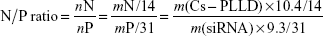Back to Journals » International Journal of Nanomedicine » Volume 13
Nanoscale polysaccharide derivative as an AEG-1 siRNA carrier for effective osteosarcoma therapy
Authors Wang F, Pang J, Huang L, Wang R, Li D, Sun K, Wang L, Zhang L
Received 30 July 2017
Accepted for publication 7 December 2017
Published 8 February 2018 Volume 2018:13 Pages 857—875
DOI https://doi.org/10.2147/IJN.S147747
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Fen Wang,1,* Jia-Dong Pang,2,* Lei-lei Huang,1 Ran Wang,1 Dan Li,3 Kang Sun,4 Lian-tang Wang,1,* Li-Ming Zhang2,*
1Department of Pathology, The First Affiliated Hospital of Sun Yat-sen University, 2PCFM Lab and GDHPPC Lab, School of Materials Science and Engineering, Sun Yat-sen University, Guangzhou, 3Guangdong Provincial Engineering Research Center of Molecular Imaging, The Fifth Affiliated Hospital of Sun Yat-sen University, Zhuhai, 4School of Engineering, Sun Yat-sen University, Guangzhou, China
*These authors contributed equally to this work
Background: Nanomedicine, which is the application of nanotechnology in medicine to make medical diagnosis and treatment more accurate, has great potential for precision medicine. Despite some improvements in nanomedicine, the lack of efficient and low-toxic vectors remains a major obstacle.
Objective: The aim of this study was to prepare an efficient and low-toxic vector which could deliver astrocyte elevated gene-1 (AEG-1) small interfering RNA (siRNA; siAEG-1) into osteosarcoma cells effectively and silence the targeted gene both in vitro and in vivo.
Materials and methods: We prepared a novel polysaccharide derivative by click conjugation of azidized chitosan with propargyl focal point poly (L-lysine) dendrons (PLLD) and subsequent coupling with folic acid (FA; Cs-g-PLLD-FA). We confirmed the complexation of siAEG-1and Cs-g-PLLD or Cs-g-PLLD-FA by gel retardation assay. We examined the cell cytotoxicity, cell uptake, cell proliferation and invasion abilities of Cs-g-PLLD-FA/siAEG-1 in osteosarcoma cells. In osteosarcoma 143B cells tumor-bearing mice models, we established the therapeutic efficacy and safety of Cs-g-PLLD-FA/siAEG-1.
Results: Cs-g-PLLD-FA could completely encapsulate siAEG-1 and showed low cytotoxicity in osteosarcoma cells and tumour-bearing mice. The Cs-g-PLLD-FA/siAEG-1 nanocomplexes were capable of transferring siAEG-1 into osteosarcoma cells efficiently, and the knockdown of AEG-1 resulted in the inhibition of tumour cell proliferation and invasion. In addition, caudal vein injecting of Cs-g-PLLD-FA/siAEG-1 complexes inhibited tumor growth and lung metastasis in tumor-bearing mice by silencing AEG-1 and regulating MMP-2/9.
Conclusion: In summary, Cs-g-PLLD-FA nanoparticles are a promising system for the effective delivery of AEG-1 siRNA for treating osteosarcoma.
Keywords: chitosan, gene delivery, astrocyte elevated gene-1, small interfering RNA, osteosarcoma
Introduction
Human cancer is a complex and intrinsically heterogeneous disease. Cancer patients may exhibit similar symptoms and appear to have the same pathological disease but may have entirely different genetic abnormalities. Precision medicine aims to explore cancer heterogeneity by incorporating diagnostic technology to characterize each cancer patient’s molecular subtype for tailored treatments.1,2 Nanomedicine, which is the application of nanotechnology in medicine to make medical diagnosis and treatment more accurate, has great potential for precision medicine.3 Among the applications of nanotechnology in medicine, utilizing small interfering RNA (siRNA) therapeutic nanoparticles is an innovative strategy to generate novel treatments for precision medicine.4,5 Thus, nanomedicine may give rise to a new era of precision medicine.
The siRNA structure with 19–21 base pairs has been regarded as a robust tool in posttranscriptional gene silencing.6,7 Targeted gene knockdown via siRNA delivery has exciting potential for the treatment of diseases, including cancer caused by aberrant gene expression.8,9 Various siRNA-based therapeutics have been developed and show great promise in cancer treatments.10,11 However, safe and efficient delivery of siRNA to the affected tissues remains a challenging obstacle to the implementation of siRNA therapies.12
With the development of nanomedicine, various synthetic nanocarriers have been constructed to deliver siRNA into cancer cells, and they show great promise in cancer treatments. These nanocarriers include lipid-based nanoparticles (LNPs),13 cationic LNPs,14 polyplexes,15 and so on. Cationic LNPs readily bind and condense nucleic acids, and have been commonly used as transfection vehicles for siRNA. Of the many cationic polymers, polyethylenimine (PEI) has been widely used for nucleic acid delivery. The “proton-sponge” effect of PEI allows siRNA to escape from endosomes and to deliver nucleic acids into the cell. However, the large amount of positive charge results in a rather high cellular toxicity and is the major limiting factor for their application in vivo.16 Lipid-based nanostructures may either contain lipids on the shell of the nanoparticle or as the core material. Neutral liposomes are characterized with low immunogenicity and toxicity. However, neutral liposomes have reduced to interact with anionic polynucleotides, resulting in poorer entrapment efficiency.17 Polyplexes are polycation-based nanoparticles (100–400 nm) formulated siRNAs. The major advantage of using polycations is their structural flexibility. Recently, poly lactic-co-glycolic acid (PLGA) has been used as a nanocarrier for plasmid DNA and siRNA delivery. The advantages conferred by PLGA-based siRNA delivery include high plasma stability and low toxicity. However, poor electrostatic attraction between PLGA and siRNA limits the use of the former as a carrier of the latter.18,19
As a polyplex nanocarrier, chitosan is a natural product of the deacetylation reaction of chitin and is widely used in the biomedical field, because it shows low cytotoxicity and is biodegradable.20,21 It can form complexes with nucleic acids via electrostatic interactions, and it effectively protects nucleic acids from nuclease degradation to increase the cellular accumulation of siRNA molecules and facilitate their release from endosomes to the cytosol in cells. Therefore, chitosan has attracted increasing attention as a nonviral nucleic acid delivery vehicle.22–25 Although, chitosan has been extensively modified and investigated for nucleic acid delivery, the efficiency of gene delivery by its derivatives is still relatively low.26–29 There is very little information available on the modification of chitosan for use as a targeted gene delivery vector. Folic acid (FA; vitamin B9) is selected for cancer-targeted therapies, because folate receptors are rapidly internalized when engaged by FA, and most malignant tumors have high folate receptor expression for rapid cell growth.30 Coating nanocarriers with FA has enhanced transfection efficiency in targeted cancer cells.
