Back to Journals » International Journal of Nanomedicine » Volume 13
Nanotopographic cues and stiffness control of tendon-derived stem cells from diverse conditions
Authors Kim SJ , Tatman PD, Song DH, Gee AO, Kim DH , Kim SJ
Received 29 July 2018
Accepted for publication 4 October 2018
Published 8 November 2018 Volume 2018:13 Pages 7217—7227
DOI https://doi.org/10.2147/IJN.S181743
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Sun Jeong Kim,1 Philip D Tatman,2 Da-Hyun Song,1 Albert O Gee,3 Deok-Ho Kim,2 Sang Jun Kim1
1Department of Physical and Rehabilitation Medicine, Stem Cell and Regenerative Medicine Institute, Samsung Medical Center, Seoul, Republic of Korea; 2Department of Bioengineering, University of Washington, Seattle, WA, USA; 3Department of Orthopedic Surgery and Sports Medicine, University of Washington, Seattle, WA, USA
Background: Tendon-derived stem cells (TDSCs) are key factors associated with regeneration and healing in tendinopathy. The aim of this study was to investigate the effects of mechanical stiffness and topographic signals on the differentiation of TDSCs depending on age and pathological conditions.
Materials and methods: We compared TDSCs extracted from normal tendon tissues with TDSCs from tendinopathic Achilles tendon tissues of Sprague Dawley rats in vitro and TDSCs cultured on nanotopographic cues and substrate stiffness to determine how to control the TDSCs. The tendinopathy model was created using a chemical induction method, and the tendon injury model was created via an injury-and-overuse method. Norland Optical Adhesive 86 (NOA86) substrate with 2.48 GPa stiffness with and without 800 nm-wide nanogrooves and a polyurethane substrate with 800 nm-wide nanogrooves were used.
Results: TDSCs from 5-week-old normal tendon showed high expression of type III collagen on the flat NOA86 substrate. In the 15-week normal tendon model, expression of type III collagen was high in TDSCs cultured on the 800 nm NOA86 substrates. However, in the 15-week tendon injury model, expression of type III collagen was similar irrespective of nanotopographic cues or substrate stiffness. The expression of type I collagen was also independent of nanotopographic cues and substrate stiffness in the 15-week normal and tendon injury models. Gene expression of scleraxis was increased in TDSCs cultured on the flat NOA86 substrate in the 5-week normal tendon model (P=0.001). In the 15-week normal tendon model, scleraxis was highly expressed in TDSCs cultured on the 800 nm and flat NOA86 substrate (P=0.043). However, this gene expression was not significantly different between the substrates in the 5-week tendinopathy and 15-week tendon injury models.
Conclusion: Development and maturation of tendon are enhanced when TDSCs from normal tendons were cultured on stiff surface, but not when the TDSCs came from pathologic models. Therapeutic applications of TDSCs need to be flexible based on tendon age and tendinopathy.
Keywords: gene expression, differentiation, animal models, tendon-derived stem cells
Introduction
Tendinopathy and tendon injuries are common ailments whose prevalence have increased over time.1 However, despite numerous investigations in pursuit of regenerative agents that enhance tendon healing, such solutions remain scarce.2–4 Platelet-rich plasma and mesenchymal stem cells, especially adipose-derived stem cells, have been used to enhance tendon healing and regeneration, but their effects are controversial.5–9
Tendon-derived stem cells (TDSCs) are key factors associated with regeneration and tendon healing in tendinopathy and tendon injury. TDSCs are also associated with the pathogenesis of calcific tendinopathy and fatty infiltration by aberrant differentiation.10,11 However, how TDSCs are controlled and induced to engage in the healing and regeneration of tendons in tendinopathy remains unclear. Investigators have attempted to use mechanical stimulation, topographic signals, biochemical factors, electrospun fibers, and combinations of these strategies to regulate stem cell differentiation into teno-lineage.11,12
Mechanical stimulation or cyclic stretching is an important factor in differentiating mesenchymal stem cells into a specific lineage, such as chondrocytes or tenocytes.13,14 This mechanical stimulation can be simulated by matrix stiffness or surface topography and influences the fate of stem cells.15
Nanotopographic cues are well-known technology for controlling and manipulating cells into becoming possible therapeutic candidates for treatment of cardiac, renal, and vascular diseases.16–18 Nanotopographic cues affect stem cell adhesion, migration, proliferation, differentiation, and apoptosis by adjusting their structures, surface stiffness, and porosity.19 Tendons are aligned structures, and decreased tendon function caused by tendinopathy and tendon injury is associated with tendon misalignment.20 Therefore, alignment of TDSCs is important in tendon regeneration and healing; well-aligned fibers commit stem cells to teno-lineage differentiation.21,22
The aim of this study was to identify the effects of mechanical stiffness and topographic signals on the differentiation of TDSCs depending on age and pathological conditions. In this in vitro study, we compared TDSCs extracted from normal tendon tissues with TDSCs from tendinopathic tendon tissues and cultured them on substrates with and without nanotopographic features and with different surface stiffnesses to determine how to control TDSCs.
