Back to Journals » Infection and Drug Resistance » Volume 13
Nanoparticles-Based Biosensor Coupled with Multiplex Loop-Mediated Isothermal Amplification for Detection of Staphylococcus aureus and Identification of Methicillin-Resistant S. aureus
Authors Jiang L, Li X, Gu R, Mu D
Received 27 December 2019
Accepted for publication 30 March 2020
Published 29 April 2020 Volume 2020:13 Pages 1251—1262
DOI https://doi.org/10.2147/IDR.S243881
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Luxi Jiang, Xiaomeng Li, Rumeng Gu, Deguang Mu
Department of Respiratory Medicine, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, Zhejiang, People’s Republic of China
Correspondence: Deguang Mu Tel +86-571-85893509
Email [email protected]
Introduction: Staphylococcus aureus (S. aureus), including methicillin-resistant S. aureus (MRSA), is a common human pathogen, which can cause a variety of infections from mild to severe. In this article, a new diagnostic method called multiplex loop-mediated isothermal amplification combined with nanoparticles-based lateral flow biosensor (mLAMP-LFB) has been developed, which was proved to be fast, reliable, and simple for detecting S. aureus, and differentiate MRSA from methicillin-susceptible S. aureus (MSSA).
Materials and Methods: We designed a set of six primers targeting the nuc gene of S. aureus, and a set of five primers targeting the mecA gene of MRSA. The lateral flow biosensor visually reported the S. aureus-LAMP results within 2 mins. S. aureus species and non-S. aureus species were used to identify the specificity and sensitivity of the assay.
Results: The best conditions for LAMP were 50 mins at 63°C, and the sensitivity was 100 fg. No cross-reactivity was shown and the specificity of this assay is 100%. This assay requires 20 mins for DNA preparation, 50 mins for isothermal amplification and 2 mins for biosensor detection. The total time is within 75 mins. Among 96 sputum samples, LAMP-LFB and traditional culture method showed the same results, 8 (8.33%) samples were MRSA-positive, and 9 (9.38%) samples were MSSA-positive. Seven (7.29%) samples were MRSA-positive and 7 (7.29%) were MSSA-positive by PCR method. Compared with the culture method, diagnostic accuracy of m-LAMP-LFB assay was 100%. The results showed that the m-LAMP-LFB method has better detection ability than the PCR method.
Discussion: In short, this m-LAMP-LFB assay is a specific and sensitive method that can quickly identify S. aureus stains, and distinguish MRSA from MSSA, and can be used as a new molecular method for detection of S. aureus in laboratories.
Keywords: S. aureus, MRSA, MSSA, limit of detection, lateral flow biosensor, loop-mediated isothermal amplification, mLAMP
Introduction
Staphylococcus aureus (S. aureus) is a common gram-positive bacterium, and nearly 30% of human population is colonized with it.1 Besides, S. aureus is also a human pathogen, which can cause a diverse range of infections from mild, such as food-borne disease, to severe, such as pneumonia with high mortality, sepsis.2 S. aureus is toxigenic, and the toxin can reduce the efficacy of antibiotics. Methicillin-resistant Staphylococcus aureus(MRSA) infection has become a serious worldwide problem, with high mortality and morbidity since the 1970s.3 As it is reported, in developed countries, MRSA has arrived in over 60% of all isolated S. aureus, and the incidence of MRSA causing hospital acquired pneumonia has reached 40% in China and the incidence of MRSA has increased to 49% in the United States.4,5 Therefore, powerful assays that can identify S. aureus, MRSA, methicillin-susceptible S. aureus (MSSA) both efficiently and rapidly are urgently needed.
Traditional methods used to identify S. aureus and MRSA include culture, microdilution resistance examinations, and colony morphology.5 These traditional methods have their advantages in identifying these pathogens. However, they usually have cannot be ignored disadvantages: these methods usually are time-consuming and laborious and have poor sensitivity. It still takes another 2 days to identify MRSA, after the culture result is positive of S. aureus.6 Because of this, until the results of antibiotic susceptibility tests are provided, vancomycin and other glycopeptide antibiotic will be used as empirical treatment in patients with suspected MRSA infections.7 Therefore, the empirical use of antibiotics increased the drug resistance of S. aureus. Thereby, the reliable and rapid identification of S. aureus, MSSA and MRSA has great clinical significance.