Osteosarcoma is the most common primary malignant bone tumor in children and adolescents, and is characterized by aggressive growth as well as poor long-term survival due to early frequent systemic metastasis, particularly lung metastasis.31,32 The standard treatment for osteosarcoma is a combination of surgery and chemotherapy. The 5-year survival rate for patients with local osteosarcoma remains ~65%–70%; for patients with metastatic disease, the 5-year survival rate is only 20%.33 Astrocyte elevated gene-1 (AEG-1), primarily identified as a late response gene induced by HIV-1 infection, plays multiple roles, including malignant transformation, chemoresistance acquisition, angiogenesis, and metastasis. As a multifunctional oncoprotein, AEG-1 has been shown to be overexpressed in multiple types of human tumor malignancies, including lung cancer, breast cancer, hepatocellular carcinoma, glioma, and prostate cancer.34–36 Our previous study suggested that AEG-1 was overexpressed in human osteosarcoma tissues and that downregulation of AEG-1 expression inhibited tumor cell proliferation and invasion in vitro.37 Following its initial identification, AEG-1 was thought to be a potential focus for targeted therapy based on its multifaceted roles in regulating malignant tumor progression.
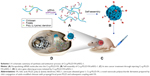
To achieve efficient systemic siRNA delivery to osteosarcoma, we developed a novel nanoscale polysaccharide derivative, which was prepared by click conjugation of azido-modified chitosan with propargyl focal point poly (L-lysine) dendrons (PLLD) and subsequent coupling with FA (Cs-g-PLLD-FA). The nanocarrier increased the solubility and stability of the nucleic acid binding capability of chitosan nanoparticles, and exerted a positive influence on the ability of FA to deliver siRNA to osteosarcoma cells. We characterized the properties and toxicity of the nanocarrier, examined its ability to deliver AEG-1 siRNA into osteosarcoma cells, and investigated the tumor suppression effect in vitro and in vivo. The schematic summary of synthesis and antitumor process of Cs-g-PLLD-FA/siAEG-1 is illustrated in Scheme 1.
Materials and methods
Materials
Chitosan (molecular weight =10 kDa, deacetylation degree =85.3%) was purchased from Haidebei Marine Bioengineering Co Ltd (Jinan, Shandong, China) and was used as received. Then, 1-azido-2,3-epoxypropane was synthesized by two-step reactions in our laboratory according to the method reported previously by Pahimanolis et al.38 Propargyl focal point PLLD-G3 (generation =3) was synthesized in our laboratory by divergent and convergent approaches, as reported in our recent publication.39 Copper sulphate (CuSO4·5H2O) and sodium ascorbate (99%) were purchased from Alfa Aesar (Haverhill, MA, USA). The human osteosarcoma cell line 143B and U2OS were purchased from the China Center for Type Culture Collection (Wuhan, Hubei, China). RPMI 1640 medium was supplied by Thermo Fisher Scientific (Waltham, MA, USA). Fetal bovine serum was supplied by Gibco (Grand Island, NY, USA). Targeting AEG-1 siRNA and fluorescein-tagged siRNAs (siAEG-1, sense 5′-GGU CUC AGA UGA UGA UAA ATT-3′ and antisense 5′-UUU AUC AUC AUC UGA GAC CTT-3′) were supplied by Ribobio (Guangzhou, Guangdong, China). The MTT kit and dimethyl sulfoxide (DMSO) were purchased from KeyGen (Nanjing, Jiangsu, China) and Sigma-Aldrich (St Louis, MO, USA), respectively. Lipofectamine 2000 and an anti-AEG-1 rabbit antibody were purchased from Thermo Fisher Scientific. Anti-matrix metalloprotease (MMP)-2 rabbit antibody, anti-MMP-9 rabbit antibody, and anti-Ki-67 rabbit antibody were purchased from Cell Signaling Technology (Danvers, MA, USA). Trizol reagent was purchased from Thermo Fisher Scientific and FastStart Universal SYBR Green Master was purchased from Hoffman-La Roche Ltd (Basel, Switzerland).
