Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
miR-7, miR-10a and miR-143 Expression May Predict Response to Bevacizumab Plus Chemotherapy in Patients with Metastatic Colorectal Cancer
Authors Romero-Lorca A, Novillo A, Gaibar M , Gilsanz MF , Galán M , Beltrán L, Antón B , Malón D, Moreno A, Fernández-Santander A
Received 2 April 2021
Accepted for publication 20 May 2021
Published 29 September 2021 Volume 2021:14 Pages 1263—1273
DOI https://doi.org/10.2147/PGPM.S313594
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Alicia Romero-Lorca,1 Apolonia Novillo,1 María Gaibar,1 María Fuencisla Gilsanz,1 Miguel Galán,1 Laura Beltrán,1 Beatriz Antón,2 Diego Malón,2 Amalia Moreno,2 Ana Fernández-Santander1
1Biomedical and Health Sciences Faculty, Universidad Europea de Madrid, Madrid, 28670, Spain; 2Department of Oncology, University Hospital of Fuenlabrada, Madrid, 28942, Spain
Correspondence: Ana Fernández-Santander
Biomedical and Health Sciences Faculty, Universidad Europea de Madrid, c/ Tajo s/n, Villaviciosa de Odón, Madrid, 28670, Spain
Tel +34 912 115 288
Fax +34 916 168 265
Email [email protected]
Purpose: Bevacizumab is a monoclonal antibody that binds to vascular endothelial growth factor A. It is currently used in combination with chemotherapy to treat metastatic colorectal cancer. This therapy is not equally effective in every patient; in some, mechanisms of resistance arise that remain poorly understood. The aim of the present work was to determine whether the expression of 26 miRNAs could be associated with the effectiveness of bevacizumab plus chemotherapy, with progression-free survival (PFS), and with overall survival (OS) in metastatic colorectal cancer.
Patients and Methods: Paraffin-embedded biopsies from 76 patients with metastatic colorectal cancer were collected to isolate miRNAs. The expression of 26 miRNAs was analyzed by quantitative RT-PCR. For the purpose of analysis, patients were classified as either “responders” (PFS ≥ 6 months since beginning treatment) or “non-responders” (PFS < 6 months). For the analysis of PFS and OS, patients were classified into two groups using the median gene expression value as the cut-off point (“high” [≥ 50% percentile] or “low” [< 50% percentile]). Time-to-event data were analyzed using the Kaplan–Meier method and compared by the log rank test. Cox regression was used to estimate hazard ratios (HR) and their 95% confidence intervals.
Results: miR-7-5p and miR-10a-5p were more strongly expressed in non-responders than responders (p=0.049 and p=0.043, respectively), and OS was poorer in patients showing these higher expression levels (HR=2.54, 95% CI 1.42– 4.55, p=0. 001, and HR=1.81, 95% CI 1.02– 3.20, p=0.039, respectively). The overexpression of miR-143-3p, however, was associated with a better prognosis and significantly better PFS (HR=0.57; 95% CI: 0.33– 0.96; p=0.033).
Conclusion: High expression values for miR-7-5p and miR-10a-5p might be considered markers of a poorer prognosis in patients with metastatic colorectal cancer treated with bevacizumab plus chemotherapy, while the same for miR-143-3p might be a marker of better outcomes.
