Back to Journals » OncoTargets and Therapy » Volume 10
MicroRNA-194 modulates epithelial–mesenchymal transition in human colorectal cancer metastasis
Authors Cai H, Chen X, Tang Y, Deng Y
Received 19 October 2016
Accepted for publication 18 December 2016
Published 28 February 2017 Volume 2017:10 Pages 1269—1278
DOI https://doi.org/10.2147/OTT.S125172
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Hong-Ke Cai,1 Xi Chen,2 Yun-Hao Tang,1 Yong-Chuan Deng3
1Department of Gastrointestinal Surgery, Second Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Center of Molecular Medicine and Cancer Research, Chongqing Medical University, Chongqing, People’s Republic of China; 3Department of Surgical Oncology, Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, People’s Republic of China
Abstract: MicroRNAs (miRNAs), as key regulators of gene expression, are closely related to tumor occurrence and progression. MiR-194 has been proved as a tumor regulatory factor in various cancers; however, the biological function and mechanism of action in colorectal cancer (CRC) have not been well explored. In the present study, we found that miR-194 expression is upregulated in CRC clinical specimens, while overexpression of miR-194 promotes cell migration and invasion in CRC cell lines. Besides, miR-194 significantly influenced the epithelial–mesenchymal transition (EMT) markers by downregulating E-cadherin expression (P<0.01) and upregulating vimentin and MMP-2 expression (P<0.001, P<0.05). Cell migration is the cell movement related to actin cytoskeleton. In this study, we found miR-194 increased cell polarization in SW480 cells. Moreover, zymography assay showed that miR-194 significantly upregulated the gelatin-degrading activity of MMP-2 (P<0.01). Collectively, our findings suggest that miR-194 functions as a tumor promoter in CRC, which may provide new insights for the study of CRC development and metastasis.
Keywords: colorectal cancer, miR-194, epithelial mesenchymal transition, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors, and also the leading cause of cancer death worldwide.1 Lymph node metastasis is not only the key route of distant metastasis, but also a vital prognostic factor in CRC patients.2,3 Consequently, further understanding of the molecular mechanism underlying lymph node metastasis is of great clinical significance.
MicroRNAs (miRNAs) are a class of small, noncoding RNAs that regulate gene expression posttranscriptionally by directly binding their 3′-untranslated region (3′-UTR) of mRNA.4 An increasing number of studies confirm that miRNAs play a central role in inflammation, invasiveness, and progression in CRC.5–9 Epithelial–mesenchymal transition (EMT) plays an essential role in cancer invasion and metastasis in CRC.10 During the EMT process, cancer cells lose the expression of adhesion proteins such as E-cadherin, and increase the expression of mesenchymal phenotype markers such as vimentin and the matrix metalloproteinases (MMPs) family.11 The downregulation of E-cadherin is normally considered to be the hallmark of EMT, which can be mediated by its transcriptional repression through the binding of EMT transcription factors (EMT-TFs),12 such as Twist, Snail, Slug, Zeb1, and Zeb2.13 MMPs have been identified as key enzymes in the EMT process as they are capable of degrading the extracellular matrix components, specifically basement membrane, proteoglycan, fibronectin, and collagen.14
In our previous study, we applied miRNA microarray analysis to two different epithelial features cell lines SW480 and SW620, which demonstrated that miR-194 and miR-192 were differentially expressed and that the results were consistent with Chen’s previous study.15 In this study, we have evaluated the functional role of miR-194 in cell lines, as well as through a direct comparison between primary CRCs and corresponding matching lymph nodes metastasis. We provide evidence that miR-194 can regulate EMT in human CRC metastasis.
Materials and methods
Tissue specimens
A total of 62 CRC tumor tissues were collected from patients who underwent surgical resection in The Second Affiliate Hospital of Chongqing Medical University and The Second Affiliate Hospital of Zhejiang University School of Medicine from August 2014 to September 2015. The 62 CRC patients had not undergone radiotherapy or chemotherapy before surgical resection. The tissues were frozen immediately after resection and stored at −80°C until this study was conducted in November 2015. Clinicopathologic characteristics of the patients are summarized in Table 1. The study was approved by the ethical review committees (Ethic Number: 2014048).
  | Table 1 Sequence of primers used in this study |
Cell lines
Human CRC cell lines SW480 with epithelial features and SW620 with mesenchymal features were donated by Cancer Research Institute of Zhejiang University. The two cell lines were cultured in L15 medium (GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) and in a humid atmosphere of 5% CO2 at 37°C.