Azidation of chitosan by 1-azido-2,3-epoxypropane
The introduction of azide groups onto the backbone of chitosan was carried out as follows: 250 mg of chitosan and 180 μL (1.5 mmol) of a freshly prepared solution of 1-azido-2,3-epoxypropane were successively added to distilled water with magnetic stirring. The clear reaction mixture obtained was stirred for 24 hours at 30°C in the dark. After the reaction, the product was dialyzed in distilled water for 3 days (molecular weight cutoff [MWCO] =8,000) and was lyophilized to obtain azido-modified chitosan (Cs-N3, yield 86%). Fourier-transform infrared spectroscopy (FTIR; Perkin–Elmer Paragon 1000 spectrometer; PerkinElmer Inc, Waltham, MA, USA) and 1H nuclear magnetic resonance (NMR; Bruker DPX-300 NMR spectrometer; Bruker Corporation, Billerica, MA, USA) analyses were used to confirm the azidation of amylose by 1-azido-2,3-epoxypropane.
Click reaction between azido-modified chitosan and propargyl focal point PLLD
The click reaction between Cs-N3 and PLLD-G3 was carried out as follows: 0.515 g of PLLD-G3 (0.29 mmol) and 0.05 g of Cs-N3 were added successively with magnetic stirring to 10 mL of DMSO. After a clear solution was obtained, 18 mg of CuSO4·5H2O and 28 mg of sodium ascorbate were added successively under a nitrogen atmosphere. The resultant reaction mixture was heated to 40°C for 48 hours. After the reaction, the product was dialyzed in distilled water for 3 days (MWCO =8,000) and lyophilized to obtain the dendronized chitosan derivative (Cs-g-PLLD, yield 73%). FTIR (Perkin–Elmer Paragon 1000 spectrometer) and 1H NMR (Bruker DPX-300 NMR spectrometer) analyses were used to confirm the click reaction between Cs-N3 and PLLD-G3. Based on elemental analysis, the degree of substitution (the number of polyamidoamine dendrons per 100 anhydroglucose units of chitosan) of PLLD-G3 on chitosan was determined to be 10.35.
Conjugation between Cs-g-PLLD and FA
Conjugation between Cs-g-PLLD and FA was carried out as follows: 10 mL of DMSO, 0.359 g of FA (0.81 mmol), 0.168 g of dicyclohexylcarbodiimide, and 0.102 g of N-hydroxysuccinimde were added successively with magnetic stirring for 5 hours at 25°C. After filtration, a clear solution was obtained, and then, 0.5 mL of solution and 30 mg of Cs-g-PLLD were added successively with magnetic stirring for 48 hours. After the reaction, the product was dialyzed in distilled water for 3 days (MWCO =8,000) and was lyophilized to obtain the dendronized chitosan derivative (Cs-g-PLLD-FA, yield 61%). FTIR (Perkin–Elmer Paragon 1000 spectrometer) and 1H NMR (Bruker DPX-300 NMR spectrometer) analyses were used to confirm this conjugation between Cs-g-PLLD and FA.
Complexation of Cs-g-PLLD and Cs-g-PLLD-FA with AEG-1 siRNA and the characterization of the complexes
Complexations of Cs-g-PLLD and Cs-g-PLLD-FA with AEG-1 siRNA in phosphate-buffered saline (PBS, pH 7.4) were carried out at various nitrogen/phosphorus (N/P) ratios. Elemental analysis showed that the mass content of nitrogen in Cs-PLLD is about 10.4%; moreover, the mass content of phosphorus in siRNA is about 9.3% according to its structure. So, we can calculate the N/P ratio by mass fraction as follows:
|
For each complexation, the resultant suspension was homogenized by gentle vortexing for 10 seconds and was then incubated at room temperature for 30 minutes before use. The particle sizes (diameter) and zeta potentials of the Cs-g-PLLD/siAEG-1 and Cs-g-PLLD-FA/siAEG-1 complexes were determined using a zeta potential analyzer instrument (ZetaPALS; Brookhaven Instruments Corporation, Austin, TX, USA). Prior to the measurements, the complexes were diluted to 1.0 mL with PBS. Morphological examination of the complexes was performed using a JEM-2010HR high-resolution transmission electron microscope after the complexes were counterstained with uranyl acetate.
Gel retardation assay
The complexation of Cs-g-PLLD or Cs-g-PLLD-FA with AEG-1 siRNA was investigated by 1% agarose gel electrophoresis. Cs-g-PLLD or Cs-g-PLLD-FA and AEG-1 siRNA were mixed at N/P ratios of 1:1, 2:1, 4:1, 8:1, 10:1, 20:1, and 30:1, forming different formulation compositions. The resulting complexes were kept at room temperature for 30 minutes to facilitate complexation. Next, the samples were loaded onto 1% agarose gels stained with ethidium bromide. The samples were electrophoresed at 100 V for 15 minutes in Tris-acetate buffer, and the bands were visualized on an ultraviolet transilluminator.
Cell culture and MTT assay
The human osteosarcoma cell lines 143B and U2OS were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 μg/μL streptomycin, and 100 μg/μL penicillin, in a 37°C incubator, containing 5% CO2. The cytotoxic effects of Cs-g-PLLD and Cs-g-PLLD-FA against osteosarcoma cells were measured by the MTT assay. In brief, cells were seeded on 96-well plates with 3×103 cells/well. The next day, cells were incubated for 48 hours with different concentrations of conjugates in serum containing culture medium. Next, MTT was added to each well. After 4 hours incubation at 37°C, the medium was discarded, and 150 μL of DMSO was added to each well. The absorbance was measured at 570 nm as the reference wavelength.
