antibody conjugated with horseradish peroxidase and subjected to plate reading at optical density of 450 nm. These data demonstrated that TIM-4 was present on neural stem cell-derived exosomes (Figure 3B). Because exosomes contain PS, Ad5 could potentially bind exosomes through interaction with anionic phospholipids,13 followed by entry of the Ad/exosome complex by means of interaction with anionic phospholipids. Of important note, we demonstrated that exosome-mediated uptake of Ad5 could be blocked with coincubation of anti-TIM-4 antibody (Figure 3C). As a negative control, we used a nonspecific antibody in this assay, and this antibody could not block exosome-mediated virus entry (data not shown). Typical PS-binding proteins can also bind phosphatidylcholine and phosphatidylethanolamine. Therefore, TIM-4 could potentially be associating with one of these phospholipids as well.
Virus trafficking is a complicated biological process. This process can be facilitated by nonclassic receptor-independent pathways such as exosomal trafficking. This pathway also allows virus to be delivered to a wide variety of cell types and organs that may not be a natural target for the viruses such as those described herein. This pathway is extremely important for retroviruses, which can be dormant and can be reactivated in various cell types and organs. The interaction of Ad5 with neural stem cell-derived exosomes in Figure 2 is possibly through interaction of the Ad protein, hexon, with the anionic phospholipids.13 Subsequent entry of the Ad/exosome complex is possible by means of interaction with anionic phospholipid. This interaction allows receptor-independent entry of Ad virus into a CAR-deficient cell line. Data in Figure 3 highlight that TIM-4 interactions are involved in exosome-mediated transport. Further identification aimed at elucidating virus/exosome pathways may be important to reduce natural virus infection. In addition, elucidation of exosome trafficking can be a potentially therapeutic target or avenue for vaccine application.
Acknowledgments
This work was supported by the National Institutes of Health (grant #5R01AI089337-03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
BS, LLG, and QLM designed the study; BS, LLG, and AK performed the experiments; BS, LLG, and QLM analyzed the data; BS, LLG, and QLM wrote the manuscript; BS, LLG, AK, and QLM critically revised the manuscript. All authors gave final approval of the article to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14(7):14240–14269. | ||
Sokolova V, Ludwig AK, Hornung S, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. | ||
Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. | ||
McDonald MK, Capasso KE, Ajit SK. Purification and microRNA profiling of exosomes derived from blood and culture media. J Vis Exp. 2013;(76):e50294. | ||
Ismail N, Wang Y, Dakhlallah D, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121(6):984–995. | ||
Naslund TI, Gehrmann U, Qazi KR, Karlsson MC, Gabrielsson S. Dendritic cell-derived exosomes need to activate both T and B cells to induce antitumor immunity. J Immunol. 2013;190(6):2712–2719. | ||
Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211(5):582–590. | ||
Persson A, Fan X, Widegren B, Englund E. Cell type- and region-dependent coxsackie adenovirus receptor expression in the central nervous system. J Neurooncol. 2006;78(1):1–6. | ||
Chivet M, Hemming F, Pernet-Gallay K, Fraboulet S, Sadoul R. Emerging role of neuronal exosomes in the central nervous system. Front Physiol. 2012;3:145. | ||
Kadiu I, Narayanasamy P, Dash PK, Zhang W, Gendelman HE. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J Immunol. 2012;189(2):744–754. | ||
Passineau MJ, Siegal GP, Everts M, et al. The natural history of a novel, systemic, disseminated model of syngeneic mouse B-cell lymphoma. Leuk Lymphoma. 2005;46(11):1627–1638. | ||
Krämer-Albers EM, Bretz N, Tenzer S, et al. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? Proteomics Clin Appl. 2007;1(11):1446–1461. | ||
Balakireva L, Schoehn G, Thouvenin E, Chroboczek J. Binding of adenovirus capsid to dipalmitoyl phosphatidylcholine provides a novel pathway for virus entry. J Virol. 2003;77(8):4858–4866. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.