RNA extraction and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instruction, and reverse transcribed using the PrimeScript RT Reagent Kit (Takara, Dalian, People’s Republic of China). Expression of miR-194 was analyzed using Hairpin-it miRNAs qPCR Quantitation Kit (GenePharma, Shanghai, People’s Republic of China). Expression of U6 snRNA (GenePharma) and GAPDH (Takara) was used as endogenous control. qRT-PCR was performed with the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The primers of candidate genes are listed in Table 1. All experiments were performed in at least triplicate, and the relative expression levels were calculated using the 2−ΔΔCt method.
Lentivirus infection and stable cell transfection
The lentivectors’ expression of miR-194 and negative control were purchased from GenePharma Company. Lentivectors and packaging vectors were co-transfected into SW480 cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells were cultured to approximate 80% confluence and then added and then added 2.0×105 transducing units (TU)/well lentivectors. Substitute the culture medium with varying concentrations of Blasticidin (0, 4, 6, 8, 10 μg/mL; Sigma-Aldrich, St Louis, MO, USA), and replenish the selective media every 3–4 days until the drug resistance colonies yield (approximately 21 days).
Cell proliferation assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was performed to measure cell proliferation. In brief, the cells were seeded in 96-well plates at a density of 5,000 per well, and grown for 24, 48, and 72 hours, and then MTT (Sigma-Aldrich) solution (5 mg/mL) was added to each well and incubated for another 4 hours at 37°C. Then the medium was removed, and 100 μL dimethyl sulfoxide (DMSO; Sigma-Aldrich) was added to each well to dissolve crystals. Next, the plates were put into the 37°C incubator for 5 minutes to dissolve air bubbles. The absorbance was measured at a wavelength of 490 nm using a microplate reader (Molecular Devices VersaMax, Remington, Madison, NC, USA).
Wound healing assays
The cells were seeded in 6-well plates at a density of 1.5×106 per well, and wounds were generated using pipette tips. Serum-free medium was then added for 48 hours in a humid atmosphere of 5% CO2 at 37°C. The wound closure was assessed by ImageJ Software (National Institutes of Health, Bethesda, MD, USA).
Invasion and migration assays
Invasion and migration assays were performed using a transwell system (BD Biosciences, Bedford, MA, USA). A total of 2×105 cells in a serum-free medium were seeded into the upper chamber with matrigel (for invasion assay) or without matrigel (for migration assay), while the medium containing 20% FBS was added to the lower chamber. After 24 hours of incubation, cells on the upper membrane were removed with cotton wool, whereas cells adhering to the lower surface were fixed in methanol for 15 minutes and then stained with 0.1% crystal violet for 30 minutes. The number of cells was counted in five random fields using an inverted microscope (Olympus, Tokyo, Japan).
Western blotting
Western blot analysis was performed according to the protocol provided by the manufacturer (Bio-Rad Laboratories Inc., Hercules, CA, USA). Briefly, total protein was extracted using radioimmunoprecipitation assay (RIPA) cell lysis buffer (Beyotime, Shanghai, People’s Republic of China), and the concentration was determined by Bicinchoninic Acid Protein Assay Kit (Beyotime). Total protein was separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 6%–10% gel), transferred onto polyvinylidene difluoride membranes, and then incubated with the specific primary antibodies against E-cadherin (Cell Signaling Technology, Beverly, MA, USA), vimentin (Cell Signaling Technology), MMP-2 (GeneTex, Inc., Irvine, CA, USA), and MMP-9 (GeneTex, Inc.) overnight at 4°C. GAPDH (Beyotime) was used as an internal reference. Then the membranes were washed with Tris Buffered Saline, with Tween-20 (TBST) and probed with a horseradish peroxidase-labeled goat anti-rabbit IgG antibody for 1 hour. Signals were visualized using electrochemiluminescent substrates (Millipore, Billerica, MA, USA). The images were analyzed by Sigma Photo Pro 6.0 software (SPSS Inc., Chicago, IL, USA).