In vitro RNAi experiments
The 143B and U2OS cells were grown in 24-well plates to 30% confluency; thereafter, the growth medium was replaced with fresh growth medium containing either Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 (siAEG-1 was labeled with cy3, and the concentration of siRNA in each well was 50 nM, the quantity of Cs-g-PLLD or Cs-g-PLLD-FA in each well was 10 μg). Next, the 24-well culture plates were incubated at 37°C for 6 hours. After 24 hours incubation in fresh growth medium, cells were analyzed for siRNA uptake using an inverted fluorescence microscope (IX71; Olympus Corporation, Tokyo, Japan). Cells were also collected by trypsinization, centrifuged, resuspended in PBS, and analyzed by flow cytometry (Beckman Coulter, Brea, CA, USA).
For confocal laser scanning microscopy observations, 143B and U2OS cells (5×104 cells/well) were seeded in a 35-mm glass-bottomed culture dish (MatTek Corporation, Ashland, MA, USA) and were incubated for 24 hours. After in vitro transfection for 4 hours (siAEG-1 was labeled with cy3, and the concentration of siRNA in each well was 50 nM, the quantity of Cs-g-PLLD or Cs-g-PLLD-FA in each well was 10 μg), the cells were stained with DAPI, directly observed using the Olympus FluoView confocal microscope and analyzed using FV10-ASW viewer software (Olympus Corporation).
Immunoblotting analysis
Protein was extracted from the cell pellet using lysis buffer, and an equal amount of protein was loaded onto 10% SDS polyacrylamide gels for separation, followed by transfer onto polyvinylidene fluoride membranes. The membranes were probed with an anti-AEG-1 rabbit antibody and incubated with horseradish peroxidase-conjugated anti-rabbit IgG. AEG-1 expression was determined using enhanced chemiluminescence. The membranes were stripped and reprobed with an anti-GAPDH antibody as an internal loading control.
RNA extraction and quantitative reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA from cells was extracted using Trizol reagent and quantitative RT-PCR was conducted using FastStart Universal SYBR Green Master kit according to the manufacturer’s instruction as previously described.37
Invasion assay
Invasive ability was measured using 24-well BioCoat cell culture inserts (Corning Costar, Cambridge, MA, USA) with an 8 μm pore polyethylene terephthalate membrane coated with Matrigel basement membrane matrix (Trevigen, Gaithersburg, MD, USA). In brief, the wells of the lower chamber were filled with medium containing 10% fetal bovine serum. Next, 5×104 cells/well were seeded in the upper compartment in serum-free medium. The invasion assay was performed at 37°C in a 5% CO2 humidified incubator for 24 hours. At the end of the invasion assay, the filters were removed, fixed with methyl alcohol, and stained with hematoxylin. Cells on the upper surface of the filters were removed by wiping with a cotton swab, and invasion was determined by counting cells that migrated to the lower side of the filter using a microscope at ×400 magnification.
In vivo xenograft tumor assays
The 143B cells were subcutaneously inoculated into the flanks of 4-week-old male nu/nu nude mice (1×106 cells/mouse, n=5 per group). Laboratory animal husbandry and in vivo experiments were approved by the Medical Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University and performed according to the guidelines of the National Laboratory Animal Center. The diameters of the resulting tumors were measured every 3 days, and the tumor volume was calculated as follows: large diameter × (small diameter)2 ×0.52. Xenograft tumors were harvested at the end point of the experiment and were sent for routine tissue processing.
Mice were anesthetized and transported to the Medical Science Experimentation Center of Sun Yat-sen University. Computed tomography (CT) scans were taken using a CT 18fluro-D-glucose positron emission computed tomography (18F-FDG PET-CT) imaging device (Inveon, Istanbul, Turkey) before the mice were sacrificed, to evaluate lung metastasis formation.
Immunohistochemical (IHC) staining
IHC staining was performed using standard streptavidin-biotin peroxidase methods. Antigen retrieval was performed by heating the slides in 10 mmol/L sodium citrate buffer (pH 6.0) for 15 minutes at 95°C. The slides were then incubated in 10% normal blocking serum for 30 minutes and then with anti-AEG-1 antibody overnight at 4°C, followed by incubation with biotin-labeled anti-rabbit secondary antibodies and streptavidin-horseradish peroxidase complex. The tissue sections were counterstained with 10% Mayer’s hematoxylin, dehydrated, and mounted in Crystal Mount. The other antibodies used were an anti-MMP-2 rabbit antibody, an anti-MMP-9 rabbit antibody, and an anti-Ki-67 rabbit antibody. Digital photomicrographs were obtained at ×400 magnification. The scores of immunohistochemically stained sections were calculated by multiplying the percentage of positive cells by the staining intensity.
Statistical analysis
All experiments were repeated at least three times. Data were analyzed using the unpaired Student’s t-test, and a P-value <0.05 was considered significant.