Molecular methods such as polymerase chain reaction (PCR) and real-time PCR have been developed for detection of S. aureus and MRSA.8–10 These methods can be used to analyze clinical specimens (sputum, blood and bronchoalveolar lavage fluid (BALF)) with high specificity and sensitivity. However, the simplicity and rapidity of these methods are relatively limited, due to its long genomic template extraction and amplification protocols. Besides, most PCR-based methods using the ThermoCycler, the Stratagene Mx3000P PCR (Stratagene, La Jolla, USA), or even the LightCycler system (Roche, Mannheim, Germany) which were quite expensive and not approachable in most clinical institutions.11 Therefore, a timely, efficient, labor-saving, simple and cost-effective assay should be established to identify S. aureus and test its resistance to methicillin.To date, many isothermal amplification methods have been developed for molecular analysis.12,13 Loop-mediated isothermal amplification (LAMP) is a kind of sequence-specific isothermal amplification method, that offers nucleic acid amplification using 4 to 6 primers and a polymerase (Bst 2.0) with chain displacement activity at a fixed temperature (usually between 60–67ºC).14,15 LAMP has displayed several traits including that it has high sensitivity and specificity and sensitivity, can be completed in a short time (less than one hour), can amplify at various pH and the temperature ranges which is advantageous for clinical diagnosis, and that the LAMP reagents are relatively inexpensive and can be stable at a room temperature.16,17 Thus far, LAMP has been used in many fields, especially in molecular diagnostics.14 This LAMP technology is currently reported used for identifying S. aureus while cannot identify MRSA.18 And lots of monitoring methods, such as gel electrophoresis, turbidimeters, colorimetric agents, nanoparticle-based lateral flow biosensors (LFBs) and lab-on-chip devices, have been used to analyze LAMP amplicons.19 In particular, because of their rapidness, low cost and simplicity, various nanoparticle-based LFBs are increasingly used as alternative tools to analyze LAMP products.20 Based on this, we aimed to develop a novel assay combining mLAMP and LFB technology to detect S. aureus, and differentiate MRSA from MSSA. Then, the sensitivity and specificity of the LAMP-LFB assay were evaluated, and 96 clinical sputum were tested using the mLAMP-LFB assay. Besides, we compared the results obtained from mLAMP-LFB with the data from the culture and traditional PCR methods.
Materials and Methods
Reagents and Apparatus
Genomic template extraction kits (QIAamp DNA minikits; Qiagen, Hilden, Germany) were purchased from Qiagen (Beijing, China). Dye (Crimson red) streptavidin-coated polymer nanoparticles (129 nm, 10 mg mL-1, 100 mM borate, pH 8.5 with 0.1% BSA, 0.05% Tween 20 and 10 mM EDTA) were purchased from Bangs Laboratories, (Indiana, USA). Biotin-BSA (biotinylated bovine serum albumin) and anti-FITC (rabbit anti-fluorescein antibody), anti-dig (sheep anti-digoxigenin antibody) were purchased from Abcam (Shanghai, China). Universal isothermal amplification kits and visual detection reagent (VDR) were purchased from HaiTaiZhengYuan (Beijing, China). LFB materials, such as conjugate pad, sample pad, backing card, nitrocellulose membrane (NC) and absorbent pad were purchased from the Jie-Yi Biotechnology (Shanghai, China).