Keywords: miRNAs, colorectal cancer, bevacizumab, paraffin-embedded biopsies, progression free survival, overall survival
Introduction
MicroRNAs (miRNAs) are a class of non-coding, short, single-stranded, and strongly conserved RNAs with an important role in regulating gene expression at the post-transcriptional level.1 They are involved in cell differentiation, proliferation, invasion, apoptosis, and in tumorigenesis, both as oncogenic elements and oncosuppressors.2 Specific miRNAs have been reported to regulate the angiogenic process via the exertion of pro- or anti-angiogenic effects.3
The inhibition of angiogenesis during tumor growth is standard treatment for metastatic colorectal cancer (mCRC). New anti-angiogenic drugs emerge every year. One such agent, bevacizumab (BVZ), is a monoclonal antibody that binds to vascular endothelial growth factor A (VEGFA). It is currently used in combination with chemotherapy (CT) in patients with mCRC. However, treatment is not equally effective in everyone; in some people, mechanisms of resistance arise that are not well understood.4 There is increasing evidence that miRNAs are important regulators of metabolic reprogramming, proliferation and chemoresistance in colorectal cancer (CRC), and that they therefore influence prognosis and survival.5 A better understanding of the use of miRNAs as biomarkers might aid in the selection of patients with mCRC who would benefit from BVZ+CT.
miR-7-5p, an oncosuppressive miRNA in melanoma and breast cancer, has been reported a possible biomarker of prognosis in patients with these conditions.6,7 However, whether it is dysregulated in mCRC remains a matter of debate, and its potential as a biomarker is uncertain. Nagano et al reported miR-7-5p expression to be significantly stronger in CRC tissue than in normal mucosae, and to be associated with a poor prognosis (highlighting its role as an oncogenic rather than a tumor-suppressing element).8 Certainly, some in vitro assays involving human colon cancer cell lines have suggested the involvement of miR-7-5p in the generation of drug-resistant phenotypes. For instance, greater cetuximab sensitivity has been observed in cells showing stronger miR-7-5p expression.9 Similarly, miR-10a-5p has been reported to play a number of important roles in different tumors, although again the results obtained have been controversial. In vitro experiments have suggested miR-10a-5p overexpression to promote cell migration and invasion in breast and cervical cancer.10,11 However, other studies suggest it to repress these processes.12 miR-10a-5p is also reported to increase gemcitabine resistance in vitro and in vivo.13 It has therefore been suggested that, given their relationship with prognosis, biopsy histopathological characteristics, age and other clinical data, some miRNAs, eg, miR-143, might provide markers that could boost the implementation of personalized medicine.14
The aim of the present work was to determine whether the expression of 26 miRNAs, analyzed by quantitative RT-PCR in paraffin-embedded biopsies from 76 patients with mCRC, is associated with the effectiveness of BVZ+CT in such patients, as well as with progression-free survival (PFS) and overall survival (OS).
Materials and Methods
Patients and Outcomes
The present patients (n=76; men=53, women=23), all of whom had mCRC, were treated at the Oncology Department of the University Fuenlabrada Hospital between 2010 and 2018. Their KRAS status was determined as part of routine diagnosis. All received BVZ+CT (mainly FOLFOX4 or FOLFIRI) and were assessed for response to treatment, PFS and OS according to RECIST (Response Evaluation Criteria In Solid Tumors) criteria. At the start of the BVZ+CT treatment, all patients had reached tumor stage IV. PFS was determined as the time from the beginning of treatment to the date of the first documented evidence of disease progression (physician assessed), last tumor assessment, or death in the absence of disease progression. OS was calculated as the time from the beginning of treatment to the date of death from any cause, or last follow-up. Tumor response after 6 months of treatment was determined by computed tomography. For the purpose of analysis, patients were classified as either “responders” (R) (ie, PFS ≥6 months since beginning treatment) or “non-responders” (NR) (PFS <6 months). The R group included patients who benefited from treatment, those who showed a partial response, and those who showed stable disease after 6 months of treatment. The NR group included patients who experienced disease progression over the same period. The study protocol was approved by the Fuenlabrada Hospital Ethics Committee in March 2015 (reference number APR 15/38). All experiments were conducted in accordance with the Declaration of Helsinki. Informed consent was given by all patients, allowing the experimental use of biopsies/tumors collected for diagnosis.
