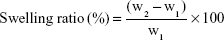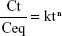Back to Journals » International Journal of Nanomedicine » Volume 13
Molecular study of wound healing after using biosynthesized BNC/Fe3O4 nanocomposites assisted with a bioinformatics approach
Authors Moniri M, Boroumand Moghaddam A, Azizi S, Abdul Rahim R, Zuhainis Saad W, Navaderi M, Arulselvan P , Mohamad R
Received 12 December 2017
Accepted for publication 1 February 2018
Published 21 May 2018 Volume 2018:13 Pages 2955—2971
DOI https://doi.org/10.2147/IJN.S159637
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Mona Moniri,1,2 Amin Boroumand Moghaddam,1,2 Susan Azizi,1 Raha Abdul Rahim,3 Wan Zuhainis Saad4,5 Mohammad Navaderi6,7 Palanisamy Arulselvan8 Rosfarizan Mohamad1,4
1Department of Bioprocess Technology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Selangor, Malaysia; 2Young Researcher and Elite Club, Sabzevar Branch, Islamic Azad University, Sabzevar, Iran; 3Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Selangor, Malaysia; 4Department of Microbiology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Selangor, Malaysia; 5Institute of Tropical Forestry and Forest Products, Universiti Putra Malaysia, Serdang, Selangor, Malaysia; 6Young Research and Elite Club, Parand Branch, Islamic Azad University, Parand, Iran; 7Department of Medical Genetics, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran; 8Muthayammal Centre for Advanced Research, Muthayammal College of Arts and Science, Rasipuram, Tamilnadu, India
Background: Molecular investigation of wound healing has allowed better understanding about interaction of genes and pathways involved in healing progression.
Objectives: The aim of this study was to prepare magnetic/bacterial nanocellulose (Fe3O4/BNC) nanocomposite films as ecofriendly wound dressing in order to evaluate their physical, cytotoxicity and antimicrobial properties. The molecular study was carried out to evaluate expression of genes involved in healing of wounds after treatment with BNC/Fe3O4 films.
Study design, materials, and methods: Magnetic nanoparticles were biosynthesized by using Aloe vera extract in new isolated bacterial nanocellulose (BNC) RM1. The nanocomposites were characterized using X-ray diffraction, Fourier transform infrared, and field emission scanning electron microscopy. Moreover, swelling property and metal ions release profile of the nanocomposites were investigated. The ability of nanocomposites to promote wound healing of human dermal fibroblast cells in vitro was examined. Bioinformatics databases were used to identify genes with important healing effect. Key genes which interfered with healing were studied by quantitative real time PCR.
Results: Spherical magnetic nanoparticles (15–30 nm) were formed and immobilized within the structure of BNC. The BNC/Fe3O4 was nontoxic (IC50>500 μg/mL) with excellent wound healing efficiency after 48 hours. The nanocomposites showed good antibacterial activity ranging from 6±0.2 to 13.40±0.10 mm against Staphylococcus aureus, Staphylococcus epidermidis and Pseudomonas aeruginosa. The effective genes for the wound healing process were TGF-B1, MMP2, MMP9, Wnt4, CTNNB1, hsa-miR-29b, and hsa-miR-29c with time dependent manner. BNC/Fe3O4 has an effect on microRNA by reducing its expression and therefore causing an increase in the gene expression of other genes, which consequently resulted in wound healing.
Conclusion: This eco-friendly nanocomposite with excellent healing properties can be used as an effective wound dressing for treatment of cutaneous wounds.
Keywords: wound healing, antibacterial activity, bioinformatics study, scratch assay, bacterial nanocellulose, green method
Introduction
Being the largest organ in the body, among other critical roles, skin serves as an impermeable insulating layer against environmental microorganisms and prevents dehydration. Loss of skin integrity after injury, surgery or illness may result in physiological imbalance and ultimately significant disability, or even death.1 Wound healing is a complex process of restoring impaired cells and tissues to their normal state. It works as a cellular response to injury and is involved in the activation of fibroblast, endothelial cells and macrophages.2 It is also involved in a well-orchestrated integration of biological and molecular events of cell migration, cell proliferation and extracellular matrix (ECM) deposition.1
Cutaneous wound healing encompasses reepithelialization, which involves migration and proliferation of keratinocytes to cover the dermal surface.3 Moreover, previous comprehensive molecular research has been carried out to evaluate gene expression and signal molecules in regards to the reepithelialization process.4,5 Detection of the key genes which play a significant role in the wound healing process, and investigating their gene expression level when unscathed and comparing to wound cases may lead to finding effective treatment and facilitate the healing of wounds.
Bacterial nanocellulose (BNC) is known as a useful and good wound dressing having the properties of novel wound dressing materials, and is applied for chronic ulcers.6 However, BNC itself does not have an antimicrobial effect, which is essential to inhibit wound infection throughout the wound healing. Several researchers integrated drug,7 polymers8 and nanoparticles (NPs)8 in BNC to make wound dressing materials with antibacterial activities, but few successful outcomes due to toxicity of applied materials in synthesis process have been reported. To overcome such a deficiency, we developed a green method to synthesize iron oxide NPs with the potential to repair tissues and induce cell therapy9 within the BNC by using Aloe vera leaf extracts to make an efficient BNC with antibacterial effects and reepithelialization properties. A. vera has hundreds of nutrients and active compounds such as vitamins, enzymes, minerals, sugars, lignin, anthraquinones, saponins, salicylic acid and different types of amino acids.10 A. vera has been proven to be a significant antimicrobial with antiviral, antifungal, anti-inflammatory immune stimulator and cell growth stimulator properties.11–13 These compounds are likely to be absorbed on the NPs’ surface and the BNC during the synthesis process, as a result, BNC/Fe3O4 could be extremely suitable for wound healing.
Finally, the molecular studies carried out to evaluate the expression of gene and signal molecules recognized. by bioinformatics method have been conducted to aid better understanding of the reepithelialization properties of BNC/Fe3O4 nanocomposites. Bioinformatics is an interdisciplinary field that develops methods and software tools to understand biological data. Common uses of bioinformatics include the identification of candidate genes. The methods of bioinformatics software have proved to be useful in detecting abundantly expressed genes that are restricted to certain tissues. Also, these tools could be useful to identify specific target genes for miRNAs.14 Bioinformatics-based tools provide detection pathways of different disorders through finding the effector genes and their regulator factors in disorder process.15
Materials and methods
Materials
D-glucose, peptone, yeast extract, Na2HPO4, citric acid, agar powder, Mueller-Hinton agar, ferrous chloride tetrahydrate (FeCl2·4H2O >99%), ferric chloride hexahydrate (FeCl3·6H2O, 99%) and the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma Chemicals (Perth, Australia). Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich), fetal bovine serum (FBS) and phosphate-buffered saline (PBS) were obtained from Thermo Fisher Scientific (Waltham, MA, USA); penicillin-streptomycin (PAA, Pasching, Austria) and methanol were from Merck (Darmstadt, Germany). Normal adult human dermal fibroblast (HDF) cells were purchased from American Type Culture Collection (Manassas, VA, USA).