Primer Design
Based on S. aureus nuc gene (GenBank accession EF529597), MRSA mecA gene (GenBank Accession No. X52593) and the Primer Explorer V4 (http://primerexplorer.jp/e/; Eiken Chemical, Tokyo, Japan), two sets of LAMP primers were designed for this assay. The Basic Local Alignment Search Tool (BLAST) was used to check the specificity of two LAMP primers. The details of the LAMP primers are shown in Figure 1 and Table 1. All primers were synthesized by TsingKe (Beijing, China) at HPLC purification grade.
 |
Table 1 The Primers Used in the Current Report |
Bacterial Strains and Template Preparation
The 39 strains used in this study were collected from clinical and environmental samples (Table 2). S. aureus (MSSA) (ATCC 25923) and S. aureus (MRSA) (ATCC 43300) were used as reference strains to optimize the LAMP-LFB assay. DNA templates were extracted by DNA extraction kit (QIAamp DNA Mini Kits, Hilden, Germany) according to the manufacturer’s instructions, and measured by a spectrophotometer (Nano drop ND-1000, Calibre, Beijing, China). The original DNA extraction concentration of ATCC 25923 is 264.1 ng/μL, and the DNA extraction concentration of ATCC 43300 is 123.2 ng/μL. The DNA templates of S. aureus ATCC 25923 and ATCC 43300 were serially diluted 10-fold (1 ng/μL, 100 pg/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL, 1 fg/μL) to optimize the temperature and test the sensitivity of the LAMP-LFB assay. And 1μL of each dilution was used as template for LAMP reaction. The DNA extraction concentration of sputum samples used in this assay ranged from 7 ng/μL to 58 ng/μL.
 |
Table 2 Bacterial Strains Used in the Current Study |
Preparation of Lateral Flow Biosensor
Based on previous publications, we use LFB to report LAMP results in this assay.18,21,22 In short, LFB was designed to detect three targets (two target amplicons and a chromatography control). Four components, including an immersion pad, a conjugate pad, an NC membrane, and an absorbent pad, were assembled onto a plastic adhesive backing card. The capture reagents were fixed in 0.01 M phosphate-buffered saline (PBS, Ph 7.4), and the reagents contained anti-FITC (0.2mg/mL), anti-Dig (0.2mg/mL) and biotin-BSA (4mg/mL). The first line (conjugated with anti-FITC) is the target amplicon I test line (T1), the second line (conjugated with anti-Dig) is the target amplicon II test line (T2), and the third line (conjugated with biotin-BSA) is the chromatography control (CL). And each line was separated by 5mm. SA-PNP (Dye streptavidin-coated polymer nanoparticles, 129 nm, 10mg mL-1, 100mM borate, pH 8.5 with 0.1% BSA, 0.05% Tween 20 and 10mM EDTA) were then dispensed onto the conjugate pad of the biosensor. Streptavidin, a tetrameric protein with four biotin-binding sites, was covalently conjugated to these functionalized nanoparticles with excellent retention of biotin-binding activity. The streptavidin-biotin bond is one of the strongest non-covalent, affinity interactions, thus streptavidin-coated polymer nano-particles can provide an efficient and facile means for binding of biotinylated amplicons for use in diagnosis.
The Singlex LAMP Assay
To check the availability of LAMP primers, reactions for nuc gene and mecA gene have been tested according to the standard LAMP assay mentioned in previous article.23 The amplification 25 µL mixtures were consist of 0.2 µM each FIP∗ and FIP primers, 12.5 µL (2 ×) reaction mix, 0.2 µM each LF* and LB primer, 0.4 µM BIP primers, 0.1 µM each F3 and B3 primers, 1 µL DNA template and 1 µL (8 U) of Bst DNA polymerase. All the mixtures were heated for 50 min at 63ºC. Streptococcus pneumonia strains (S. pneumonia, ATCC700674) and Listeria monocytogenes strains (L. monocytogenes, Isolated strains) were used as negative controls (NC), and double distilled water (DW) were used as a blank control.
The Multiplex LAMP Assay
The 25 µL mixtures of the multiplex LAMP (m-LAMP) assay were consist of 12.5 µL (2 ×) reaction mix, 0.2 µM nuc-BIP primers, 0.05 µM each nuc-F3 and nuc-B3 primers, 0.1 µM each nuc-FIP∗ and nuc-FIP primers, 0.1 µM each nuc-LF∗ and nuc-LB primer, 0.2 µM mecA-BIP primers, 0.05 µM each mecA -F3 and mecA -B3 primers, 0.1 µM each mecA -LF∗ and mecA -LB primer, 0.1 µM each mecA-FIP∗ and mecA -FIP primers, 1 µL DNA template and 1 µL (8 U) of Bst DNA polymerase. All the mixtures were heated for 50 min at 63ºC. Streptococcus pneumonia strains (S. pneumonia, ATCC700674) and Listeria monocytogenes strains (L. monocytogenes, Isolated strains) were used as negative controls (NC), and double distilled water (DW) was used as a blank control.