Materials and methods
Animal care
Sprague Dawley (SD) rats were obtained from Orient Bio Inc (Seongnam, Republic of Korea). Rats were housed in Samsung Biomedical Research Institute and fed a standard chow diet for the duration of each experiment. Animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Samsung Medical Center (Approval No 20130328001 and the guideline name was Samsung Medical Center IACUC guideline).
Tendinopathy model
For the tendinopathy model, a previously demonstrated chemical induction method was utilized.23 Chemically induced tendinopathy was evoked by oral administration of 900 mg/kg ofloxacin (Tarivid®; Jeil Pharm, Seoul, Republic of Korea) to 5-week-old SD rats. To confirm the development of tendinopathy, the left Achilles tendon was extracted for histological examination using H&E staining. Development of tendinopathy was defined by disordered alignment of collagen fibers and increase in vascularity; the right Achilles tendon was extracted for primary culture of TDSCs. For comparison, Achilles tendons were also cultured from normal 5-week-old SD rats.
Tendon injury model
For the tendon injury model, we adopted a model of injury followed by overuse modified from a previously reported method.24 Achilles tendon injuries were surgically induced by cutting the Achilles tendons of 4-week-old SD rats, followed by primary suture repair. After 1 day of recovery, the animals were subjected to a running protocol to induce overuse after injury. The animals ran on treadmills at a rate of 17 m/min for 1 hour, five times per week, for 11 weeks. Then, at 15 weeks of age, the left Achilles tendon of these rats was extracted for histological examination to confirm the tendon injury. Development of tendinopathy was defined by disordered alignment of collagen fibers and increase in vascularity. Concurrently, the right Achilles tendon was extracted for primary culture of TDSCs. For comparison, the Achilles tendons were cultured from normal 15-week-old SD rats. All animals were sacrificed by CO2 inhalation.
Isolation of TDSCs
TDSCs were isolated from the left Achilles tendon as previously described.25,26 The Achilles tendons were carefully removed from the rats to ensure a minimum margin of 0.5 mm between tendon and muscle or enthesis. The tissue was sectioned into 1×1 mm pieces and digested in saline using collagenase (C0130; Sigma-Aldrich Co., St Louis, MO, USA) and dispase (D4693; Sigma-Aldrich Co.). The digest was incubated at 37°C for 2 hours, centrifuged at 2,000 rpm for 15 minutes, and resuspended in DMEM with 20% FBS and 1% penicillin/streptomycin. Cells were cultured until approximately 80% confluence. To isolate TDSCs, single-cell suspensions were cultured in 96-well plates with DMEM with 20% FBS and 1% penicillin/streptomycin.
TDSCs were identified based on colony formation. Identification of TDSCs from selected colonies was verified using positive immunofluorescence staining for OCT4, nucleostemin, SSEA4, and tenomodulin in addition to verification of multipotent differentiation. Multipotency of TDSCs was confirmed based on the differentiation of TDSCs cultured in adipogenic, chondrogenic, and osteogenic media. Isolated TDSCs cultured in osteogenic medium (A10072-01; Gibco; Thermo Fisher Scientific, Waltham, MA, USA) were evaluated for osteogenic differentiation using Alizarin Red S assay. Those cultured in adipogenic medium (A10070-01; Gibco) were evaluated for adipogenic differentiation using Oil Red O assay. TDSCs to be cultured in chondrogenic medium (A10071-01; Gibco) were first pelleted into a micromass culture by centrifugation and were evaluated using a NovaUltra Safranin O Stain Kit (IW-3011; IHC World, Ellicott City, MD, USA).