miRNA Selection
The miRNAs for analysis were selected based on their role in tumor growth (proliferation, loss of apoptosis, chemoresistance) and the angiogenic process as reported in the literature. Twenty six miRNAs were finally selected: hsa-miR-7-5p, hsa-miR-10a-5p, hsa-miR-20b-5p, hsa-miR-27b-3p, hsa-miR-29b-3p, hsa-miR-34a-5p, hsa-miR-92b-3p, hsa-miR-101-3p, hsa-miR-124-3p, hsa-miR-125a-5p, hsa-miR-126-3p, hsa-miR-143-3p, hsa-miR-145-5p, hsa-miR-155-5p, hsa-miR-192-5p, hsa-miR-196b-5p, hsa-miR-199a-5p, hsa-miR-200b-3p, hsa-miR-204-5p, hsa-miR-222-3p, hsa-miR-363-3p, hsa-miR-376a-3p, hsa-miR-455-5p, hsa-miR-497-5p, hsa-miR-663a and hsa-miR-664-3p (Table 1). UniSp6 was used to monitor cDNA synthesis.
 |
Table 1 Sequences and Code Numbers of the 26 miRNAs Analyzed (http://Www.mirbase.org) |
Gene Expression Analysis
miRNAs were extracted from 2.5 mm3 paraffin-embedded tumor biopsies or whole tumors as required using the miRNeasy FFPE Kit (Qiagen, Germantown, MD, USA). Reverse transcriptase reactions were performed using the EXIQON RT microRNA PCR miRCURY LNA Kit (Qiagen, Germantown, MD, USA). The 26 selected miRNAs were spotted on custom array plates. The U6 housekeeping gene was used as an internal control. Assays were run in a C1000 Touch Thermal Cycler (Bio-Rad, Madrid, Spain). The reaction conditions were 95°C for 5 min, followed by 40 cycles of 95°C for 5 s, and 60°C for 15 s. All reactions were performed in duplicate. Expression levels were determined using CFX Maestro™ Software (Bio-Rad, Madrid, Spain) according to the ∆∆Ct method.
Statistical Analysis
The Shapiro–Wilk test was used to examine the normality of distribution of miRNA gene expression among the R and NR patients. Accordingly, the Mann–Whitney or Student t test was used to compare the expression of each miRNA between these responder status groups. For the analysis of PFS and OS, patients were classified into two groups using the median gene expression value as the cut-off point (“high” [≥50% percentile] or “low” [<50% percentile]). Time-to-event data were analyzed using the Kaplan–Meier method and compared by the log rank test; significance was set at p<0.05. Cox regression was used to estimate hazard ratios (HR) and their 95% confidence intervals. miRNA expressions significantly associated with PFS, OS and HR values were adjusted by multivariate Cox regression including gender, age at diagnosis, KRAS status, and type of chemotherapy regimen as covariables. All calculations were performed using Program Stata v.14.2 software (Stata Corp, LLC TX, USA).
Results
Patient Clinical Characteristics
Table 2 shows the clinical characteristics of the enrolled patients (n=76; men=53, women= 23). The median age of the patients was 62 years (range 27–84 years). Most tumors were left-sided (73.3% compared to 26.7% for right-sided). Some 75.7% of patients possessed the KRAS mutation. Most patients had stage IV (metastatic) disease at diagnosis; 100% were at this stage by the beginning of treatment. The number of responders (R) and non-responders (NR) to BVZ+CT was similar.
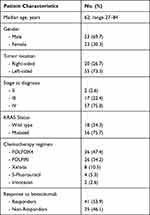 |
Table 2 Patient Characteristics |
No correlation was detected between response to therapy and the clinical characteristics of the patients. No miRNA expression was significantly associated with KRAS status or tumor localization. Disease progressed in 95.8% of patients. The median PFS and OS values were 8.7 months (95% CI 7.1–12.2 months) and 23.5 months (95% CI 20–29.3 months) respectively (Figure 1).