LAMP-LFB Assay
1 μL of LAMP products (haptens (FITC or/and Dig) and biotin-labeled LAMP products) and 60 μL of running buffer were added into the sample area. Then, the LAMP products and SA-PNPs can be simultaneously transferred by capillary flow from the conjugate region to TL (T1, T2) and CL. Biotin-labeled LAMP products form complex with SA-PNPs through the biotin-streptavidin-biotin interactions in the conjugated region, and biotin/LAMP complexes were fixed on the test strip through the interaction of hapten (FITC or/and Dig) and antibody (anti-FITC or/and anti-dig). Through the interaction of streptavidin and biotin, SA-PNPs that did not construct complexes were captured at CL. Therefore, SA-PNPs/LAMP/FITC complexes, SA-PNPs/LAMP/Dig complexes and non-complexed SA-PNPs were reported by visible line at TL (T1, T2) and CL, respectively. LFB has been used as an alternative for analyzing LAMP products.15 Besides, other monitoring methods, such as real-time turbidity (LA-320C) and VDR are used to determine and optimize the nuc-LAMP, mecA-LAMP, and m-LAMP products.
Optimizing the Temperature of LAMP-LFB Assay
During the amplification stage, the optimal temperatures of LAMP primers (nuc-LAMP primers and mecA-LAMP primers) were tested. We compared reaction temperatures from 60 to 67ºC. We used S. pneumonia (ATCC700674) and L. monocytogenes (Isolated strains) as negative controls (NC), and double distilled water (DW) as blank control (BC).
Sensitivity of LAMP-LFB Assay
We prepared serial dilution of S. aureus (ATCC 43300) (1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg per microliter), and added 1μL of genomic DNA into the amplification mixtures. We also examined the LoD (limit of detection) of the Singlex (nuc-LAMP and mecA-LAMP) and m-LAMP reactions as mentioned above. The LoD of singlex and multiplex reactions is considered to be the dilution gradients that finally show a positive result.
Optimizing the Reaction Time of LAMP-LFB Assay
We evaluated the LAMP amplification at different times (from 30 mins to 60 mins, with 10 mins intervals), and reported the results by LFB.
Specificity of LAMP-LFB Assay
We proved the specificity of this m-LAMP-LFB assay by genomic DNA (at least 10 ng per microliters) from 24 S. aureus species strains and 15 Non-S. aureus species strains (Table 2). And we also used LFB to show the results of all m-LAMP assays. We also repeated all the examinations for three times.
Evaluation of the Feasibility of m-LAMP-LFB Assay
A total of 96 sputum samples, which were suspected from human S. aureus, were collected from Zhejiang province, China (Ethic approval: The research protocol for the current study has been approved by The Ethics Committee of the Zhejiang provincial people’s Hospital, Hangzhou, Zhejiang, China). These samples were used for S. aureus, MRSA, MSSA diagnosis using culture-based method, traditional PCR method, and this new m-LAMP-LFB assay. Traditional cultures were conducted with blood agar plate. In short, about 1 mL sputum samples were inoculated into a blood plate to culture at 37ºC, 5% CO2 for 3–5 days, and then the bacteria strain was selected for culture. Other methods, such as serum agglutination test and Gram stain were also used to identify S. aureus isolates. Besides, DNA templates of 500 μL samples were extracted by QIAamp method for PCR and LAMP-LFB assays. PCR was performed with S. aureus specific primers for nuc gene with an amplicon size of 270 bp24 and MRSA specific primers for mecA gene with an amplicon size of 310 bp.25 And finally, we compared the results of m-LAMP-LFB method with those of the culture and PCR methods.