Isolation and identification of bacteria cellulose
In this study, Gluconacetobacter was isolated from rotten pineapple fruit, using the method described by Park et al.16 First, 10 g of rotten fruit was transferred into 90 mL of a modified Hestrin and Schramm (HS) medium in a 250 mL flask containing 2.0% D-glucose (w/v), 0.5% peptone (w/v), 0.5% yeast extract (w/v), 0.27% Na2HPO4 (w/v), 0.115% citric acid (w/v) and agar powder 1.5% (w/v).17 Then, the pH of culture medium was adjusted to 5.5 by HCL or NaOH (1M). The growth of the colonies was observed during incubation at 30°C for 3 days. After incubation, the flask with white pellicle covering the surface of the liquid medium was selected. The culture broth of the selected flask was serially diluted with 0.85% NaCl (w/v), and 0.1 mL of each dilution was spread on HS-agar and then incubated at 30°C for 24 h. Afterward, the morphological (Gram staining test) and physiological characteristics (catalase tests) of isolated strain were evaluated. The most efficient cellulose-producing bacteria were examined through 16S rRNA gene sequence analysis using the method described by Yukphan et al.18 A specific fragment for 16S rRNA gene-coding regions was amplified using PCR amplification. Universal primers 27F and 1492R were used. The positions in the rRNA gene fragment were based on the Escherichia coli numbering system (accession number UPMC 1188).19 A phylogenetic tree was constructed by the neighbor-joining method of Saitou and Nei20 using MEGA program (version 7.0). The pure colonies were obtained and the isolation was detected as Gluconacetobacter xylinus RM1 based on the molecular characterization through 16S rRANA sequencing and it has 1,394 bp (Figure 1). G. xylinus RM1 was deposited in the Microbial Culture Collection Unit (UNICC, Institute of Bioscience, Universiti Putra Malaysia, under the accession number UPMC 1188).
To investigate BNC-producing capacity, one loop of a cellulose-producing isolate was transferred to 100 mL of HS medium in a 250 mL flask and incubated under stirred culture at 30°C for 24 days. A typical purification procedure of BNC-containing captured cells includes treatment with NaOH (1M) at 100°C for 15 min to remove the microbial cells and then frequently washing off the pellicle with distilled water until its pH becomes neutral.
Extraction of A. vera leaf
A. vera leaves were collected from Institute of Bioscience, Universiti Putra Malaysia. Leaf extract was obtained by taking 10 g of leaves. Leaves were washed thoroughly with distilled water, dried at room temperature and cut into fine pieces. The pieces were boiled in 100 mL distilled water for 20 min at 60°C in a glass beaker. After boiling, the extract was filtered using Whatman No 1.
Biosynthesis of BNC/Fe3O4 nanocomposites
A facile, rapid and green approach is also carried out to prepare magnetite (Fe3O4) NPs in a one-step reaction. In this method, iron oxide was prepared with BNC by immersing BNC (2 g) pellicles in different concentrations of FeCl3 and FeCl2 solution with a 2:1 M ratio under nitrogen atmosphere with continuous magnetic string at 45°C. After thorough dispersing of Fe2+ and Fe3+ in BNC, 30 mL of A. vera extract was added to a mixture suspension and the reaction process was continued until the color of the mixture changed from yellowish to a black color, followed by ultrasonication for 30 min. Afterward, the samples were washed with distilled water several times and poured into plate dishes and subsequently dried in a freeze drier. The obtained sample films based on the percentage of applied iron precursors (1.0, 4.0, 8.0 and 16.0 wt%) were called BNC/Fe3O4 (1.0%), BNC/Fe3O4 (4.0%), BNC/Fe3O4 (8.0%) and BNC/Fe3O4 (16.0%), respectively.
Characterization of BNC/Fe3O4 nanocomposites
Crystalline structure and phase purity of BNC/Fe3O4 nanocomposites were determined by X-ray diffraction (XRD) analysis recorded by diffractometer (X’pert PXRD; Philips, Almelo, the Netherlands), with Cu-Ka radiation at 40 kV in the scan range of 2θ from 2° to 80°. FTIR spectra were achieved on an FTIR spectrometer (1,725×; PerkinElmer, Waltham, MA, USA) in the 4,000–400 cm−1 range. The morphology of the nanocomposite samples was viewed under a field emission scanning electron microscopy (FESEM, JSM-6360LA; Philips).
Swelling behavior of BNC/Fe3O4 nanocomposites
Kinetic test of swelling was assessed by periodically measuring weight increase of the sample films according to the method by Lavorgna et al.21 The swelling ratio was performed by soaking of 100 mg of BNC/Fe3O4 nanocomposites in PBS 0.01 M, pH 7.4, at 37°C. At various time points, the samples were taken from the solution and additional water was removed from the surface by blotting on wet filter paper before being weighed. The swelling ratio of BNC/Fe3O4 nanocomposites was calculated according to Equation 1
|
|
where w1 is the initial weight of the sample and w2 is the weight of swollen sample. The equilibrium swelling ratio was determined at the point the BNC/Fe3O4 nanocomposites reached a constant weight value. The swelling capacities were measured in triplicate.
In vitro release assessment of BNC/ Fe3O4 nanocomposites
The Fe ions release profile from nanocomposites was determined using inductively coupled plasma atomic emission spectrometry (1,000×; PerkinElmer). Firstly, the samples were cut to a size of 1×1×0.1 cm3 and put in falcon tubes containing 10 mL of PBS (pH 7.4) at 37°C. The PBS solutions were collected at specific time points (6, 12, 24, 48, 72 and 96 h) and shaken before measurement, and then the same volume of fresh PBS solution was added. The release profile of magnetic NPs from BNC nanocomposites was studied by release kinetics Ritger-Peppas model,22 a semi-empirical power law described by Equation 2
|
|
where Ct and Ceq are the cumulative concentrations of iron released from the BNC at a specified time and at equilibrium, respectively, k is a characteristic constant of the BNC and n is the diffusional coefficient used to interpret the release mechanism.
Antibacterial activity assessment
Antibacterial activity of samples was investigated by the agar diffusion method on gram-negative (Pseudomonas aeruginosa) and gram-positive (methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis) pathogenic bacteria. The freeze-dried BNC/Fe3O4 nanocomposites in different concentrations (1.0%, 4.0%, 8.0% and 16.0%) and the freeze-dried BNC/A. vera nanocomposite (as control) were cut into disk shapes of 1.5 mm diameters. The disk-shaped samples were sterilized by autoclaving for 20 min at 121°C, put on the agar plates, which were already inoculated with 100 μL spore suspensions of bacteria, and finally incubated for 24 h at 37°C. After incubation, the zone of whole inhibition was measured. All tests were replicated three times.