Fabrication of nanogrooved polymers for nanotopographic signals and TDSC culture
The substrates used in this study were chosen because they have mechanical properties similar to those of native tendons. The Young’s modulus of native tendon ranges from 200 kPa to 8 GPa from embryonic development to adulthood.27–30 To simulate these conditions, a Norland Optical Adhesive 86 (NOA86) substrate, with a Young’s modulus of 2.48 GPa (Minuta Tech, Osan, Republic of Korea), was chosen to provide an environment with mechanical properties comparable to a developing tendon. The NOA86 was coated with a 20 nm layer of 100% silicone and carved using forced capillary lithography31,32 to create 800 nm-wide nanogrooves that would provide nanotopographic cues to the TDSCs (Figure 1). For comparison, NOA86 with the same stiffness was prepared without nanogrooves to determine the impact of the nanotopographic cues and a polyurethane (PUA) substrate with 19.8 MPa stiffness was prepared with 800 nm-wide nanogrooves to determine the effect of substrate stiffness.
TDSCs were seeded at a density of 20,000 cells per substrate for each substrate type, covered with a 20 nm-diameter coverslip, and cultured in DMEM with 20% FBS and 1% penicillin/streptomycin in an incubator at 37°C and with 5% CO2 for 1 week.
Immunofluorescence staining
The primary antibodies used in this study were rabbit polyclonal anti-collagen I (1:500, ab292; Abcam, Cambridge, UK) and mouse monoclonal anti-collagen III (1:100, NBP1-05119; Novus Biologicals, Littleton, CO, USA). Secondary antibodies used in this study were donkey polyclonal rabbit IgG antibody (Alexa Fluor® 488) (1:1,000, ab150073; Abcam) and donkey polyclonal mouse IgG antibody (Alexa Fluor® 594) (1:1,000, ab150108; Abcam).
One week after culture, cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature, quenched with 0.25% Triton X-100 in PBS for 10 minutes at room temperature, blocked with 1% BSA in PBS for 30 minutes at room temperature, and incubated with primary antibody overnight at 4°C. The cells were washed with PBS and incubated with a secondary antibody for 2 hours at room temperature in the dark. The cells were rinsed three times in PBS and mounted with fluoroshield mounting medium with DAPI (ab104139; Abcam). Confocal microscopy was performed using a Zeiss Lsm 700 (Carl Zeiss Meditec, Jena, Germany).
Image analysis and alignment
The degree of cell alignment on nanogrooved substrates was determined as previously described.32 Images of cells were obtained after 1 week in culture and processed with ImageJ (The National Institutes of Health). Cell perimeters were outlined, and the angle of the major axis was calculated.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated by lysis in TRIzol (15596-018; Thermo Fisher Scientific). cDNA libraries were made from 4 μg of RNA using superscript II reverse transcriptase (18064-014; Invitrogen) in 40 μL of reaction buffer at 42°C for 2 hours. Real-time PCR was performed with 30 ng of cDNA using a QuantiTect SYBR Green PCR Kit (204143; Qiagen NV, Venlo, the Netherlands) in a total volume of 25 μL with specific primers on a Light Cycler apparatus (ABI 7000; Thermo Fisher Scientific). The PCR procedure used was denaturation at 95°C for 15 minutes, 40 cycles at 95°C for 15 seconds, optimal annealing temperature for 30 seconds, and 72°C for 30 seconds. The following primer sequences were used (Genotech Corp., Daejeon, Republic of Korea): scleraxis (forward 5′-AACACGGCCTTCACTGCGCTG-3′ and reverse 5′-CAGTAGCACGTTGCCCAGGTG-3′) and GAPDH (forward 5′-GAGTCCACTGGCGTCTCCAC-3′ and reverse 5′-GGTGCTAAGCAGTTGGTGGT-3′).33,34
The relative expression level of each target gene was calculated using the 2−ΔΔCt method.35 The gene was standardized to GAPDH. Significant changes in gene expression were calculated with respect to stem cell injury, and response to nanotopographic cues and substrate stiffness was evaluated using a Kruskal–Wallis test with post hoc Mann–Whitney U test.
Results
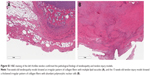
H&E staining of the left Achilles tendon confirmed the pathological findings of tendinopathy and tendon injury models. Upon histological examination, the 5-week-old tendinopathy model showed an irregular pattern of collagen fibers with multiple lipid vacuoles, and the 15-week-old tendon injury model showed a thickened irregular pattern of collagen fibers with abundant polymorphic nuclear cells (Figure S1).