Results
Confirmation and Demonstration of LAMP Products
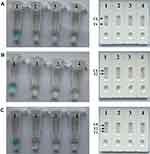
When the assay was conducted at a fixed temperature of 63ºC for 1 h, an appreciable S. aureus-LAMP reaction was observed. The S. aureus-LAMP amplified products were light green, while colorlessness was remained in the negative ones (Figure 2A (left)-2C (left)). For nuc test (Figure 2A (right)), the crimson bands were seen in both T1 and CL, and for mecA test (Figure 2B (right)), the T2 and CL were visible. In addition, T1, T2 and CL appeared in the multiplex LAMP products (Figure 2C (right)). The results showed that the two sets of primers for nuc and mecA were good for establishing the LAMP-LFB method.
Optimal Temperature of LAMP Primer Set
The detection temperature for the m-LAMP-LFB assay is very important, so we use target DNA (10 pg/mL) at different reaction temperature (60–67ºC, with 1ºC interval) to optimize the amplification temperature of nuc- and mecA-LAMP methods. The reactions of nuc- and mecA-LAMP were analyzed by real-time turbidity detection, and the kinetic curves at different temperatures were obtained. However, for the nuc-LAMP reactions, robust amplification were obtained at an analysis temperature of 62–65ºC, and for the mecA-LAMP reactions, robust amplification were obtained at 62°C-64°C (Figure 3). Finally, 63ºC was chosen as the optimal temperature for the singlex and m-LAMP reactions.
Sensitivity of nuc- and mecA-LAMP Assays
Serially diluted genomic DNA templates (1 ng/μL, 100 pg/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL, and 1 fg/μL) were used to determine the sensitivity of nuc- and mecA-LAMP. As shown in Figure 4, the LoD of nuc-LAMP assay was 100 fg per reaction, and two crimson lines (T1, and CL) appeared on the LFB, indicating that the nuc gene was positive. The mecA-LAMP assay was also 100 fg per vessel, and two crimson lines (T2, and CL) appeared on the LFB, indicating that the mecA gene was positive. In addition, the LoD of VDR detection (Figure 4A (bottom), B (bottom)) for nuc- and mecA-LAMP was consistent with LFB analysis (Figure 4A (top) and B (top)).
Sensitivity of m-LAMP Assay
The amplified m-LAMP products were showed by LFB. As shown in Figure 5, three crimson lines (T1, T2, and CL) showed on the LFB, which indicated that the nuc and mecA genes are positive. Only a crimson band (CL) appeared on the LFB, indicating negative results. Such as a blank control or the target genomic DNA content is less than 100 fg. The sensitivity of m-LAMP to detect both nuc and mecA genes was also 100 fg per vessels, which was fully consistent with singlex LAMP assay (Figures 4 and 5).
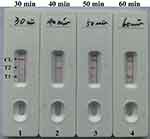
Optimal Duration Time of m-LAMP Assay
We evaluated the effect of different durations (from 30 mins to 60 mins, with an interval of 10 mins) at optimal amplification temperature (63ºC), and the target DNA at the LoD level (100 fg) was detected when m-LAMP reaction lasted only for 50 mins (Figure 6). Therefore, the 50-min amplification time was taken as the optimal amplification time for m-LAMP. So this new m-LAMP-LFB detection process takes only 75 mins, including 20 mins of genomic template preparation, 50 mins of LAMP reaction, and 2 mins of LFB analysis.
Specificity of m-LAMP Method
In this paper, MRSA, MSSA, and non-S. aureus bacterial pathogens were used to determine the specificity of the m-LAMP method. As shown in Figure 7 and Table 2, m-LAMP assay can specifically detect all S. aureus strains, and non-S. aureus bacterial pathogens are negative. With biosensors, T1, T2 and CL simultaneously appeared in the LFB detection area, indicating positive results of MRSA (Figure 7, biosensor 1). In addition, T1 and CL simultaneously appeared in the LFB detection area, indicating the positive results for MSSA (Figure 7, biosensor 2). Only one crimson band (CL) appeared in the LFB detection area, indicating the negative results for non- S. aureus strains and negative control (DW) (Figure 7, biosensor 3–18).