Cytotoxicity assay
The in vitro cytotoxicity of BNC/Fe3O4 nanocomposites (1.0%, 4.0%, 8.0% and 16.0%) was evaluated on HDF by a method using MTT assay to determine the range of concentrations of nanocomposites to be used for scratch assay and further analysis. HDFs belong to a dermis cell type with mesenchymal origin and are found in all connective tissues. HDFs could be obtained from adult normal dermis or neonatal foreskin.23–25 These cells have an appropriate kinetic of growth for culturing easily in vitro. Briefly, HDF cells were seeded on DMEM media with 5% FBS and 1% penicillin–streptomycin at a density of 1×106 cells/mL in 96-well microplates and incubated in CO2 incubator with 5% humidity at 37°C for 24 h. Afterward, the medium was replaced with 100 μL of BNC/Fe3O4 nanocomposite solution with different concentrations (31.25, 62.5, 125, 250 and 500 μg/mL) for 72 h. Subsequently, MTT powder was dissolved in PBS at a concentration of 5 mg/mL. MTT solution was added to each well (20 μL) and plates were incubated at 37°C for 3 h. The medium was replaced with 100 μL DMSO to dissolve the formazan crystal formed by live cells. The absorbance for each well was measured at 570 nm on a microplate reader. The cell viability was calculated as the percentage of absorbent compared to control. The IC50 value, defined as the amount of sample that inhibits 50% of cell growth, was calculated from the concentration-response curves.
In vitro wound healing scratch assay
HDF cells were seeded in 24-well plates at a concentration of 3×105 cells/mL cultured in a DMEM media containing 5% FBS and grown to attach cells for 24 h at 37°C and 5% CO2. Subsequently, the media were pipetted out and discarded. A small area was then scratched using sterile 200 μL pipette tip, before adding different concentrations of nanocomposites (1.0, 4.0, 8.0 and 16.0 wt%); the cells were extensively rinsed with PBS to remove the loosened debris of the cells. After 24 h of incubation at 37°C, the distance between two layers of cells, which was scratched by pipette tip, was then inspected microscopically at 0, 24 and 48 h, respectively. For observation of wound area closure, the migration of HDF cells after filling scratched area, was captured by a digital camera attached to an inverted microscope and computer system.
Gene expression study
Bioinformatics study
The most effective genes that are involved in healing are identified (Figure 2) through information obtained from literature mining, the PubMed database. To date, 280 articles have been published since 2008, 146 of which had specific criteria just on wound healing. Around 80 genes that were differentially expressed were recognized. The most effective genes for wound healing were investigated via databases such as DisGeNET (http://www.disgenet.org), Gene Ontology Annotation (http://www.informatics.jax.org/vocab/gene_ontology), Genetics home research (https://ghr.nlm.nih.gov/), UniProtKB (http://www.uniprot.org/help/uniprotkb), Expression Atlas (https://www.ebi.ac.uk/gxa/home/) and Gene expression profiling were analyzed from GEO datasets (https://www.ncbi.nlm.nih.gov/gds). In addition, gene ontology (GO) analysis was used by AmiGO2 tool (http://amigo.geneotology.org/amigo/landing) for biological function.
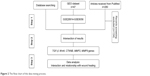  | Figure 2 The flow chart of the data mining process. |
To investigate physical and genetic interactions between genes–genes and miRNA–gene, they were mapped by Cytoscape 3.4 and miRTargetLink Human (http://ccb-web.cs.uni-saarland.de/mirtargetlink) software, respectively. Finally, the effect of miRNAs in cellular processes was used by DIANA tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php).
Gene expression analysis in wound healing of BNC/Fe3O4 nanocomposites by quantitative real-time reverse transcriptase PCR (qRT-PCR)
RNA extraction was carried out by RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) for HDF cell line, untreated (negative control) or treated with 8.0% (250 μg/mL) of BNC/Fe3O4 nanocomposites in different periods of time (6, 12 and 24 h). The procedure was performed according to the manufacturer’s instructions. In addition, small RNA (≤200 nucleotides) isolation from total RNA was performed using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. Then small RNA polyadenylation was accomplished using Poly (A) polymerase (Takara, Shiga, Japan) enzyme reaction.
The cDNA was synthesized from purified RNA and poly(A)-tailed small RNA with an RT2 First Strand Kit (Qiagen, Inc.) according to the manufacturer’s guidelines as the template for qRT-PCR. Corbett Rotor-Gene 6000 (Qiagen, Inc.) was used to perform qRT-PCR. A final volume of 25 μL premix was prepared containing 12.5 μL of RT2 SYBR®Green ROX™ FAST mastermix (Qiagen, Inc.), 1.2 μL of primers (RT2 qPCR Primer Assays, Qiagen, Inc.), 1 μL of cDNA and 10.3 μL RNase-free water to make the final volume. The primer pairs for target genes and β-actin chosen from the Primer Bank website are shown in Table 1.
  | Table 1 Genes used in qRT-PCR |
The default PCR conditions were as follows: the PCR plate was run at 95°C for 5 min to activate the enzyme, 35 and 40 cycles of 30 s at 95°C (denaturation) followed by 30 s at 60°C (annealing and synthesis) and 30 s at 72°C for RNA and miRNA, respectively. Finally, the dissociation curve was constructed immediately after the PCR run to check and verify results. In relative quantification, all samples were normalized to a constantly expressed housekeeping mRNA (reference mRNA) β-actin and 5srRNA. Only one reference gene was used in the current study because of limitation for interpretations. Relative gene expression was calculated for each mRNA marker using the 2−ΔΔCt method: 2−ΔΔCt = 2Ct (treated cells)-Ct (control cells), where 2 = the amplification efficiency and the template doubles in each cycle during exponential amplification.
Statistical analysis
Experimental results are presented as mean ± SD, and all measurements and analyses were carried out in triplicate. GraphPad Prism software version 6.01 (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analysis. At the level of *p≤0.05, **p<0.01, ***p<0.001 and ****p<0.0001, one-way analysis of variance and differences were found to be significant.
Results and discussion
Characterization of BNC/Fe3O4 nanocomposites
In this study, Fe3O4 NPs were prepared using A. vera extract as a reducing agent in BNC hydrogel, as a matrix for immobilization of the magnetic NPs without using any toxic materials. The formation of magnetic NPs was confirmed with the color change of hydrogel suspensions from yellow to dark brown after 1 h of reaction time. A. vera was used as a reducing agent in NP production due to having plentiful reducing groups such as hydroxyl, carbonyl and amide groups. However, the exact mechanism to synthesize metal NP has not been reported, but it seems by transforming of free electron from carbonyl, hydroxyl or amide groups to the free orbital of metal ions in a Red/Ox system, metal ions converted to metal oxide NPs after heat treatment. Furthermore, due to having abundant hydroxyl groups and the anionic character, BNC was also used as a matrix to immobilize NPs. The synthesis of Fe3O4 NPs is composed of two steps: first is the molecule creation (reducing of metal ions by the assistance of reducing groups of A. vera) and second the molecules combine causing the formation of metal oxide clusters. Probably the interaction with BNC prevents further coalescence by stabilizing the process.
XRD analysis
The XRD patterns for BNC/Fe3O4 nanocomposites, pure Fe3O4 NPs and pure BNC are shown in Figure 3. BNC/Fe3O4 nanocomposites show two sets of diffraction peaks corresponding to BNC and Fe3O4 NPs, in the 2θ range of 10°–80°.26,27 Two main diffraction peaks appeared at 2θ=14.7° and 22.16° that can be attributed to BNC. The characteristic peaks of Fe3O4 NPs are marked by their indices (220), (311), (400), (422), (511) and (440). These peaks prove the synthesis of cubic inverse spinal structure of magnetite Fe3O4 NPs.28 Due to the similar characteristic peaks that can be detected in BNC/Fe3O4 nanocomposites, it can be confirmed that the formation of Fe3O4 NPs in BNC did not result in any phase change of magnetite Fe3O4 NPs. In addition, the diffraction peaks of Fe3O4 became gradually intense and strong as the content of Fe3O4 increased, suggesting the increase of crystalline structure of the magnetic NPs.