Isolation of TDSCs was validated by identifying cells that positively stained for nucleostemin, OCT4, SSEA4, and tenomodulin (Figure S2). Each TDSC cell line was successfully differentiated into osteogenic, adipose, and chondrogenic cell lines, demonstrating their multipotent capacity. These results have been described previously.36
Effects of nanotopographic cues and substrate stiffness on the TDSCs
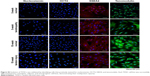
Irrespective of age or pathological status, TDSCs cultured on the 800 nm NOA86 and 800 nm PUA were well aligned along the grooves while cells cultured on the flat NOA86 were not aligned (Figure 2).
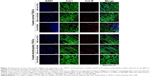
Expression of type I and type III collagen was observed in the TDSCs extracted from 5-week normal and 5-week tendinopathy models cultured on each of the substrates to understand the impact of nanotopographic cues and substrate stiffness on the cells (Figure 3). In the 5-week normal and 5-week tendinopathic conditions there was no difference in expression of type I collagen among the different substrates. However, expression of type III collagen in the 5-week normal condition was slightly higher on the flat NOA86 than on either of the grooved substrates, while its expression in the 5-week tendinopathic condition was slightly higher on 800 nm NOA86 than on 800 nm PUA or on flat NOA86. Furthermore, higher levels of type III collagen were also found in the 5-week tendinopathic condition than in the normal condition (either with 800 nm NOA86 or with 800 nm PUA).
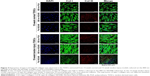
TDSCs in the 15-week tendon injury model showed higher cell densities and higher type I collagen expression than TDSCs in the 15-week normal tendon model irrespective of nanotopographic features or surface stiffness.
In the 15-week normal tendon model, expression of type III collagen was high in TDSCs cultured on the 800 nm NOA86 substrates (Figure 4). However, expression of type III collagen in the 15-week tendon injury model was independent of nanotopographic cues and substrate stiffness, as was the expression of type I collagen in the 15-week normal and tendon injury models.
Gene expression in the TDSCs based on nanotopographic cues and substrate stiffness
Results from qRT-PCR showed increased gene expression of scleraxis in TDSCs cultured on the flat NOA86 substrate in the 5-week normal tendon model (P=0.001). In the 15-week normal tendon model, scleraxis was highly expressed in TDSCs cultured on the 800 nm and flat NOA86 substrate (P=0.043). However, this gene expression was not significantly different between the substrates in the 5-week tendinopathy and 15-week tendon injury models (Figure 5).
Discussion
In the present study, TDSCs extracted from normal tendons showed similar expression of type I and III collagens irrespective of nanotopographic cues or substrate stiffness. However, scleraxis, a major protein related to development and maturation of tendon, increased under the stiff surface condition.
Expression of type I collagen in TDSCs extracted from the 5-week and 15-week normal and tendinopathic models was observed to be independent of nanotopographic cues and substrate stiffness. However, expression of type III collagen in the 5-week normal condition was slightly higher on flat NOA86 than on either of the grooved substrates, while its expression in the 5-week tendinopathic condition was slightly higher on 800 nm NOA86 than on 800 nm PUA or on flat NOA86. This finding is contradictory to Islam et al’s study that demonstrated increased collagen expression due to increased substrate stiffness by induction of tenogenic differentiation of mesenchymal stem cells.37,38 This contradictory finding might be due to the groove width and depth. The researchers in the previous study used 10 μm groove width and 3 μm depth, whereas we used 800 nm groove width and depth, which was shown to have maximal effects in another prior study.39 We selected 800 nm groove width and depth because this produced optimal regeneration in cardiac stem cells in our previous study.40
The difference of expression between type I and type III collagens according to pathological condition can be explained by type I collagen being the most abundant component of the tendon matrix and exhibiting relatively stable expression, while type III collagen is easily affected by diverse environments.41 Higher levels of type III collagen found in the 5-week tendinopathic condition compared to the normal condition can be explained by increased regenerative capacity in tendinopathic condition.
TDSCs in the 15-week tendon injury model showed higher cell densities than TDSCs in the 15-week normal tendon model irrespective of nanotopographic signals or surface stiffness. We thought that this was due to the increased proliferative properties of stem cells after injury to aid in healing.42 Relatively high expression of type I collagen in the 15-week tendon injury model compared to that in the 15-week normal tendon model might be due to these increased cell densities.