 |
Figure 1 Progression-free survival (A) and overall survival (B). |
miRNA Gene Expression in Relation to Treatment Effectiveness
Figure 2 shows the mean expression values for the 26 miRNAs analyzed, normalized against the U6 gene. miR-7-5p-5p and miR-10a-5p showed significantly higher expression values in the NR patients than in the R patients (p=0.049 and p=0.043, respectively; Table 3). The higher expression values recorded for miR-20b-5p, miR-29b-3p and miR-204-5p in the NR patients compared to the R patients did not reach significance (0.05<p≤0.01, Table 3). Indeed, no significant differences were detected between these response groups for any of the other miRNAs.
 |
Table 3 Correlation Between miRNAs Expression with Respect to Responder Status (R= Responders, NR= Non-Responders). Significant Results are Shown in Bold Type |
 |
Figure 2 Average relative gene expression (2∆∆Ct) of the 26 miRNAs with respect to responder status: responders (light grey, n=41) and non-responders (dark grey, n=35). |
miRNA Gene Expression in Relation to Progression-Free Survival and Overall Survival
The PFS values with respect to gene expression were very similar, except for miR-143-3p; a significantly higher PFS was recorded for those with high gene expression values (HR=0.57, 95% CI 0.33–0.96, p=0.033, Table 4, Figure 3; adjusted HR=0.48, 95% CI 0.26–0.89, p=0.019). In addition, OS was significantly worse among those showing high miR-7-5p expression values (HR=2.54, 95% CI 1.42–4.55, p=0.001, Table 5, Figure 4; adjusted HR=2.95, 95% CI 1.58–5.52, p=0.001). Similar results were obtained for miR-10a-5p, with poorer OS seen for the group of patients with high median gene expression values (HR=1.81, 95% CI 1.02–3.20, p=0.039; Table 5, Figure 5; adjusted HR=1.92, 95% CI 1.05–3.52, p=0.035).
 |
Table 4 Progression-Free Survival with Respect to miRNA Expression. Significant Results are Shown in Bold Type |
 |
Table 5 Overall Survival in Relation to miRNA Expression. Significant Results are Shown in Bold Type |
Discussion
Colorectal cancer is one of the most common cancers; most patients develop metastatic disease. Drug development efforts in recent decades have been directed towards preventing angiogenesis given its importance in mCRC biology, the therapeutic target being VEGFA. BVZ, which came out of these research efforts, was initially approved for the treatment of mCRC in the United States and the European Union (in 2004 and 2005, respectively), but it is now approved for treating a range of solid tumors.15 Several randomized studies on BVZ in patients with mCRC suggest that it does provide benefit when administered in conjunction with some newer CT regimens.16 However, this was not the case for all patients; resistance developed in some, the mechanism for which remains poorly understood. Biomarkers that would allow the selection of patients likely to respond in terms of treatment effectiveness and survival prior to beginning treatment would be very advantageous.
The alterations in cell metabolism produced by miRNAs might contribute towards tumor cell proliferation and metastasis and, therefore influence resistance to treatment and consequently PFS and OS. miR-7-5p is recognized as a tumor suppressor in certain types of human cancer, eg, melanoma and breast cancer, in which it is frequently found downregulated. When present, it is a potent inhibitor of the growth and progression of melanoma via the inactivation of the transcription factor p65/nuclear factor-κB signaling pathway.17 The literature contains a few studies on miR-7-5p expression and the effectiveness of drugs, but these were only performed in vitro. Suto et al reported how treatment with miR-7-5p in different human colon cancer cell lines alters cetuximab sensitivity,9 while Ge et al showed that miR-7-5p may be differentially expressed in gefitinib-resistant human lung cancer cell lines, suggesting its potential involvement in the generation of the drug-resistant phenotype.18 In the present work, which was performed with tissues from patients, the results indicate miR-7-5p expression to be significantly greater among the NR patients, suggesting a potential clinical effect of its overexpression related to BVZ+CT resistance (p=0.049, Table 3).