The size of the mean crystal was calculated using the equation of Debye-Scherrer (D = Kλ/βcosθ), where D is the mean crystal size, K is the Scherrer constant (0.9), λ is the XRD wavelength (0.15418 nm), β is the peak width of half maximum intensity and θ is the Bragg diffraction angle. Thus, the average crystal size of the Fe3O4 NPs was estimated in the range of 15–30 nm.
Infrared spectroscopy
Figure 4 shows the FTIR spectra of pure BNC (Figure 4A), A. vera extract (Figure 4B) and BNC/Fe3O4 nanocomposites (Figure 4C–F). The absorption peak at around 570 cm−1 in BNC/Fe3O4 spectra attributes to the stretching of Fe–O confirming the presence of magnetite in nanocomposites.29 The characteristic band of pure BNC appeared at 3,327 and 2,887 cm−1 (Figure 4A), which can be assigned to O−H and C−H stretching vibrations, respectively, that were shifted to 3,354 and 2,910 cm−1 in BNC/Fe3O4 nanocomposites. Another three bands at 1,677, 1,402 and 1,040 cm−1 belonging to the O−H bending absorbed water, C−H bending vibration and C−O−H stretching vibrations, respectively,30 were shifted to higher wavenumbers after the formation of magnetic NPs in BNC. This shift of the bands can be attributed to the binding of the corresponding functional groups with the NPs. Similar results were reported in the synthesis of Ag NPs using marine alga Sargassum muticum31 and Chlorophyceae,32 where shifting of stretching vibration of the (NH) C=O group was signified to the involvement of secondary amines in the stabilization of NPs.
The FTIR spectra of A. vera extract (Figure 4B) shows two intense absorption bands at around 3,317 and 1,627 cm−1 (Figure 4A), which can be assigned to the stretching vibration of O−H and (NH) C=O groups of amino acids present in A. vera. After synthesis of magnetic NPs, these peaks became shorter in the nanocomposite samples indicating the participation of these functional groups in the formation of Fe3O4 NPs. This reduction is more obvious with the increase in the concentration of formed magnetic NPs.
FESEM analysis
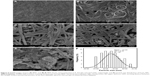
FESEM images of pure BNC and BNC containing various concentrations of magnetic NPs are shown in Figure 5A–E. The structure of BNC is three-dimensional nonwoven network and consists of large number of pores (Figure 5A). Such 3D network allowed guest molecules to penetrate throughout its inner space easily. After BNC was soaked in Fe2+ and Fe3+ solution for several hours, Fe3O4 seeds and NPs emerged to adhere to the surface and inside of BNC fibers (Figure 5B–E), indicating affinity between magnetic particles and BNC. As shown in Figure 5B–E, the spherical shape of magnetic NPs with the mean size of 33±4.26 nm and the narrow size distribution (Figure 5F) was synthesized at the different concentrations of Fe2+: Fe3+ in BNC matrices. In Figure 5B, it can be seen that the magnetic NPs dispersed on the BNC substrate homogeneously. Magnetic NPs become more apparent when Fe3O4 concentration in BNC increases. In Figure 5C and D, Fe3O4 NPs were well relatively distributed with small NP agglomerations. In Figure 5E, when the Fe3O4 concentration is 16.0 wt%, a large agglomeration of magnetic NPs can be clearly observed. Therefore, the distribution of NPs decreased by increasing the molar concentration of formed Fe3O4 NPs.
Swelling property
Swelling behavior and structural stability of composite materials play a significant role in their practical use in wound dressing applications. The swelling behavior of BNC and BNC/Fe3O4 nanocomposites is shown in Figure 6, and as shown, swelling rate was rapidly increased in the first 50 min, followed by a slower swelling rate up to 120 min. This is due to both chemical and physical structures; in the chemical structure, BNC is a hydrophilic polymer that is expected to absorb molecules of water,33 and in the physical structure, BNC is a three-dimensional non-woven network with an enormous quantity of pores preserved by freeze-drying technique. This physical structure is expected to make a capillary force within the network of BNC and suck up the molecules of water.34 Furthermore, the results illustrate that BNC/Fe3O4 nanocomposites showed a higher swelling capacity compared to that of the pure BNC. The enhancement of the swelling capacity of the magnetic nanocomposites may be due to the existence of magnetic NPs with dissimilar morphologies, sizes and surface charges. The charged Fe3O4 NPs result in the diffusion of more water molecules in order to neutralize the build up ion osmotic pressure.35,36 The degree of osmotic swelling depends on the number of charges present in the nanocomposite and is proportional to the surface charge and quantity of the NPs.37 Therefore, nanocomposites with high load of Fe3O4 NPs swell more and these results agree with previous observation in hydrogel containing green synthesized Ag37 and Fe3O4 NPs.38 On the other hand, the hydrophilic nature of compounds of plant extract present in nanocomposites with high capacity to absorb water molecules can be considered as another reason for swelling enhancement of BNC/Fe3O4 nanocomposites as compared to the pure BNC.
It was also observed that maximum swelling occurs within 24–48 h in the cases of BNC network and loaded hydrogels Fe3O4 NPs and then it reached equilibrium almost after 48 h.
In vitro Fe ions releasing properties of BNC/Fe3O4 nanocomposites
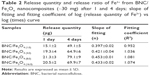
Fe ions releasing profiles of BNC/Fe3O4 nanocomposites in PBS solution are presented in Figure 7A and B. The power law exponent (n) from slopes of the logarithmical curves of Ct/Ceq as a function of time and fitting coefficient (R2) was calculated and given in Table 2. The results demonstrate that (n) is approximately a similar value of samples indicative of analogous transport mechanism. When n = ~0.45, it means that the ion release mechanism is controlled by Fickian diffusion.39,40 The power law of ion releasing rate of all the tested nanocomposite samples is nearly 0.45 with R2 ≥0.952, which shows that the Fe ions release mechanism is controlled by Fickian diffusion.
It was obvious that Fe ions was released from the BNC in the range of 50–70 μg after 96 h, and the rest of the Fe3O4 might have stayed within the BNC due to physical interactions with BNC nanofibers.
Antibacterial activity
The high level of bacteria causing infection was a major reason of prolonged wound healing. In this research, the BNC/Fe3O4 nanocomposites were used since pure BNC materials do not have antimicrobial activity and therefore cannot prevent wound infection. In addition, BNC with a porous network structure is suitable to transfer antibiotics and other medicines to the wound acting as an effective obstacle against any external infection.41
The mixture of antimicrobial agents with BNC using physical, chemical and green methods can help to produce BNC-based antimicrobial composites. Research also suggests that the action of NPs on microbes cause the antimicrobial effect.