Cytoskeleton staining and confocal microscopy
A total of 1×104 cells were seeded on Millicell EZ Slide (Millipore) and cultured in a humid atmosphere of 5% CO2 at 37°C. After 8 hours, the cells were washed twice with phosphate-buffered saline (PBS), and then fixed with 3.7% paraformaldehyde for 10 minutes at room temperature. Then the cells were washed for another 10 minutes with 0.1% TritonX-100 in PBS and blocked with 5% bovine serum albumin (BSA). Cells were incubated with Actin-Tracker Green (Beyotime) for 60 minutes to stain the F-actin filaments. The cell nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 minutes. Finally, fluorescence images were collected using a Carl-Zeiss LSM 710 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Zymography assay
Gelatin zymography was used to analyze the enzymatic activity of the secreted collagenases MMP-2 and MMP-9. In brief, the cells were seeded in 6-well plates at a density of 2×106 per well. After 12 hours, the cells were washed twice with PBS and then cultured in a serum-free medium for 48 hours. The medium was collected, and each sample was so adjusted as to contain the same quantity of protein, and then separated by 8% SDS-PAGE containing 0.1% gelatin. The gels were washed twice in 2.5% Triton X-100 for 45 minutes and incubated in renaturing buffer at 37°C for 42 hours, then stained for 3 hours with 0.05% Coomassie Blue in 10% acetic acid/30% methanol. Then the gels were destained with 10% acetic acid/30% methanol for 0.5 hour, 10% acetic acid/20% methanol for 1 hour, and 5% acetic acid/10% methanol for 2 hours. The band intensities were quantitated using the Image-Pro Plus 5.1 software (Medium Cybernetics Inc., Bethesda, MD, USA).
Statistical analysis
Each assay was performed at least in triplicate, and data were analyzed using the Statistical Product and Service Solutions (SPSS) 19.0 software (SPSS Inc.). Experimental data were expressed as the mean ± standard deviation (SD). Statistical significance was analyzed using Student’s t-test. Pearson’s correlation coefficient was used to measure correlation. P<0.05 was considered statistically significant.
Ethical statement
The study was approved by the ethical review committees of The Second Affiliate Hospital of Chongqing Medical University and The Second Affiliate Hospital of Zhejiang University School of Medicine. The patients gave written informed consent for their tissue samples to be collected and used in the study.
Results
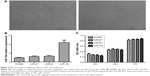
MiR-194 is significantly upregulated in lymph nodes metastasis tissues from CRC patients
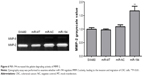
The expression levels of miR-194 were evaluated by qRT-PCR in 62 CRC tumor tissues. As shown in Figure 1A, miR-194 expression was significantly upregulated in lymph nodes metastasis compared with lymph nodes negative. Correlation analysis revealed that miR-194 upregulation was significantly correlated with lymph nodes metastasis (P<0.05), while no significant correlation was found in other clinicopathologic characteristics (Table 2). We next investigated miR-194 expression in SW480 with epithelial features and SW620 with mesenchymal features by qRT-PCR and found significant differences in the miR-194 expression levels in the two cell lines (eightfold, Figure 1B, P<0.01).
MiR-194 did not significantly influence proliferation in CRC cells
Lentivirus system was performed to establish stable SW480 cell lines overexpressing miR-194, mock transfection (MT), and negative control (NC) as the control groups. Compared with SW480 cells, the morphology of overexpressing miR-194 cells changed obviously with visible interstitial characteristic (Figure 2A). Overexpression of miR-194 was validated by qRT-PCR (fivefold, Figure 2B, P<0.01). MTT assays showed that miR-194 overexpression did not significantly influence cell proliferation ability of SW480 cells (Figure 2C, P>0.05).
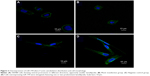
MiR-194 increases cell polarization in CRC cells
In order to investigate the effects of miR-194 on cell morphology and actin cytoskeletal reorganization, we stained the F-actin filaments in CRC cell lines using fluorescent phalloidin, as shown in Figure 3. We observed that SW480 cells showed several protrusions in different directions, apparently smaller lamellipodia, but that cells overexpressing miR-194 were elongated, suggesting reestablished polarization, featuring one or two predominant lamellipodia. By time lapse, we observed that cells overexpressing miR-194 did translocate effectively owing to this strongly polarized phenotype.
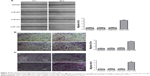
MiR-194 promotes the invasion and migration of CRC cells
Effects of miR-194 on CRC cell migration and invasion were evaluated through wound healing assays and transwell with or without matrigel assays. As indicated in Figure 4, SW480 cells treated with lentivirus coding for miR-194 significantly promoted cell invasion and migration ability compared with the negative control group and mock transfection group (P<0.001).