Demonstrating the Feasibility of m-LAMP-LFB Assay by Sputum Samples
We further confirmed the feasibility of m-LAMP-LFB method. We used culture, PCR and LAMP-LFB assay to test 96 sputum samples collected from patients. The results are summarized in Table 3. Among 96 sputum samples, 8 (8.33%) and 9 (9.38%) samples were MRSA-positive and MSSA-positive by LAMP-LFB and traditional culture biotechnology, respectively. Therefore, compared with the culture-biotechnical method, the diagnostic accuracy of m-LAMP-LFB method is 100%. However, only 7 (7.29%) samples were MRSA-positive and 7 (7.29%) were MSSA-positive by PCR method. These results showed that the m-LAMP-LFB method was a good detection method and has better detection ability than PCR method.
 |
Table 3 Comparison of Conventional LAMP-LFB, Culture Biotechnical and PCR Methods for the Detection of S. Aureus in Sputum Samples of Human |
Discussion
Staphylococcus aureus, including MRSA and MSSA, is an important pathogen, which can produce a variety of infections, including skin infection, lung infection, infective endocarditis and septic shock.1,26 The rapid detection of clinical specimens for S. aureus and MRSA is very important. In this report, we have successfully established m-LAMP assay which can distinguish S. aureus strains and differentiate MRSA from MSSA according to nuc and mecA genes. The mLAMP primers of this new assay are designed for 7 or 8 regions of the target genes with high selectivity (Figure 1). We used nuc gene to design the sequences of nuc-LAMP primer set, which is specific for S. aureus. In addition, the resistance of S. aureus to methicillin is mainly mediated by low affinity penicillin binding protein 2a or 2ʹ (PBP2a or PBP2ʹ), which is specifically encoded by mecA gene. And in this assay, we used mecA gene (Figure 1) which is related to the methicillin-resistance of S. aureus strains to design the sequences of mecA-LAMP primer set.26 The specificity of this assay was performed using genomic templates extracted from 24 strains of S. aureus (5 MRSA and 19 MSSA) and 15 strains of non-S. aureus. All S. aureus strains were tested positive, while non-S. aureus strains were tested negative. This m-LAMP method for identification of S. aureus targeting the nuc gene has 100% specificity, and the m-LAMP method for identification of MRSA targeting the mecA gene related to methicillin resistance has 100% specificity (Figure 7 and Table 2). In addition, the most important thing is that this m-LAMP method can distinguish MRSA from MSSA while detecting S. aureus strains.
In this experiment, we use LFB to detect the experimental results of LAMP. LFB is intuitive, fast, convenient and easy-to-use (Figures 2–7). Compared with other monitoring technologies, such as real-time turbidity and colorimetric indicator (Figures 2–3), LFB method is not only faster, simpler, but also has a lower error rate. As LFB method does not involve the use of special instruments, reagents, and processes, LFB is more appropriate for simple, rapid, and sensitive detection of LAMP products than other analytical methods. Besides, the LFB can be used to detect two targets visually and simultaneously in one single test in our assay.27
In the LAMP assay, we found that when the nuc and mecA genes were tested independently, the 100 fg DNA templates per tube was the LoD. The LoD of the LAMP experimental results displayed by LFB is consistent with that of the VDR (Figure 4). In the m-LAMP assay, we found that when performing simultaneous detection of nuc and mecA genes, 100 fg of target DNA templates per tube was still the LoD. The test results are consistent with the singlex nuc-LAMP and mecA-LAMP detection (Figures 4 and 5). In addition, the results of m-LAMP can be displayed by simple instruments, such as conventional heating instruments or laboratory water bath instruments. As long as the simple instrument can provide a constant temperature of 63◦C, there is no need to use other complex ones.