High expression values for miR-7-5p in the present patients were related to significantly poorer OS (p=0.001, Figure 4). This agrees with miR-7-5p acting as a tumor progression-associated element in CRC, and thus being associated with a poor prognosis, as previously reported in studies of paraffin-embedded samples from patients with this disease.8 Similarly, Yang et al showed miR-7-5p expression to be significantly increased in samples from patients who experienced early relapse compared to those who experienced non-early relapse. In agreement with the present results, patients in the higher miR-7-5p expression level subgroup showed significantly poorer OS than those in the lower miR-7-5p expression level subgroup.19 A study involving serum miR-7-5p quantification also showed significantly increased levels in patients with epithelial ovarian cancer compared to healthy women.20 In contrast, some authors report miR-7-5p to have a role as an inhibitor of the angiogenic process in CRC via its role in ERK signaling.21 For instance, Shi et al reported miR-7-5p to be a tumor suppressor element in breast cancer cells.7 Given these contrasting observations, the impact of overexpression of miR-7-5p on the progression of different tumor tissues needs to be further assessed if we are to clarify its role in the prognosis of different cancer types and whether it has a future as a therapeutic target in CRC.22
Hassel et al identified a number of miRNAs, including miR-10-a, as regulators of angiogenesis.23 The overexpression of miR-10-a in zebrafish embryos and cultured endothelial cells increased blood vessel outgrowth, suggesting this miRNA to be a potential target for the selective modulation of this process.24 In the present work, miR-10-a overexpression was seen in the NR patients (p=0.043, Table 3) and was related to resistance to antiangiogenic therapy. In vitro studies have also shown an association between miR-10 and resistance to cisplatin in lung cancer,25 and to gemcitabine in human pancreatic ductal adenocarcinoma cell lines.13 The overexpression of miR-10 in hepatocellular carcinoma has also been implicated in resistance to sorafenib therapy, although the authors of that study warned of the need for more investigation.26 The present results disagree, however, with reports of miR-10a-5p upregulation in tumor tissues of patients with mCRC who responded to treatment with BVZ + FOLFOX.27 These results indicate the need for larger studies if miR-10a-5p is to be confirmed as a biomarker able to predict the response of BVZ+CT in patients with mCRC.
Chemoresistance is a major factor underlying poor prognosis. The present patients with significantly poorer OS showed the overexpression of miR-7-5p (p=0.001, Figure 4) and miR-10a-5p (p=0.039, Figure 5). Similar results were obtained in patients with low-grade gliomas, they showed upregulated miR-10a-5p expression to be associated with a higher mortality rate and shorter OS.28 The latter in silico study, which used a bioinformatic methodology, identified HSP90AA1 as a target gene of miR-10a-5p and identified the dysregulation of the PI3K-AKT pathway in low-grade gliomas when miR-10a-5p is upregulated.28 Other reports too have shown miR-10a-5p to be aberrantly increased in metastatic tumors, eg, in pancreatic cancer29 and non-small cell lung cancer,30 indicating its possible function as a proto-oncogenic element in some types of tumor. Zhi et al indicated miR-10a-5p to be a biomarker of poor prognosis in acute myeloid leukemia via its association with poor OS; significantly increased miR-10a-5p expression was detected in patients who relapsed compared to those who enjoyed complete remission.31
In contrast to the poor prognosis associated with high miR-7-5p and miR-10a-5p expression levels, the present results indicate high miR-143-3p expression levels to be associated with better outcomes in patients with mCRC (Figure 3), in whom it might function as a tumor suppressor. It has been shown that miR-143-3p impairs tumor growth and angiogenesis by inhibiting the PI3K/AKT/HIF-1/VEGF pathway.32 Other authors report the overexpression of miR-143-3p to significantly suppress cell invasion compared with miR-controls in human colorectal cancer cell lines HCT116 and SW480.33 They also predicted ERBB3 to be a target of miR-143-3p, and concluded that miR-143-3p overexpression significantly reduces ERBB3 protein expression. Thus, ERBB3 may be negatively regulated by miR-143-3p in colorectal cancer cells. These results agree with the significant association between better PFS in the group of patients with high miR-143-3p expression levels (p=0.033, Figure 3) seen in the present work, and as reported by yet other authors.14 The study suffers from the limitation of a small sample size. However, the earlier studies reported did not involve much larger samples. Future work will require samples that are not only larger but also homogeneous in terms of the chemotherapeutic agents involved. Such retrospective observational studies could form the basis of larger clinical trials designed to assess the clinical use of the studied miRNAs as predictors of BVZ efficacy.