They can attach to the bacterial cell wall and subsequently penetrate in that causing structural changes in the cell membrane like the permeability of cell membrane and eventually the death of the cell. Sondi and Salopek-Sondi state that “pits” are formed on the surface of cell where accumulation of the NPs occurs.42
The BNC/Fe3O4 nanocomposites showed inhibition zone ranging from 6±0.2 to 13.40±0.10 mm against gram-negative (P. aeruginosa) and gram-positive (S. aureus and S. epidermidis) pathogenic bacteria (Table 3). The antibacterial activity of nanocomposites improved with the increase in magnetic NP loading. The strong antibacterial activity was observed to be BNC/Fe3O4 (8.0 wt%) nanocomposite with maximum inhibition zones of 13.40±0.10 mm and 10±0.40 mm against S. aureus and S. epidermidis bacteria, respectively. Beyond this level of NP loading, the antibacterial activity of nanocomposites (16.0 wt%) decreased probably due to the formation of bulky agglomerates of magnetic NPs, which were less possible to penetrate into the cell wall to destroy the bacteria from the inside. The nanocomposites showed the lower inhibition zones against S. epidermidis compared to P. aeruginosa and S. aureus bacteria.
  | Table 3 Antibacterial activity of BNC/Fe3O4 nanocomposites against bacteria |
A reason for BNC/Fe3O4 to have the bactericidal activity is the significant ability of magnetic NPs to release iron ions. The high specific surface-to-volume ratio of iron oxide NPs increases their contact with microorganisms, likely to be promoting the dissolution of iron ions, thereby improving biocidal effectiveness.43 Another reason is due to the presence of reactive oxygen species generated by different NPs.44 The presence of Fe3O4 NPs can interact with outer bilayer of bacteria and interrupt the chemical reaction of hydrogen peroxide and membrane proteins; thus, the hydrogen peroxide produced enters the cell membrane of bacteria and kills them.45 On the other hand, the chemical surface of magnetic NPs coated with biomolecules of A. vera extract with antibacterial ability can be effective on antibacterial activity of magnetic NPs.
The antibacterial activity of BNC/A. vera as control was also assessed to evaluate the antibacterial efficiency of A. vera extract. The results showed that the nanocomposite was effective in killing of the tested bacteria with inhibition zones of 4.8±0.11 mm and 5.5±0.15 mm against P. aeruginosa and S. aureus bacteria, respectively. These results indicate the contribution of plant extract in antibacterial activity of BNC/Fe3O4.
In vitro cell viability assay
The viability of HDF cells was determined by the treatment of BNC/Fe3O4 nanocomposites with various concentrations (31.25, 62.5, 125, 250 and 500 μg/mL) through MTT assay after 72 h of incubation (Figure 8). It can be suggested that the BNC/Fe3O4 nanocomposites are less toxic and biocompatible. The cell viability increased with the increase in magnetic NP concentration. Moreover, the cell viability of HDF treated with different concentrations of BNC/Fe3O4 nanocomposites was more than 60% in all the tested samples. In other words, none of the BNC/Fe3O4 nanocomposites substantially inhibited the cell growth indicating that IC50 is higher than 500 μg/mL. Based on the MTT results, BNC/Fe3O4 nanocomposites are biocompatible and non-toxic, and therefore it is useful for in vivo applications.
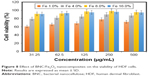  | Figure 8 Effect of BNC/Fe3O4 nanocomposites on the viability of HDF cells. |
In vitro scratch assay
Dermal wound healing is a complicated process that involves proper harmony between several skin components to allow the repair of damaged tissues and to restore the normal skin functions.46 In this study, in vitro wound healing activity of BNC/Fe3O4 nanocomposites was determined in a concentration (250 μg/mL) that had lower cytotoxicity in MTT assay. The migration of HDFs to cover the scratch created to mimic wound was captured utilizing light microscope attached to a camera at intervals of 0, 24 and 48 h after the scratch. All the images showed the progression of wound closure on scratch wounded HDF cells. Enhanced migration and wound closure were observed in BNC/Fe3O4 nanocomposites, revealing a slower pace of migration in untreated cells compared to treated ones. Figure 9 shows the rapid migration of cell and wound closure rate of BNC/Fe3O4 nanocomposite-treated HDF cells.
BNC/Fe3O4 nanocomposites (8.0 wt%) with a concentration of 250 μg/mL had a better reaction than other concentrations; therefore, this concentration was used for further analysis. The current research finding of potential wound healing capacity of bacterial cellulose-based green synthesized Fe3O4 NPs will be a positive additive for the biomedical applications.
Gene expression
Bioinformatics study
Based on what was found in the literature, the most effective genes in wound healing process were recognized to be TGF-β1, CTNNB1, MMP2, MMP9, WNT4, hsa-miR-29b-3p and hsa-miR-29c-3p. In addition, some databases reported that collagens have similar effect in wound healing compared to selected genes, because they are one of the main components in connective tissues and are involved in cellular matrix formation.
GEO2R analysis of GSE28914 and GSE8056 showed the comparison array profiling of wound-related genes in normal (intact) situation demonstrating the effect of genes in healing process.
GO analysis reveals that the aforementioned genes have relationships with different metabolic and cellular processes (Table 4). Obtained gene ontologies showed the involvement of these genes in wound healing-related process.
  | Table 4 GO analysis of the most effective genes in wound healing |
Figure 10 shows that network interaction of healing genes with each other. CTNNB1 gene has the most interactions with other genes.
  | Figure 10 Interaction network of effective in wound healing. |
Co-localization interaction of genes confirmed that there are some pivotal genes at the same side of tissues that may coordinate to each other during wound healing process. Based on obtained results from database, it was found that collagens have a significant role in wound healing and the network interaction genes with collagens were mapped. COL1A1 gene has a strong interaction with those genes playing remarkable roles in healing process of wound (Figure 11).
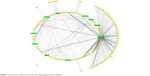  | Figure 11 Network interaction of wound healing genes with collagens. |
ECM pathway and pivotal genes may play important roles in migration of keratinocytes and fibroblasts, which cause to accelerate wound healing process (Figure 12).
  | Figure 12 Extracellular matrix (ECM) receptor interaction pathway. |
In this study, the most effective miRNAs that interacted with the wound healing process were identified by DIANA tools (Figure 13). The expression of hsa-miR-744-5P, hsa-miR-29b-3p, hsa-miR-21-5p, hsa-miR-29c-3p, hsa-miR-143-3p can affect TGF-β1, Wnt4, CTNNB1, MMP2, MMP9, COL5A1, COL1A1, COL1A2, COL5A2 and ECM pathway.
  | Figure 13 Network interaction of wound healing with microRNAs. |
The focus of this research was on some important genes that may lead to find a new effective molecular-based treatment for wound healing. Investigation of regulating factors that control the expression of these genes could be imperative to facilitate the balance of gene expression followed by acceleration of healing.