This new LAMP-LFB detection process takes only 75 mins, including 20 mins of genomic template preparation, 50 mins of LAMP reaction, and 2 mins of LFB analysis. In order to evaluate the availability of this new LAMP-LFB assay, 96 sputum specimens were tested by culture, PCR diagnostic method, and LAMP-LFB detection. This new LAMP-LFB method targets MRSA and MSSA with high detection efficiency. One of the sputum specimens was tested positive for MRSA by LAMP-LFB and culture tests, but tested negative by traditional PCR test. Two other sputum specimens were tested positive for MSSA by LAMP-LFB and culture tests, but negative by traditional PCR test. The above results indicate that the diagnostic rate of the traditional PCR method is lower than that of the new LAMP-LFB detection method, which may be because the target gene templates copy numbers in the sample is lower than the detection limit of the traditional PCR test. In addition, compared with the PCR and culture methods, the m-LAMP-LFB method only requires an instrument which can provide a constant temperature of 63ºC, without using expensive molecular detection equipment. And this m-LAMP-LFB method also has a faster detection speed. However, this new assay also has its limitation. Results of the LAMP detection can only be qualitatively displayed as red bands.
Conclusion
In conclusion, we have successfully established a m-LAMP-LFB method that targeted the nuc and mecA genes. This method showed high specificity sensitivity, which could successfully detect S. aureus strains, distinguish MRSA from MSSA, and had the LoD of 100 fg genomic template per tube. Moreover, the clinical feasibility of the m-LAMP-LFB also was successfully demonstrated using clinical samples. Particularly, the LAMP results were indicated using biosensor, which was simple, fast, objective and easy-to-operate. Thus, the m-LAMP-LFB established in this report could be used as valuable tool for rapid, simple, sensitive and reliable detection of S. aureus and identification of MRSA in basic and clinical laboratories.
Acknowledgments
This study was supported by grants from Medical Science and Technology Project of Zhejiang province (No. 2020KY400) and (No. 2020372703).
Disclosure
The authors declare that they have no conflict of interest in this work.
References
1. Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG
2. Kuroda M, Ohta T, Uchiyama I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225–1240. doi:10.1016/S0140-6736(00)04403-2
3. Fakoya ECBVPTECCADAOEOAOJ. Methicillin-resistant Staphylococcus aureus: a mini review. Int J Med Res Health Sci. 2018;7(1):122–127.
4. Chinese Society of Respiratory Diseases isg. Guidelines for diagnosis and treatment of ventilator-associated pneumonia in Chinese adult hospital. Chin J Tubere Respir Dis. 2018;41(4):255–280.
5. Misawa Y, Saito R, Moriya K, et al. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J Infect Chemother. 2007;13(3):134–140. doi:10.1007/s10156-007-0508-9
6. Chen C, Zhao Q, Guo J, Li Y, Chen Q. Identification of methicillin-resistant Staphylococcus aureus (MRSA) using simultaneous detection of mecA, nuc, and femB by Loop-Mediated Isothermal Amplification (LAMP). Curr Microbiol. 2017;74(8):965–971. doi:10.1007/s00284-017-1274-2
7. Paule SM, Pasquariello AC, Thomson RB, Kaul KL, Peterson LR. Real-time PCR can rapidly detect methicillin-susceptible and methicillin-resistant staphylococcus aureus directly from positive blood culture bottles. Am J Clin Pathol. 2005;124(3):404–407. doi:10.1309/6EA3U9V8NCLLGKQN
8. Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44(1):124–131. doi:10.1128/JCM.44.1.124-131.2006
9. Mangold KA, Santiano K, Broekman R, et al. Real-time detection of blaKPC in clinical samples and surveillance specimens. J Clin Microbiol. 2011;49(9):3338–3339. doi:10.1128/JCM.00268-11
10. Kalinga A, Post RJ. An apparent halt to the decline of Simulium woodi in the Usambara foci of onchocerciasis in Tanzania. Ann Trop Med Parasitol. 2011;105(3):273–276. doi:10.1179/136485911X12899838683403