Conclusion
The present findings suggest that high expression levels of miR-7-5p and miR-10a-5p might be considered biomarkers that predict a poor prognosis and resistance to BVZ+CT in patients with mCRC, whereas high miR-143-3p expression levels might indicate a better prognosis and longer PFS. These findings could form the basis of larger studies, including clinical trials, designed to examine the use of these miRNAs as predictors of prognosis and survival in patients with mCRC who receive treatment with BVZ+CT.
Acknowledgments
Thanks are expressed to Cristina Andreu for helpful discussion and statistical support. This research was funded by the Universidad Europea de Madrid (grant number 2016/UEM13) and by the Foundation of the Universidad Europea (grant numbers FGUE001804 and FGUE001805).
Disclosure
The authors have no conflicts of interest for this work to declare.
References
1. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524.
2. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi:10.1038/nature10888
3. Peng W, Liu YN, Zhu SQ, et al. The correlation of circulating pro-angiogenic miRNAs’ expressions with disease risk, clinicopathological features, and survival profiles in gastric cancer. Cancer Med. 2018;7:3773–3791. doi:10.1002/cam4.1618
4. Azam F, Mehta S, Harris AL. Mechanisms of resistance to antiangiogenesis therapy. Eur J Cancer. 2010;46:1323–1332. doi:10.1016/j.ejca.2010.02.020
5. Hon KW, Abidin SAZ, Othman I, et al. Insights into the role of microRNAs in colorectal cancer (CRC) metabolism. Cancers. 2020;12:1–17.
6. Giles KM, Brown RAM, Epis MR, et al. miRNA-7-5p inhibits melanoma cell migration and invasion. Biochem Biophys Res Commun. 2013;430:706–710. doi:10.1016/j.bbrc.2012.11.086
7. Shi Y, Luo X, Li P, et al. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett. 2015;358:27–36. doi:10.1016/j.canlet.2014.12.014
8. Nagano Y, Toiyama Y, Okugawa Y, et al. MicroRNA-7 is associated with malignant potential and poor prognosis in human colorectal cancer. Anticancer Res. 2016;36:6521–6526. doi:10.21873/anticanres.11253
9. Suto T, Yokobori T, Yajima R, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36:338–345. doi:10.1093/carcin/bgu242
10. Chang C-H, Fan T-C, Yu J-C, et al. The prognostic significance of RUNX2 and miR-10a/10b and their inter-relationship in breast cancer. J Transl Med. 2014;12:257. doi:10.1186/s12967-014-0257-3
11. Long M-J, Wu F-X, Li P, et al. MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett. 2012;324(2):186–196. doi:10.1016/j.canlet.2012.05.022
12. Jia H, Zhang Z, Zou D, et al. MicroRNA-10a is down-regulated by DNA methylation and functions as a tumor suppressor in gastric cancer cells. PLoS One. 2014;9:e88057. doi:10.1371/journal.pone.0088057
13. Xiong G, Huang H, Feng M, et al. MiR-10a-5p targets TFAP2C to promote gemcitabine resistance in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2018;37:1–15. doi:10.1186/s13046-018-0739-x
14. Moreno EC, Pascual A, Prieto-Cuadra D, et al. Novel molecular characterization of colorectal primary tumors based on miRNAs. Cancers. 2019:11. doi:10.3390/cancers11030346