Scleraxis is an important transcription factor that influences the development and maturation of tendon-to-bone attachment.43 Scleraxis was prominently expressed in TDSCs which were extracted from the 5-week normal tendon model and cultured on the flat NOA86 and in TDSCs which were extracted from the 15-week normal tendon model and cultured on the 800 nm and flat NOA86 substrates. These results could be explained by induction of tenogenic differentiation by substrate stiffness, but not by nanotopographic cues.37,38 This finding contradict the results of a previous study, but this might be due to the different depth and width of the grooves used in that study.38 However, in the chemically induced tendinopathy model, scleraxis was not highly expressed regardless of the substrate used. We hypothesize that quinolone caused toxicity in the TDSCs via oxidative stress,44 and this oxidative stress was attenuated by the nanotopographic cues and substrate stiffness. The failure to induce tenogenic differentiation via nanotopographic cues or substrate stiffness in injury-induced tendinopathy might also be due to attenuation of oxidative or mechanical stress.
The results of our study show that therapeutic applications need to be adaptive because the effects of substrate stiffness and nanotopographic cues on the TDSCs were different based on the mechanism of tendinopathy. However, independent of the specific strategy, how to provide the stiffness and nanotopographic cues to the TDSCs in patients with tendinopathy remains a problem. Injection or implantation of TDSCs or mesenchymal stem cells that have been cultured on substrates with adequate nanotopographic cues and stiffness to pathological areas is one possible therapeutic option. However, poor retention and low survival of injected or implanted cells in the pathological areas are major obstacles to this approach.45 Based on our results, further in vivo studies should be conducted to determine how stiffness and nanotopographic cues can be delivered to the TDSCs in the tendinopathy animal model or in patients with tendinopathy.
The present study had several limitations. We used 800 nm width and depth for the nanotopographic cues because these were shown to have optimal effects in the cells in a previous study.39 However, stem cells show different reactions based on the width and depth of grooves,46 and the reactions can differ depending on the source of stem cells. To determine the maximal effects of TDSCs, diverse conditions should be tested. We did not perform this study using TDSCs extracted from human tissues because we could not acquire TDSCs from normal human tendons of different ages due to ethical issues. To extrapolate these results to clinical studies, it will be necessary to investigate characteristics of TDSCs extracted from diverse human pathological tendon tissues.
Conclusion
Our study revealed that scleraxis, a major protein related to development and maturation of tendon, increased when TDSCs from normal tendons were cultured on stiff surface (NOA86), but not when the TDSCs came from pathological models. This suggests that therapeutic applications need to be flexible because the effects of substrate stiffness and nanotopographic cues on the TDSCs were different based on tendon age and tendinopathy.
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI16C1104) and the National Research Fund (2018R1D1A1B07047084).
Author contributions
Sun Jeong Kim, Philip D Tatman, Albert O Gee, Deok-Ho Kim, and Sang Jun Kim contributed to conception and design. Sun Jeong Kim, Da-Hyun Song, Deok-Ho Kim, and Sang Jun Kim contributed to acquisition of data. All authors contributed to interpretation of data, drafting of the article, gave final approval of the manuscript version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Maffulli N, Wong J, Almekinders LC. Types and epidemiology of tendinopathy. Clin Sports Med. 2003;22(4):675–692. | ||