Results and discussion
Synthesis of Cs-g-PLLD and Cs-g-PLLD-FA
Figure 1 illustrates the synthesis of azido-modified chitosan (Cs-N3), Cs-g-PLLD, and Cs-g-PLLD-FA. Figure S1A shows the FTIR spectra of chitosan, azido-modified chitosan (Cs-N3), Cs-g-PLLD, and Cs-g-PLLD-FA. Compared with the spectrum of chitosan, the spectrum of Cs-N3 showed a new strong peak at 2,113 cm−1, confirming the introduction of an azide group into the chitosan backbone. After click conjugation, the spectra of Cs-g-PLLD and Cs-g-PLLD-FA did not show the characteristic absorption bands of the azido group (2,113 cm−1),40 but exhibited the main characteristic bands of PLLD-G3 (2,925 cm−1, 2,848 cm−1 (νC-H), 1,623 cm−1 (νC=O), and 1,574 cm−1 (νCO-NH)) and Cs-N3. Further analyses were conducted on the 1H NMR spectra shown in Figure S1B and C. The 1H NMR spectrum of Cs-g-PLLD showed not only the characteristic peaks of Cs-N3 at 2.60–5.40 ppm and characteristic peaks of PLLD-G3 at 1.10–1.60 and 3.0–3.60 ppm, but also a new signal at 8.26 ppm, which could be attributed to the presence of the triazole proton.41 These results indicate the success of the click conjugation between the PLLD-G3 dendron and Cs-N3. Moreover, Cs-g-PLLD-FA showed more characteristic peaks of FA at 8.44 ppm (s, −CH− of pyrazine on FA), 7.51 ppm (d, −CH− of the phenyl ring on FA), and 6.79 ppm (d, −CH− of the phenyl ring on FA), indicating the success of conjugation between Cs-g-PLLD and FA.
Characterization of complexes
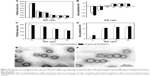
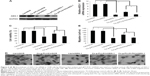
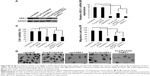
One prerequisite of Cs-g-PLLD-FA as a nonviral gene vector is the ability to condense AEG-1 siRNA into particulate structures.42 Therefore, we carried out particle size and zeta potential measurements as well as transmission electron microscopy (TEM) observations for aqueous Cs-g-PLLD-FA/siAEG-1 mixed systems. Figure 2 shows the particle size and zeta potentials of Cs-g-PLLD-FA/siAEG-1 complexes formed when the N/P ratio was ≥2 as well as TEM images of Cs-g-PLLD-FA/siAEG-1 complexes formed at an N/P ratio of 20. From Figure 2A, the mean particle size was observed to decrease from 800 to 400 nm with an increase in N/P ratios from 2 to 10 and then it remained in the size range of 270–300 nm with a further increase in the N/P ratio. This is because Cs-g-PLLD-FA and siRNA form complexes by through ionic interactions. At a low N/P ratio of 2, the complexes could not form completely and therefore had a large hydrodynamic size. At high N/P ratios, there were net electrostatic repulsive forces to prevent aggregation among the complexes. From Figure 2B, the zeta potential was found to increase from −20 to +2 with an increase in the N/P ratio from 2 to 10 and then it remained at about +9 with a further increase in the N/P ratio. In this case, the Cs-g-PLLD-FA/siAEG-1 complexes had a more negative charge than Cs-g-PLLD/siAEG-1 because of the FA on the surface of the complexes. Moreover, the binding ability of Cs-g-PLLD-FA to AEG-1 siRNA was maintained after complexation for 24 hours in the dark, as shown in Figure 2C and D. In addition, the morphologies of the Cs-g-PLLD/siAEG-1 and Cs-g-PLLD-FA/siAEG-1 complexes were investigated by TEM at an N/P ratio of 20 and were found to have a spherical shape and compacted structure, as shown in Figure 2E and F.
Gel retardation and cell viability analysis
Agarose gel electrophoresis was performed to confirm the complexation between AEG-1 siRNA and Cs-g-PLLD or Cs-g-PLLD-FA. The complexation of cationic polymers with AEG-1 siRNA is due to the electrostatic neutralization and size of the polymers, by which AEG-1 siRNA partially or completely loses its negative charge, resulting in the retardation of its mobility in an electric field. Therefore, the capacity of the polymer to complex with AEG-1 siRNA can be measured by the retardation of AEG-1 siRNA mobility in gel electrophoresis. As shown in Figure 3A and B, the brightness of ethidium bromide (EB)-stained AEG-1 siRNA bands decreased with the increasing N/P ratio of non-targeting and targeting complexes; both Cs-g-PLLD and Cs-g-PLLD-FA started to form complexes with AEG-1 siRNA at low N/P ratios, and complete retardation of AEG-1 siRNA migration occurred at an N/P ratio of 8.
As a good gene carrier, the low cytotoxicity of the vector itself was essential for practical applications. The MTT assay was performed to evaluate the cytotoxicities of the complexes in 143B and U2OS cells. Cells were treated with Cs-g-PLLD or Cs-g-PLLD-FA at different concentrations. As shown in Figures 3C and S2, even at a high concentration of Cs-g-PLLD or Cs-g-PLLD-FA (100 μg/mL), which was 10-fold higher than that required for efficient transfection, neither Cs-g-PLLD nor Cs-g-PLLD-FA showed any obvious negative effects on cell viability. In contrast, Lipofectamine 2000 itself showed some cytotoxicity at the dose recommended by the protocol supplied. Moreover, there was no significant difference between Cs-g-PLLD and Cs-g-PLLD-FA at the different concentrations tested (P<0.05), illustrating that the addition of FA did not increase the cytotoxicity of Cs-g-PLLD-FA. All of these experiments indicated that neither Cs-g-PLLD nor Cs-g-PLLD-FA was cytotoxic.