qRT-PCR assay
Based on bioinformatics research, we selected pivotal genes and miRNAs that promote wound healing for further study with qRT-PCR. The expressions of several genes known to be regulators of fibroblast cell migration during normal wound repair47 were studied. The effect of BNC/Fe3O4 nanocomposites (8.0 wt%) with a concentration of 250 μg/mL on expression genes such as TGF-β1, MMP2, MMP9, CTNNB1, Wnt4, hsa-miR-29b-3p and hsa-miR-29c-3p in HDF cell lines at different times (6, 12 and 24 h) was determined by qRT-PCR. TGF-β1 is a multifunctional cytokine known to be involved in a number of human diseases. Roberts et al found that TGF-β1 played a central role in wound healing. It influenced the inflammatory response, angiogenesis, granulation tissue formation, reepithelialization, extra-cellular matrix deposition and remodeling promoting healing and contributing to scar formation.47 During the wound healing, platelets release TGF-β; this factor stimulates the matrix metalloproteinase (MMP) production followed by activation of macrophage and neutrophils infiltration toward the wound site.48 TGF-β plays an important role in the migration and proliferation of fibroblasts. qRT-PCR analyses showed that TGF-β1 was less affected at 6 h (5.2-fold) (p<0.01) in cultures treated with BNC/Fe3O4 nanocomposites 8.0% in comparison with other hours. However, it was found to be dramatically upregulated 8.9 and 14.4-fold at 12 and 24 h, respectively, (p<0.0001) in comparison with control. Loss or absence of TGF-β1 signaling leads to non-healing wounds.49,50 In addition, El Gazaerly et al conducted research to evaluate wound healing in both normal and diabetic rats. They found that TGF-β1-treated diabetic rats showed significant healing improvement compared with diabetic rats.51
MMPs are present in both acute and chronic wounds. They play a pivotal role, with their inhibitors, in regulating ECM degradation and deposition that is essential for wound reepithelialization.52 In the normal tissue, MMPs are expressed at basal levels, if at all. When tissue remodeling is required (as in wound healing), MMPs can be rapidly expressed and activated. Therefore, MMPs are upregulated during wound healing in epidermal cells, dermal cells, fibroblasts and blood cells in mammals.53 The role of MMPs in wound healing was initially identified by the presence of active MMP2 and MMP9 in wound fluids.54 MMPs play significant role in stimulation of angiogenesis in the proximity of wounds to accelerate recovery.52 Moreover, MMP9 is expressed in several injured epithelia, including the eye, skin, gut and lung, and plays a role in wound healing and cell signaling.55–58 It also plays an important role in keratinocyte migration; it is expressed at the leading edges of migrating keratinocytes during wound closure. MMP9 knockout mice display delayed wound closure highlighting the importance of MMP9 in wound healing.59 The expression of the gelatinases MMP2 and MMP9, which are known to be implicated in regulating epithelial cell migration,59 were also investigated. Both presented an upregulated expression in all treatments was tested. Particularly, maximal upregulation for MMP9 was observed at 24 h (7.4-fold) (p<0.0001), while upregulation of MMP2 after 24 h was 5.1-folds (p<0.0001) (Figure 14). The BNC/Fe3O4 nanocomposite samples had a low MMP2 expression at the early stage (6 h), followed by upregulation (ie, at 12 and 24 h).
  | Figure 14 Gene expression analysis. |
Cell proliferation and migration are key events in reepithelialization during corneal wound healing. These processes may be mediated by the activation of MMPs because several MMPs are differentially expressed during corneal wound repair and wound closure is delayed by MMP inhibitors.58,59
The Wnt signaling pathway plays a key role in the regulation of migration, proliferation and differentiation of cells functionally relevant to skin tissue repair.60 It diversifies into three main branches: 1) the β-catenin pathway (canonical Wnt pathway), which activates target genes in the nucleus; 2) the planar cell polarity pathway, which involves Jun N-terminal kinase and cytoskeletal rearrangements; and 3) the Wnt/Ca2+ pathway, which is thought to function as a nuclear export kinase. In this research, the canonical/Wnt pathway was investigated. The Wnt4 is expressed in the dermis, although reports vary with respect to the time course of its expression.61 WNT4 and CTNNB1 through the progression of Wnt/B-catenin signaling pathway can enhance wound healing and stimulate production of MMPs.62 Recent data have shed further light on the interactions between TGF-β and CTNNB1 in cutaneous wound healing. Injuries could induce transient TGF-β signaling as observed during wound healing and this stimulation subsequently increases the level of CTNNB1.63 Based on this result, the upregulation of Wnt4 gene was observed at different hours (6, 12 and 24) with 5.6, 9.8 and 12.9-fold, respectively. The canonical Wnt signaling pathway, CTNNB1, is an important regulator of fibroblast behavior during the proliferative phase of dermal wound repair. Increasing of CTNNB1 activity during the proliferative phase is crucial for successful wound repair.5 Our results demonstrate that CTNNB1 expression was significantly increased in comparison with a control (Figure 14). The mRNA expression level was from 4.2 to 9.8-fold after treatment with BNC/Fe3O4 nanocomposites 8.0%. Chen et al have shown that CTNNB1 protein is highly expressed during the entire period of fracture repair.64 They used loss-of-function and gain-of-function approaches and found that in the early stage of healing, CTNNB1 controls the differentiation of mesenchymal cells into osteoblasts and chondrocytes. In another study, CTNNB1 protein levels and transcriptional activity were shown to be elevated in dermal fibroblasts during the proliferative phase of healing in murine cutaneous wounds and return to baseline during the remodeling phase.65
MiRNAs are endogenous, non-coding and small RNA molecules of 18–22 nucleotides in length that regulate gene expression by complementary binding to the 3′-untranslated regions of specific mRNA targets.66,67 Main function of miRNAs seems to be to downregulate gene expression by various mechanisms, including translation repression, mRNA cleavage and deadenylation. MiRNAs participate in a series of important life processes, and are important molecules in cell proliferation, development and differentiation.68
Recent studies have also shown that the miR-29 family of miRNAs is involved in heart, liver, lung and skin fibrosis.69–72 MiR-29 downregulates the expression of ECM components, collagens, fibrillin and elastin, while its downregulation in the dystrophic muscles of mdx mice not only results in impaired regeneration, but also directly contributes to muscle fibrosis.73 MiR-29 also inhibits TGF-β1 induced-transdifferentiation of myoblasts into myofibroblasts.74
Our findings show that miR-29b and miR-29c expression is reduced in HDF cells after treatment with BNC/Fe3O4 nanocomposites 8.0% at different hours (6, 12 and 24 h). The fold change of miR-29b at 6, 12 and 24 h was -6, -10.6 and -13.4-fold respectively. In addition, fold change of miR-29c was -5.5, -8.5 and -11.6 at 6, 12 and 24 h, respectively. Thus, it can be demonstrated that the compound significantly reduced the expression of miRNAs in a time-dependent manner.
Conclusion
In situ synthesis and deposition of Fe3O4 NPs into new isolated BNC fibrous membrane have been successfully developed through a completely green approach. The resulting spherical Fe3O4 NPs were uniformly dispersed in the network of BNC membranes creating a robust hybrid nanostructure. BNC/Fe3O4 nanocomposite offered excellent and sustainable controllability of Fe ion release, following an exponential power law, with exponent n dependent of Fe content. Regardless the slow Fe ion release, BNC/Fe3O4 nanocomposite still presented important antibacterial activity against S. aureus, S. epidermidis as well as P. aeruginosa. In addition, BNC/Fe3O4 nanocomposite exhibited excellent wound healing properties after 48 h. The effective genes for wound healing process were TGF-β1, MMP2, MMP9, Wnt4, CTNNB1, hsa-miR-29b and hsa-miR-29c with time-dependent manner. This study showed that BNC/Fe3O4 nanocomposite membrane is promising as an antibacterial wound dressing with desirable biocompatibility to promote wound healing.