Rabago D, Nourani B. Prolotherapy for Osteoarthritis and Tendinopathy: a Descriptive Review. Curr Rheumatol Rep. 2017;19(6):34. | ||
Navani A, Li G, Chrystal J. Platelet Rich Plasma in Musculoskeletal Pathology: A Necessary Rescue or a Lost Cause? Pain Physician. 2017;20(3):E345–E356. | ||
Reeves KD, Sit RW, Rabago DP. Dextrose Prolotherapy: A narrative review of basic science, clinical research, and best treatment recommendations. Phys Med Rehabil Clin N Am. 2016;27(4):783–823. | ||
Filardo G, Di Matteo B, Kon E, Merli G, Marcacci M. Platelet-rich plasma in tendon-related disorders: results and indications. Knee Surg Sports Traumatol Arthrosc. 2018;26(7):1984–1999. | ||
Nourissat G, Ornetti P, Berenbaum F, Sellam J, Richette P, Chevalier X. Does platelet-rich plasma deserve a role in the treatment of tendinopathy? Joint Bone Spine. 2015;82(4):230–234. | ||
Veronesi F, Salamanna F, Tschon M, Maglio M, Nicoli Aldini N, Fini M. Mesenchymal stem cells for tendon healing: what is on the horizon? J Tissue Eng Regen Med. 2017;11(11):3202–3219. | ||
Akyol E, Hindocha S, Khan WS. Use of stem cells and growth factors in rotator cuff tendon repair. Curr Stem Cell Res Ther. 2015;10(1):5–10. | ||
Usuelli FG, D’Ambrosi R, Maccario C, Indino C, Manzi L, Maffulli N. Adipose-derived stem cells in orthopaedic pathologies. Br Med Bull. 2017;124(1):31–54. | ||
Rui YF, Lui PP, Chan LS, Chan KM, Fu SC, Li G. Does erroneous differentiation of tendon-derived stem cells contribute to the pathogenesis of calcifying tendinopathy? Chin Med J (Engl). 2011;124(4):606–610. | ||
Chen JL, Zhang W, Liu ZY, Heng BC, Ouyang HW, Dai XS. Physical regulation of stem cells differentiation into teno-lineage: current strategies and future direction. Cell Tissue Res. 2015;360(2):195–207. | ||
Hakimi O, Murphy R, Stachewicz U, Hislop S, Carr AJ. An electrospun polydioxanone patch for the localisation of biological therapies during tendon repair. Eur Cell Mater. 2012;24:344–357. | ||
Fahy N, Alini M, Stoddart MJ. Mechanical stimulation of mesenchymal stem cells: Implications for cartilage tissue engineering. J Orthop Res. 2018;36(1):52–63. | ||
Burk J, Plenge A, Brehm W, Heller S, Pfeiffer B, Kasper C. Induction of tenogenic differentiation mediated by extracellular tendon matrix and short-term cyclic stretching. Stem Cells Int. 2016;2016:7342379. | ||
Li D, Zhou J, Chowdhury F, Cheng J, Wang N, Wang F. Role of mechanical factors in fate decisions of stem cells. Regen Med. 2011;6(2):229–240. | ||
MacGregor-Ramiasa M, Hopp I, Bachhuka A, Murray P, Vasilev K. Surface nanotopography guides kidney-derived stem cell differentiation into podocytes. Acta Biomater. 2017;56:171–180. | ||
Carson D, Hnilova M, Yang X, et al. Nanotopography-Induced Structural Anisotropy and Sarcomere Development in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. ACS Appl Mater Interfaces. 2016;8(34):21923–21932. | ||
Xing Q, Qian Z, Tahtinen M, Yap AH, Yates K, Zhao F. Aligned Nanofibrous Cell-Derived Extracellular Matrix for Anisotropic Vascular Graft Construction. Adv Healthc Mater. Epub 2017 Feb 9. | ||
Zhang Y, Gordon A, Qian W, Chen W. Engineering nanoscale stem cell niche: direct stem cell behavior at cell-matrix interface. Adv Healthc Mater. 2015;4(13):1900–1914. | ||
Wang JH, Guo Q, Li B. Tendon biomechanics and mechanobiology – a minireview of basic concepts and recent advancements. J Hand Ther. 2012;25(2):133–141. | ||
Zhang C, Yuan H, Liu H, et al. Well-aligned chitosan-based ultrafine fibers committed teno-lineage differentiation of human induced pluripotent stem cells for Achilles tendon regeneration. Biomaterials. 2015;53:716–730. | ||
Teh TK, Toh SL, Goh JC. Aligned fibrous scaffolds for enhanced mechanoresponse and tenogenesis of mesenchymal stem cells. Tissue Eng Part A. 2013;19(11–12):1360–1372. | ||
Kato M, Takada S, Kashida Y, Nomura M. Histological examination on Achilles tendon lesions induced by quinolone antibacterial agents in juvenile rats. Toxicol Pathol. 1995;23(3):385–392. | ||