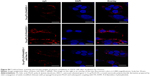
In vitro siRNA transfection
To initiate the gene silencing process, functional siRNA molecules must be transported into the targeted cells. Therefore, we sought to investigate whether Cs-g-PLLD or Cs-g-PLLD-FA could serve as carriers to deliver AEG-1 siRNA into osteosarcoma cells. Lipofectamine 2000/siAEG-1 was used as a reference. As shown in Figures 4A and S3A, cy3-siRNA was observed with an inverted fluorescence microscope, and the transfection efficiency of the Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 complex was higher at the optimal N/P ratio of 20 than at an N/P ratio of 10. At the same N/P ratio, the transfection efficiency of Cs-g-PLLD-FA/siAEG-1 was significantly higher than that of Cs-g-PLLD/siAEG-1. The results indicated that folate-modified Cs-g-PLLD indeed enhanced the transfection efficiency in osteosarcoma cells.
To elucidate how the composition of the formulations affected the cell uptake of siRNA, we incubated Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 (AEG-1 siRNA was labeled with cy3) with 143B cells for 24 hours and subsequently analyzed the cells by flow cytometry. As shown in Figure 4B, the transfection efficiency of the Cs-g-PLLD-FA/siAEG-1 complex at an N/P ratio of 20 reached the highest value at 98.7%±3.1%, while Cs-g-PLLD-FA/siAEG-1 at an N/P ratio of 10, Cs-g-PLLD/siAEG-1 at an N/P ratio of 20, and Cs-g-PLLD/siAEG-1 at an N/P ratio of 10 reached their highest transfection efficiencies at 61.6%±2.9%, 75.1%±2.6%, and 27.3%±4.1%, respectively. In addition, a similar efficiency of transfection (98.7%±3.1%) was observed for cells incubated with the Cs-g-PLLD-FA/siAEG-1 (N/P =20) complexes and cells incubated with Lipofectamine 2000/siAEG-1 complexes (93.0%±3.8%). As shown in Figure S3B, similar results were also observed in U2OS cells. We concluded that Cs-g-PLLD-FA is a highly efficient nonviral vector for gene delivery to osteosarcoma cells.
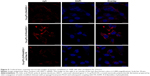
Cellular uptake of complexes
We characterized the cellular uptake and intracellular distribution of Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 (in the absence or presence of free FA) into osteosarcoma cells (the AEG-1 siRNA was labeled with cy3). Confocal microscopy images (Figures 5 and S4) showed a high degree of colocalization of the red fluorescence distributed in the cytoplasm after 4 hours of incubation, from which the simultaneous delivery of siRNA was confirmed. In addition, cells incubated with Cs-g-PLLD-FA/siAEG-1 showed much stronger red fluorescence than cells incubated with Cs-g-PLLD/siAEG-1, while the red fluorescence was weaker in the presence of free FA, suggesting that FA improved the cellular uptake of the complexes into osteosarcoma cells and that cellular uptake decreased with free FA competition.
Cell proliferation and invasion analysis in vitro
At first, we detected the AEG-1 protein and mRNA expression levels by Western blot quantitative reverse transcription-polymerase chain reaction analysis to confirm the knockdown of AEG-1 in osteosarcoma cells after transfection. As shown in Figures 6A, B and S5A, B, the AEG-1 protein and mRNA levels were reduced after transfection with Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 for 48 hours compared with Cs-g-PLLD/ scrambled small interfering RNA (siSCR) (siSCR has the same basic groups but a different sequence compared with siAEG-1), and the AEG-1 protein and mRNA levels were reduced more significantly (similar to the Lipofectamine 2000 [Lip]/siAEG-1 group) in the cells transfected with Cs-g-PLLD-FA/siAEG-1 than in those transfected with Cs-g-PLLD/siAEG-1.
Next, the cell proliferation and invasion activities were evaluated in vitro, as measured by MTT and invasion assays, respectively. The MTT assay showed that the cells transfected with Cs-g-PLLD-FA/siAEG-1 were more significantly inhibited (similar to the Lip/siAEG-1 group) than those transfected with Cs-g-PLLD/siAEG-1, resulting in a 50% growth inhibition of osteosarcoma cells compared with cells transfected with Cs-g-PLLD/siSCR (Figures 6C and S5C). Invasion assays showed that the Cs-g-PLLD-FA/siAEG-1 cells also exhibited decreased invasion ability (similar to the Lip/siAEG-1 group) compared with the Cs-g-PLLD/siAEG-1 or Cs-g-PLLD/siSCR control cells (Figures 6D, E and S5D, E). The results suggested that Cs-g-PLLD/siAEG-1 and Cs-g-PLLD-FA/siAEG-1 complexes can successfully knockdown AEG-1 protein and mRNA levels as well as reduce cell proliferation and invasion ability in osteosarcoma cells. These results are similar to those obtained in our previous study,37 in which Cs-g-PLLD-FA/siAEG-1 was found to be more efficient than Cs-g-PLLD/siAEG-1 in knocking down AEG-1.
Tumor suppression in vivo
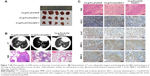
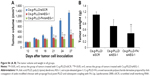
Intravenous injection of drugs is a widely accepted treatment option for malignant tumors. We examined the antitumor growth effects of Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 complexes on tumor suppression in vivo. To that end, caudal vein injections of Cs-g-PLLD/siSCR, Cs-g-PLLD/siAEG-1, or Cs-g-PLLD-FA/siAEG-1 at an siRNA dose of 2 nmol and quantity of Cs-g-PLLD or Cs-g-PLLD-FA at 100 μg per injection were administered in nude mice at predetermined days 12, 15, 18, 21, 24, and 27. As shown in Figures 7A and S6, Cs-g-PLLD-FA/siAEG-1 and Cs-g-PLLD/siAEG-1 inhibited tumor growth more effectively than Cs-g-PLLD/siSCR, and Cs-g-PLLD-FA/siAEG-1 was more effective than Cs-g-PLLD/siAEG-1. Figure 7B shows that the Cs-g-PLLD/AEG-1-transfected 143B cell line formed 91% fewer lung nodules than control-transfected cells (Student’s t-test; P=0.034). In addition, histological examination showed that the size of the nodules formed by Cs-g-PLLD/AEG-1-transfected cells was smaller than the size of those formed by control-transfected cells. There were no lung nodules detected either by CT scan or histological examination in the Cs-g-PLLD-FA/siAEG-1 group. All the results demonstrated marked anti-growth and anti-metastasis effects of AEG-1 expression in a clinically relevant mouse model and showed that siAEG-1 delivered by Cs-g-PLLD-FA showed favorable tumor targeting in vivo.