15. Garcia J, Hurwitz HI, Sandler AB, et al. Bevacizumab (Avastin®) in cancer treatment: a review of 15 years of clinical experience and future outlook. Cancer Treat Rev. 2020;86:102017. doi:10.1016/j.ctrv.2020.102017
16. Bennouna J, Sastre J, Arnold D, et al. Continuation of bevacizumab after first progression in metastatic colorectal cancer (ML18147): a randomised Phase 3 trial. Lancet Oncol. 2013;14:29–37. doi:10.1016/S1470-2045(12)70477-1
17. Giles KM, Brown RAM, Ganda C, et al. microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-κB. Oncotarget. 2016;7:31663–31680. doi:10.18632/oncotarget.9421
18. Ge X, Zheng L, Huang M, et al. MicroRNA expression profiles associated with acquired gefitinib-resistance in human lung adenocarcinoma cells. Mol Med Rep. 2015;11:333–340. doi:10.3892/mmr.2014.2757
19. Yang I-P, Tsai H-L, Miao Z-F, et al. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J Transl Med. 2016;14:108. doi:10.1186/s12967-016-0856-2
20. Meng X, Joosse SA, Müller V, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113:1358–1366. doi:10.1038/bjc.2015.340
21. Fan X, Liu M, Tang H, et al. MicroRNA-7 exerts antiangiogenic effect on colorectal cancer via ERK Signaling. J Surg Res. 2019;240:48–59. doi:10.1016/j.jss.2019.02.035
22. Chen W-Q. Role of microRNA-7 in digestive system malignancy. World J Gastrointest Oncol. 2016;8:121. doi:10.4251/wjgo.v8.i1.121
23. Hassel D, Cheng P, White MP, et al. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ Res. 2012;111:1421–1433. doi:10.1161/CIRCRESAHA.112.279711
24. Wang X, Ling CC, Li L, et al. MicroRNA-10a/10b represses a novel target gene Mib1 to regulate angiogenesis. Cardiovasc Res. 2016;110:140–150. doi:10.1093/cvr/cvw023
25. Sun W, Ma Y, Chen P, et al. MicroRNA-10a silencing reverses cisplatin resistance in the A549/cisplatin human lung cancer cell line via the transforming growth factor-β/Smad2/STAT3/STAT5 pathway. Mol Med Rep. 2015;11(5):3854–3859. doi:10.3892/mmr.2015.3181
26. Pratama MY, Pascut D, Massi MN, et al. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med. 2019;7(20):577. doi:10.21037/atm.2019.09.142
27. Kiss I, Mlčochová J, Součková K, et al. MicroRNAs as outcome predictors in patients with metastatic colorectal cancer treated with bevacizumab in combination with FOLFOX. Oncol Lett. 2017;14:743–750. doi:10.3892/ol.2017.6255
28. Son JC, Jeong HO, Park D, et al. MiR-10a and miR-204 as a potential prognostic indicator in low-grade gliomas. Cancer Inform. 2017:16. doi:10.1177/1176935117702878
29. Weiss FU, Marques IJ, Woltering JM, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2137. doi:10.1053/j.gastro.2009.08.065
30. Yu T, Liu L, Li J, et al. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6:30239–30250. doi:10.18632/oncotarget.4972
31. Zhi Y, Xie X, Wang R, et al. Serum level of miR-10-5p as a prognostic biomarker for acute myeloid leukemia. Int J Hematol. 2015;102:296–303. doi:10.1007/s12185-015-1829-6
32. Qian X, Yu J, Yin Y, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12:1385–1394. doi:10.4161/cc.24477
33. Bai JW, Xue HZ, Zhang C. Down-regulation of microRNA-143 is associated with colorectal cancer progression. Eur Rev Med Pharmacol Sci. 2016;20:4682–4687.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