Acknowledgments
The authors are grateful to the Department of Bioprocess Technology, the Faculty of Biotechnology and Biomolecular Sciences, and the Institute of Bioscience, Universiti Putra Malaysia for the laboratory facilities.
Disclosure
The authors report no conflicts of interest in this work.
References
Broughton G 2nd, Janis JE, Attinger CE. Wound healing: an overview. Plast Reconstr Surg. 2006;117(7 Suppl):1e-S–32e-S. | ||
Clark RAF, Singer AJ. Wound repair: basic biology to tissue engineering. In: Lanza RP, Langer R, Vacanti J, editors. Principles of Tissue Engineering. San Diego, CA: Academic Press; 2000:857–878. | ||
Xie Y, Upton Z, Richards S, Rizzi SC, Leavesley DI. Hyaluronic acid: evaluation as a potential delivery vehicle for vitronectin: growth factor complexes in wound healing applications. J Control Release. 2011;153(3):225–232. | ||
Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453(7193):314–321. | ||
Bielefeld KA, Amini-Nik S, Alman BA. Cutaneous wound healing: recruiting developmental pathways for regeneration. Cell Mol Life Sci. 2013;70(12):2059–2081. | ||
Czaja W, Krystynowicz A, Bielecki S, Brown RM Jr. Microbial cellulose – the natural power to heal wounds. Biomaterials. 2006;27(2):145–151. | ||
Shao W, Liu H, Liu X, et al. Development of silver sulfadiazine loaded bacterial cellulose/sodium alginate composite films with enhanced antibacterial property. Carbohydr Polym. 2015;132:351–358. | ||
Wu J, Zheng Y, Song W, et al. In situ synthesis of silver-nanoparticles/bacterial cellulose composites for slow-released antimicrobial wound dressing. Carbohydr Polym. 2014;102:762–771. | ||
Guo J, Wang R, Tjiu WW, Pan J, Liu T. Synthesis of Fe nanoparticles@ graphene composites for environmental applications. J Hazard Mater. 2012;225–226:63–73. | ||
Park YI, Jo TH. Perspective of industrial application of Aloe vera. In: Park YI, Lee SK, editors. New Perspectives on Aloe. New York: Springer Verlag; 2006:191–200. | ||
Afzal M, Ali M, Hassan RA, Sweedan N, Dhami MS. Identification of some prostanoids in Aloe vera extracts. Planta Med. 1991;57(1):38–40. | ||
Ramamoorthy L, Tizard IR. Induction of apoptosis in a macrophage cell line RAW 264.7 by acemannan, a beta-(1, 4)-acetylated mannan. Mol Pharmacol. 1998;53(3):415–421. | ||
Tizard I, Busbee D, Maxwell B, Kemp MC. Effects of acemannan, a complex carbohydrate, on wound-healing in young and aged rats. Wounds-A Compend Clin Res Pract. 1994;6(6):201–209. | ||
Del Vecchio F, Gallo F, Di Marco A, et al. Bioinformatics approach to predict target genes for dysregulated microRNAs in hepatocellular carcinoma: study on a chemically-induced HCC mouse model. BMC Bioinformatics. 2015;16(1):408. | ||
Guingab-Cagmat JD, Cagmat EB, Hayes RL, Anagli J. Integration of proteomics, bioinformatics, and systems biology in traumatic brain injury biomarker discovery. Front Neurol. 2013;4:61. | ||
Park JK, Park YH, Jung JY. Production of bacterial cellulose by Gluconacetobacter hansenii PJK isolated from rotten apple. Biotechnol Bioprocess Eng. 2003;8(2):83–88. | ||
Hestrin S, Schramm M. Synthesis of cellulose by Acetobacter xylinum. II. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem J. 1954;58(2):345–352. | ||
Yukphan P, Malimas T, Takahashi M, et al. Re-identification of Gluconobacter strains based on restriction analysis of 16S–23S rDNA internal transcribed spacer regions. J Gen Appl Microbiol. 2004;50(4):189–195. | ||
Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148(2):107–127. | ||
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. | ||
Lavorgna M, Piscitelli F, Mangiacapra P, Buonocore GG. Study of the combined effect of both clay and glycerol plasticizer on the properties of chitosan films. Carbohydr Polym. 2010;82(2):291–298. | ||
Ritger PL, Peppas NA. A simple equation for description of solute release I. Fickian and non-Fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs. J Control Release. 1987;5(1):23–36. | ||
Chang HY, Chi JT, Dudoit S, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99(20):12877–12882. | ||
French MM, Rose S, Canseco J, Athanasiou KA. Chondrogenic differentiation of adult dermal fibroblasts. Ann Biomed Eng. 2004;32(1):50–56. | ||
Wong T, McGrath JA, Navsaria H. The role of fibroblasts in tissue engineering and regeneration. Br J Dermatol. 2007;156(6):1149–1155. | ||
Shahwan T, Sirriah SA, Nairat M, et al. Green synthesis of iron nanoparticles and their application as a Fenton-like catalyst for the degradation of aqueous cationic and anionic dyes. Chem Eng J. 2011;172(1):258–266. | ||
Azizi S, Ahmad MB, Hussein MZ, Ibrahim NA, Namvar F. Preparation and properties of poly (vinyl alcohol)/chitosan blend bionanocomposites reinforced with cellulose nanocrystals/ZnO-Ag multifunctional nanosized filler. Int J Nanomedicine. 2014;9:1909–1917. | ||
Calmon MF, de Souza AT, Candido NM, et al. A systematic study of transfection efficiency and cytotoxicity in HeLa cells using iron oxide nanoparticles prepared with organic and inorganic bases. Colloids Surf B Biointerfaces. 2012;100:177–184. | ||
Huang L, Weng X, Chen Z, Megharaj M, Naidu R. Green synthesis of iron nanoparticles by various tea extracts: comparative study of the reactivity. Spectrochim Acta A Mol Biomol Spectrosc. 2014;130:295–301. | ||
Azizi S, Ahmad MB, Mahdavi M, Abdolmohammadi S. Preparation, characterization, and antimicrobial activities of ZnO nanoparticles/cellulose nanocrystal nanocomposites. Bioresources. 2013;8(2):1841–1851. | ||
Azizi S, Namvar F, Mahdavi M, Ahmad MB, Mohamad R. Biosynthesis of silver nanoparticles using brown marine macroalga, Sargassum muticum aqueous extract. Materials (Basel). 2013;6(12):5942–5950. | ||
Kannan RRR, Stirk WA, Van Staden J. Synthesis of silver nanoparticles using the seaweed Codium capitatum PC Silva (Chlorophyceae). South African J Bot. 2013;86:1–4. | ||
Klemm D, Schumann D, Udhardt U, Marsch S. Bacterial synthesized cellulose – artificial blood vessels for microsurgery. Prog Polym Sci. 2001;26(9):1561–1603. | ||
Iguchi M, Yamanaka S, Budhiono A. Bacterial cellulose – a masterpiece of nature’s arts. J Mater Sci. 2000;35(2):261–270. | ||