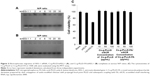
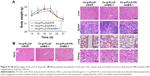
MMPs are a family of zinc-dependent endopeptidases that remodel and degrade the extracellular matrix and are considered to play important roles for tumor invasion in matrix degradation.43,44 Several reports and our previous study found that MMPs (MMP-2 and MMP-9) were involved in the AEG-1-mediated invasion of malignant tumors. Ki-67 is a nuclear protein that is associated with ribosomal RNA transcription and is a recognized marker of cell proliferation.45 We analyzed the subcutaneous tumor tissues by immunohistochemistry assays after the animals were sacrificed. As shown in Figure 7C, AEG-1 displayed a high level of expression in the Cs-g-PLLD/siSCR group but low levels of expression in the Cs-g-PLLD-FA/siAEG-1 and Cs-g-PLLD/siAEG-1 groups, and the AEG-1 protein level was lower in the Cs-g-PLLD-FA/siAEG-1 group than in the Cs-g-PLLD/siAEG-1 group. Consistent with the change in the levels of AEG-1 protein, IHC staining showed that osteosarcoma tumor tissues with high levels of AEG-1 expression exhibited strong Ki-67, MMP-2, and MMP-9 staining signals, whereas osteosarcoma tumor tissues with low AEG-1 expression exhibited low Ki-67, MMP-2, and MMP-9 expression. The results suggested that the tumor proliferation and invasion abilities were inhibited in the Cs-g-PLLD-FA/siAEG-1 and Cs-g-PLLD/siAEG-1 groups compared with the Cs-g-PLLD/siSCR group, and that the effects in the Cs-g-PLLD-FA/siAEG-1 group were more significant than in the Cs-g-PLLD/siAEG-1 group.
Moreover, Figure S7A shows that the weights of the nude mice from the different groups did not change significantly; a decrease was only noted after 21 days following the injection of 143B cells, and was a result of the tumor burden in all the groups. Figure S7B shows no significant histological damage in the representative H&E staining of the heart, liver, spleen, lung, and kidney in any group. All the results indicated that Cs-g-PLLD and Cs-g-PLLD-FA were safe materials, thus confirming the earlier results regarding the absence of toxicity.
Conclusion
We developed Cs-g-PLLD and Cs-g-PLLD-FA nanoparticles that were able to deliver siRNA into human osteosarcoma cells with no obvious cytotoxicity. The resulting AEG-1 knockdown inhibited osteosarcoma tumor proliferation and invasion in vitro. Moreover, caudal vein injection of Cs-g-PLLD/siAEG-1 or Cs-g-PLLD-FA/siAEG-1 complexes inhibited tumor growth and lung metastasis by silencing AEG-1 and regulating MMP-2/9 in a 143B-cell subcutaneous xenograft murine model. The Cs-g-PLLD-FA/siAEG-1 complexes displayed a better effect in vitro and in vivo than the Cs-g-PLLD/siAEG-1 complexes. All these findings showed that Cs-g-PLLD-FA nanoparticles have good potential as siRNA carriers for osteosarcoma therapy.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no 81402221), the Research Fund for the Doctoral Program of Higher Education of China (grant no 20130171120077), the Science and Technology Program of Guangzhou, China (grant no 201707010108), and the Science and Technology Planning Project of Guangzhou (grant no 201610010006).
Disclosure
The authors report no conflicts of interest in this work.