11. Wellinghausen N, Wirths B, Franz AR, Karolyi L, Marre R, Reischl U. Algorithm for the identification of bacterial pathogens in positive blood cultures by real-time LightCycler polymerase chain reaction (PCR) with sequence-specific probes. Diagn Microbiol Infect Dis. 2004;48(4):229–241. doi:10.1016/j.diagmicrobio.2003.11.005
12. Asiello PJ, Baeumner AJ. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip. 2011;11(8):1420–1430. doi:10.1039/c0lc00666a
13. Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–2486. doi:10.1039/c2lc40100b
14. Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother. 2013;19(3):404–411. doi:10.1007/s10156-013-0590-0
15. Prusty BR, Chaudhuri P, Chaturvedi VK, Saini M, Mishra BP, Gupta PK. Visual detection of Brucella spp. in spiked bovine semen using Loop-Mediated Isothermal Amplification (LAMP) assay. Indian J Microbiol. 2016;56(2):142–147. doi:10.1007/s12088-015-0563-3
16. Soleimani M, Shams S, Majidzadeh-A K. Developing a real-time quantitative loop-mediated isothermal amplification assay as a rapid and accurate method for detection of Brucellosis. J Appl Microbiol. 2013;115(3):828–834. doi:10.1111/jam.12290
17. Perez-Sancho M, Garcia-Seco T, Arrogante L, et al. Development and evaluation of an IS711-based loop mediated isothermal amplification method (LAMP) for detection of Brucella spp. on clinical samples. Res Vet Sci. 2013;95(2):489–494. doi:10.1016/j.rvsc.2013.05.002
18. Wang Y, Li H, Wang Y, Zhang L, Xu J, Ye C. Loop-mediated isothermal amplification label-based gold nanoparticles lateral flow biosensor for detection of enterococcus faecalis and Staphylococcus aureus. Front Microbiol. 2017;8:192.
19. Zhang X, Lowe SB, Gooding JJ. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens Bioelectron. 2014;61:491–499. doi:10.1016/j.bios.2014.05.039
20. Quesada-Gonzalez D, Merkoci A. Nanoparticle-based lateral flow biosensors. Biosens Bioelectron. 2015;73:47–63. doi:10.1016/j.bios.2015.05.050
21. Wang Y, Wang Y, Li D, Xu J, Ye C. Detection of nucleic acids and elimination of carryover contamination by using loop-mediated isothermal amplification and antarctic thermal sensitive uracil-DNA-glycosylase in a lateral flow biosensor: application to the detection of Streptococcus pneumoniae. Mikrochim Acta. 2018;185(4):212.
22. Wang Y, Liu D, Deng J, Wang Y, Xu J, Ye C. Loop-mediated isothermal amplification using self-avoiding molecular recognition systems and antarctic thermal sensitive uracil-DNA-glycosylase for detection of nucleic acid with prevention of carryover contamination. Anal Chim Acta. 2017;996:74–87. doi:10.1016/j.aca.2017.10.022
23. Wang Y, Wang Y, Luo L, et al. Rapid and sensitive detection of Shigella spp. and Salmonella spp. by multiple endonuclease restriction real-time loop-mediated isothermal amplification technique. Front Microbiol. 2015;6:1400. doi:10.3389/fmicb.2015.01400
24. Jiang L, Ren H, Zhou H, Qin T, Chen Y. Simultaneous detection of nine key bacterial respiratory pathogens using luminex xTAG((R)) technology. Int J Environ Res Public Health. 2017;14:3. doi:10.3390/ijerph14030223
25. Jonas D, Speck M, Daschner FD, Grundmann H. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J Clin Microbiol. 2002;40(5):1821–1823. doi:10.1128/JCM.40.5.1821-1823.2002
26. Hanberger H, Walther S, Leone M, et al. Increased mortality associated with meticillin-resistant Staphylococcus aureus (MRSA) infection in the intensive care unit: results from the EPIC II study. Int J Antimicrob Agents. 2011;38(4):331–335. doi:10.1016/j.ijantimicag.2011.05.013
27. Wang Y, Wang Y, Zhang L, Xu J, Ye C. Visual and multiplex detection of nucleic acid sequence by multiple cross displacement amplification coupled with gold nanoparticle-based lateral flow biosensor. Sens Actuators B Chem. 2017;241:1283–1293. doi:10.1016/j.snb.2016.10.001
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.