Gils PS, Ray D, Sahoo PK. Designing of silver nanoparticles in gum arabic based semi-IPN hydrogel. Int J Biol Macromol. 2010;46(2):237–244. | ||
Vimala K, Sivudu KS, Mohan YM, Sreedhar B, Raju KM. Controlled silver nanoparticles synthesis in semi-hydrogel networks of poly (acrylamide) and carbohydrates: a rational methodology for antibacterial application. Carbohydr Polym. 2009;75(3):463–471. | ||
Azizi S, Mohamad R, Abdul Rahim R, Mohammadinejad R, Bin Ariff A. Hydrogel beads bio-nanocomposite based on Kappa-Carrageenan and green synthesized silver nanoparticles for biomedical applications. Int J Biol Macromol. 2017;104(Pt A):423–431. | ||
Daniel-da-Silva AL, Moreira J, Neto R, Estrada AC, Gil AM, Trindade T. Impact of magnetic nanofillers in the swelling and release properties of κ-carrageenan hydrogel nanocomposites. Carbohydr Polym. 2012;87(1):328–335. | ||
Lima-Tenório MK, Tenorio-Neto ET, Garcia FP, et al. Hydrogel nanocomposite based on starch and Co-doped zinc ferrite nanoparticles that shows magnetic field-responsive drug release changes. J Mol Liq. 2015;210:100–105. | ||
Korsmeyer RW, Gurny R, Doelker E, Buri P, Peppas NA. Mechanisms of solute release from porous hydrophilic polymers. Int J Pharm. 1983;15(1):25–35. | ||
Andresen M, Stenstad P, Møretrø T, et al. Nonleaching antimicrobial films prepared from surface-modified microfibrillated cellulose. Biomacromolecules. 2007;8(7):2149–2155. | ||
Sondi I, Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J Colloid Interface Sci. 2004;275(1):177–182. | ||
Nithya R, Ragunathan R. Synthesis of silver nanoparticle using Pleurotus sajor caju and its antimicrobial study. Dig J Nanomater Biostruct. 2009;4(4):623–629. | ||
Yamamoto O. Influence of particle size on the antibacterial activity of zinc oxide. Int J Inorg Mater. 2001;3(7):643–646. | ||
Padmavathy N, Vijayaraghavan R. Enhanced bioactivity of ZnO nanoparticles-an antimicrobial study. Sci Technol Adv Mater. 2008;9(3):035004. eCollection 2008. | ||
Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci. 2004;9(1):283–289. | ||
Roberts AB, Sporn MB, Assoian RK, et al. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci U S A. 1986;83(12):4167–4171. | ||
Penn JW, Grobbelaar AO, Rolfe KJ. The role of the TGF-β family in wound healing, burns and scarring: a review. Int J Burns Trauma. 2012;2(1):18–28. | ||
Pastar I, Stojadinovic O, Krzyzanowska A, et al. Attenuation of the transforming growth factor beta–signaling pathway in chronic venous ulcers. Mol Med. 2010;16(3–4):92–101. | ||
Kim BC, Kim HT, Park SH, et al. Fibroblasts from chronic wounds show altered TGF-beta-signaling and decreased TGF-beta Type II receptor expression. J Cell Physiol. 2003;195(3):331–336. | ||
El Gazaerly H, Elbardisey DM, Eltokhy HM, Teaama D. Effect of transforming growth factor Beta 1 on wound healing in induced diabetic rats. Int J Health Sci (Qassim). 2013;7(2):160–172. | ||
Caley MP, Martins VL, O’Toole EA. Metalloproteinases and wound healing. Adv Wound Care (New Rochelle). 2015;4(4):225–234. | ||
Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. Int J Biochem Cell Biol. 2008;40(6–7):1334–1347. | ||
Salo T, Mäkelä M, Kylmäniemi M, Autio-Harmainen H, Larjava H. Expression of matrix metalloproteinase-2 and -9 during early human wound healing. Lab Invest. 1994;70(2):176–182. | ||
Fini ME, Parks WC, Rinehart WB, et al. Role of matrix metalloproteinases in failure to re-epithelialize after corneal injury. Am J Pathol. 1996;149(4):1287–1302. | ||
Betsuyaku T, Fukuda Y, Parks WC, Shipley JM, Senior RM. Gelatinase B is required for alveolar bronchiolization after intratracheal bleomycin. Am J Pathol. 2000;157(2):525–535. | ||
Mohan R, Chintala SK, Jung JC, et al. Matrix metalloproteinase gelatinase B (MMP-9) coordinates and effects epithelial regeneration. J Biol Chem. 2002;277(3):2065–2072. | ||
Castaneda FE, Walia B, Vijay–Kumar M, et al. Targeted deletion of metalloproteinase 9 attenuates experimental colitis in mice: central role of epithelial-derived MMP. Gastroenterology. 2005;129(6):1991–2008. | ||
Hattori N, Mochizuki S, Kishi K, et al. MMP-13 plays a role in keratinocyte migration, angiogenesis, and contraction in mouse skin wound healing. Am J Pathol. 2009;175(2):533–546. | ||
Shi Y, Shu B, Yang R, et al. Wnt and Notch signaling pathway involved in wound healing by targeting c-Myc and Hes1 separately. Stem Cell Res Ther. 2015;6(1):120. | ||
Houschyar KS, Momeni A, Pyles MN, Maan ZN, Whittam AJ, Siemers F. Wnt signaling induces epithelial differentiation during cutaneous wound healing. Organogenesis. 2015;11(3):95–104. | ||
Whyte JL, Smith AA, Helms JA. Wnt signaling and injury repair. Cold Spring Harb Perspect Biol. 2012;4(8):a008078. | ||
Cheon SS, Wei Q, Gurung A, et al. Beta-catenin regulates wound size and mediates the effect of TGF-beta in cutaneous healing. FASEB J. 2006;20(6):692–701. | ||
Chen Y, Whetstone HC, Lin AC, et al. Beta-catenin signaling plays a disparate role in different phases of fracture repair: implications for therapy to improve bone healing. PLoS Med. 2007;4(7):e249. | ||
Cheon SS, Cheah AY, Turley S, et al. Beta-Catenin stabilization dysregulates mesenchymal cell proliferation, motility, and invasiveness and causes aggressive fibromatosis and hyperplastic cutaneous wounds. Proc Natl Acad Sci U S A. 2002;99(10):6973–6978. | ||
Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229(5):545–550. | ||
Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–469. | ||
Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–154. | ||
Cushing L, Kuang PP, Qian J, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45(2):287–294. | ||
He Y, Huang C, Lin X, Li J. MicroRNA-29 family, a crucial therapeutic target for fibrosis diseases. Biochimie. 2013;95(7):1355–1359. | ||
Roderburg C, Urban GW, Bettermann K, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53(1):209–218. | ||
van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105(35):13027–13032. | ||
Wang L, Zhou L, Jiang P, et al. Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol Ther. 2012;20(6):1222–1233. | ||
Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7(3):e33766. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.