Glazebrook MA, Wright JR Jr, Langman M, Stanish WD, Lee JM. Histological analysis of achilles tendons in an overuse rat model. J Orthop Res. 2008;26(6):840–846. | ||
Bi Y, Ehirchiou D, Kilts TM, et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13(10):1219–1227. | ||
Rui YF, Lui PP, Li G, Fu SC, Lee YW, Chan KM. Isolation and characterization of multipotent rat tendon-derived stem cells. Tissue Eng Part A. 2010;16(5):1549–1558. | ||
An KN, Sun YL, Luo ZP. Flexibility of type I collagen and mechanical property of connective tissue. Biorheology. 2004;41(3–4):239–246. | ||
Kato YP, Christiansen DL, Hahn RA, Shieh SJ, Goldstein JD, Silver FH. Mechanical properties of collagen fibres: a comparison of reconstituted and rat tail tendon fibres. Biomaterials. 1989;10(1):38–42. | ||
Marturano JE, Arena JD, Schiller ZA, Georgakoudi I, Kuo CK. Characterization of mechanical and biochemical properties of developing embryonic tendon. Proc Natl Acad Sci U S A. 2013;110(16):6370–6375. | ||
Silver FH, Christiansen D, Snowhill PB, Chen Y, Landis WJ. The role of mineral in the storage of elastic energy in turkey tendons. Biomacromolecules. 2000;1(2):180–185. | ||
Jiao A, Trosper NE, Yang HS, et al. Thermoresponsive nanofabricated substratum for the engineering of three-dimensional tissues with layer-by-layer architectural control. ACS Nano. 2014;8(5):4430–4439. | ||
Kim DH, Lipke EA, Kim P, et al. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A. 2010;107(2):565–570. | ||
Tan Q, Lui PP, Rui YF, Wong YM. Comparison of potentials of stem cells isolated from tendon and bone marrow for musculoskeletal tissue engineering. Tissue Eng Part A. 2012;18(7–8):840–851. | ||
Wagenhäuser MU, Pietschmann MF, Sievers B, et al. Collagen type I and decorin expression in tenocytes depend on the cell isolation method. BMC Musculoskelet Disord. 2012;13:140. | ||
Chang S, Chen W, Yang J. Another formula for calculating the gene change rate in real-time RT-PCR. Mol Biol Rep. 2009;36(8):2165–2168. | ||
Kim SJ, Song DH, Kim SJ. Characteristics of tendon derived stem cells according to different factors to induce the tendinopathy. J Cell Physiol. 2018;233(8):6196–6206. | ||
Islam A, Mbimba T, Younesi M, Akkus O. Effects of substrate stiffness on the tenoinduction of human mesenchymal stem cells. Acta Biomater. 2017;58:244–253. | ||
Shi Y, Zhou K, Zhang W, et al. Microgrooved topographical surface directs tenogenic lineage specific differentiation of mouse tendon derived stem cells. Biomed Mater. 2017;12(1):015013. | ||
Liliensiek SJ, Wood JA, Yong J, Auerbach R, Nealey PF, Murphy CJ. Modulation of human vascular endothelial cell behaviors by nanotopographic cues. Biomaterials. 2010;31(20):5418–5426. | ||
Kim DH, Kshitiz, Smith RR, et al. Nanopatterned cardiac cell patches promote stem cell niche formation and myocardial regeneration. Integr Biol (Camb). 2012;4(9):1019–1033. | ||
Güngörmüş C, Kolankaya D. Gene expression of tendon collagens and tenocyte markers in long-term monolayer and high-density cultures of rat tenocytes. Connect Tissue Res. 2012;53(6):485–491. | ||
Rennert RC, Sorkin M, Garg RK, Gurtner GC. Stem cell recruitment after injury: lessons for regenerative medicine. Regen Med. 2012;7(6):833–850. | ||
Killian ML, Thomopoulos S. Scleraxis is required for the development of a functional tendon enthesis. Faseb J. 2016;30(1):301–311. | ||
Pouzaud F, Bernard-Beaubois K, Thevenin M, Warnet JM, Hayem G, Rat P. In vitro discrimination of fluoroquinolones toxicity on tendon cells: involvement of oxidative stress. J Pharmacol Exp Ther. 2004;308(1):394–402. | ||
Rossi CA, Pozzobon M, De Coppi P. Advances in musculoskeletal tissue engineering: moving towards therapy. Organogenesis. 2010;6(3):167–172. | ||
Mengsteab PY, Uto K, Smith AS, et al. Spatiotemporal control of cardiac anisotropy using dynamic nanotopographic cues. Biomaterials. 2016;86:1–10. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.