References
Jain KK. Role of biological therapies in the development of personalized medicine. Expert Opin Biol Ther. 2012;12(1):1–5. | ||
Koo KM, Wee EJH, Mainwaring PN, Wang YL, Trau M. Toward precision medicine: a cancer molecular subtyping nano-strategy for RNA biomarkers in tumor and urine. Small. 2016;12(45):6233–6242. | ||
Sunderland KS, Yang MY, Mao CB. Phage-enabled nanomedicine: from probes to therapeutics in precision medicine. Angew Chem Int Ed Engl. 2017;56(8):1964–1992. | ||
Young SW, Stenzel M, Yang JL. Nanoparticle-siRNA: a potential cancer therapy? Crit Rev Oncol Hematol. 2016;98:159–169. | ||
Peer D. Harnessing RNAi nanomedicine for precision therapy. Mol Cell Ther. 2014;2:5. | ||
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. | ||
Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. | ||
Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. 2007;120(5):953–960. | ||
Yadav S, van Vlerken LE, Little SR, Amiji MM. Evaluations of combination MDR-1 gene silencing and paclitaxel administration in biodegradable polymeric nanoparticle formulations to overcome multidrug resistance in cancer cells. Cancer Chemother Pharmacol. 2009;63(4):711–722. | ||
Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. | ||
Wei J, Cheang T, Tang B, et al. The inhibition of human bladder cancer growth by calcium carbonate/CaIP6 nanocomposite particles delivering AIB1 siRNA. Biomaterials. 2013;34(4):1246–1254. | ||
Dash TK, Konkimalla VB. Nanoformulations for delivery of biomolecules: focus on liposomal variants for siRNA delivery. Crit Rev Ther Drug Carrier Syst. 2013;30(6):469–493. | ||
Guo P, Coban O, Snead NM, et al. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev. 2010;62(6):650–666. | ||
Ma Z, Li J, He F, Wilson A, Pitt B, Li S. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem Biophys Res Commun. 2005;330(3):755–759. | ||
Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61(9):710–720. | ||
Brunot C, Ponsonnet L, Lagneau C, Farge P, Picart C, Grosgogeat B. Cytotoxicity of polyethyleneimine (PEI), precursor base layer of polyelectrolyte multilayer films. Biomaterials. 2007;28(4):632–640. | ||
Lee JM, Yoon TJ, Cho YS. Recent developments in nanoparticle-based siRNA delivery for cancer therapy. Biomed Res Int. 2013;2013:782041. | ||
Mujokoro B, Adabi M, Sadroddiny E, Khosravani M. Nano-structures mediated co-delivery of therapeutic agents for glioblastoma treatment: a review. Mater Sci Eng C Mater Biol Appl. 2016;69:1092–1102. | ||
Singha K, Namgung R, Kim WJ. Polymers in small-interfering RNA delivery. Nucleic Acid Ther. 2011;21(3):133–147. | ||
Skaugrud O, Hagen A, Borgersen B, Dornish M. Biomedical and pharmaceutical applications of alginate and chitosan. Biotechnol Genet Eng Rev. 1999;16:23–40. | ||
Lee M, Nah JW, Kwon Y, Koh JJ, Ko KS, Kim SW. Water-soluble and low molecular weight chitosan-based plasmid DNA delivery. Pharm Res. 2001;18(4):427–431. | ||
Dang JM, Leong KW. Natural polymers for gene delivery and tissue engineering. Adv Drug Deliv Rev. 2006;58(4):487–499. | ||
Guang Liu W, De Yao K. Chitosan and its derivatives – a promising non-viral vector for gene transfection. J Control Release. 2002;83(1):1–11. | ||
Koping-Hoggard M, Varum KM, Issa M, et al. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11(19):1441–1452. | ||
Hashimoto M, Morimoto M, Saimoto H, Shigemasa Y, Sato T. Lactosylated chitosan for DNA delivery into hepatocytes: the effect of lactosylation on the physicochemical properties and intracellular trafficking of pDNA/chitosan complexes. Bioconjug Chem. 2006;17(2):309–316. | ||
Zhao X, Yu SB, Wu FL, Mao ZB, Yu CL. Transfection of primary chondrocytes using chitosan-pEGFP nanoparticles. J Control Release. 2006;112(2):223–228. | ||
Lai WF, Lin MCM. Nucleic acid delivery with chitosan and its derivatives. J Control Release. 2009;134(3):158–168. | ||
Moreira C, Oliveira H, Pires LR, Simoes S, Barbosa MA, Pego AP. Improving chitosan-mediated gene transfer by the introduction of intracellular buffering moieties into the chitosan backbone. Acta Biomater. 2009;5(8):2995–3006. | ||
Riva R, Ragelle H, des Rieux A, Duhem N, Jerome C, Preat V. Chitosan and chitosan derivatives in drug delivery and tissue engineering. Chitosan for Biomaterials Ii. 2011;244:19–44. | ||
Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal Biochem. 2005;338(2):284–293. | ||
Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004 data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. | ||
Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. | ||
Bacci G, Briccoli A, Ferrari S, et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities with synchronous lung metastases: treatment with cisplatin, adriamycin and high dose of methotrexate and ifosfamide. Oncol Rep. 2000;7(2):339–346. | ||
Su ZZ, Kang DC, Chen Y, et al. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21(22):3592–3602. | ||
Yoo BK, Emdad L, Su ZZ, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119(3):465–477. | ||
Ying Z, Li J, Li M. Astrocyte elevated gene 1: biological functions and molecular mechanism in cancer and beyond. Cell Biosci. 2011;1(1):36. | ||
Wang F, Ke ZF, Sun SJ, et al. Oncogenic roles of astrocyte elevated gene-1 (AEG-1) in osteosarcoma progression and prognosis. Cancer Biol Ther. 2011;12(6):539–548. | ||
Pahimanolis N, Sorvari A, Luong ND, Seppala J. Thermoresponsive xylan hydrogels via copper-catalyzed azide-alkyne cycloaddition. Carbohydr Polym. 2014;102:637–644. | ||
Ma D, Zhang HB, Chen YY, Lin JT, Zhang LM. New cyclodextrin derivative containing poly(L-lysine) dendrons for gene and drug co-delivery. J Colloid Interface Sci. 2013;405:305–311. | ||
Mehnert W, Mader K. Solid lipid nanoparticles: production, characterization and applications. Adv Drug Deliv Rev. 2001;47(2–3):165–196. | ||
Zeng YF, Hu X, Liu FC, Bu XH. Azido-mediated systems showing different magnetic behaviors. Chem Soc Rev. 2009;38(2):469–480. | ||
Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. | ||
Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89(17):1260–1270. | ||
Ortega N, Behonick D, Stickens D, Werb Z. How proteases regulate bone morphogenesis. Ann N Y Acad Sci. 2003;995:109–116. | ||
Cuylen S, Blaukopf C, Politi AZ, et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535(7611):308–312. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.