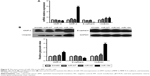
Overexpression of miR-194 induces EMT in CRC cells
We applied qRT-PCR and western blot to validate the relationship between miR-194 and the expression of EMT-related genes on CRC cell lines. Expression levels of mesenchymal markers vimentin and MMP-2 were significantly increased; in contrast, epithelial marker E-cadherin was significantly decreased (Figure 5).
MiR-194 increases MMP-2 activity, leading to the EMT process of CRC cells
MMP-2 is a key enzyme for the invasion and migration of cancer cells. To evaluate whether miR-194 regulates MMP-2 activity, we performed zymography assay. According to the zymography data, miR-194 significantly increased the gelatin-degrading activity of MMP-2. These data suggest that miR-194 increases MMP-2 activity, leading to the invasion and migration of CRC cells (160%, Figure 6, P<0.01). Taken together, these findings have important biological and clinical implications, as they suggest that upregulation of miR-194 is involved in CRC metastasis, during which the colonic cells increasingly gain mesenchymal characteristics and simultaneously lose epithelial features, which facilitates their invasion and migration abilities.
Discussion
Recently, more evidence has emerged showing that miRNAs play a key role in various types of cancer, including lung cancer,16 breast cancer,17 pancreatic cancer,18 ovarian cancer,19 and colorectal cancer.20 In CRC, miRNAs are involved in the pathogenesis of cellular differentiation, proliferation, apoptosis, and migration.21 In this study, we demonstrated that miR-194 was significantly upregulated in a control study of 62 CRC clinical specimens. In cell lines with different epithelial and mesenchymal features, SW480 cells show a lower expression of miR-194 in comparison with SW620 cells. These results suggest that miR-194 may play an important role as a tumor promoter in the progression of CRC.
The involvement of miR-194 has been demonstrated in the origination and progression in certain varieties of cancer.22,23 To investigate the functional roles of miR-194 in CRC cell lines, we constructed miR-194 stably expressing SW480 cells. Wound healing assays and transwell assays revealed that miR-194 promotes cell invasion and migration of SW480 cells; however, miR-194 is not significantly influenced by proliferation in CRC cells. This was identical with a previous finding that miR-194 expression influenced tumor progression and survival in CRC liver metastases.24 Additionally, Liang’s study25 confirmed that miR-194 promoted the migration and invasion of ovarian carcinoma cells. However, another report revealed that low expression of miR-194 contributes to malignant progression, suggesting miR-194 as a tumor suppressor in bladder cancer and non-small cell lung cancer.26,27 All of these studies indicated that miR-194 has different effects on different types of cancers, which are regulated by many cytokines and involved in different signal transduction pathways.
Cell migration is the cell movement related to actin cytoskeleton. Many molecular and genes are involved in the regulation of actin cytoskeleton and cell movement. In Karen’s study, ROCK2 attenuates Rac1 activity at the leading edge and dendritic spine head, which may provide the motive power for migration.28 The Wiskott–Aldrich syndrome family of proteins are involved in cell migration and degradation of extracellular matrix by inducing the formation of the protrusive membrane structures.29 In this study, we found miR-194 increased cell polarization in SW480 cells. However, miR-194 had no effect on the expression of Wiskott-Aldrich syndrome like (WASL) gene (data have not shown). Thus, we propose that the WASL gene is not a target gene of miR-194 in SW480 cells. Our future studies will focus on other target genes of miR-194 that are related to cell migration.
EMT is a process by which cells modify the adhesion molecules expressed, allowing them to adapt a migratory and invasive behavior.30 During EMT, downregulation of E-cadherin diminishes cell–cell adhesion, permitting CRC cells to move freely and eventually settle at distant metastatic sites,31 which also involves multiple MMPs that direct the interactions of tumor cells with the surrounding matrix environment.32 In this study, we found miR-194 was significantly upregulated in CRC with lymph nodes metastasis, suggesting miR-194 might be a regulator of EMT. Our study confirms that miR-194 inhibited epithelial marker E-cadherin and increased mesenchymal markers vimentin and MMP-2. Moreover, zymography assay was performed in our study, which allows one to measure the relative amounts of active and inactive enzymes,33 and results showed that miR-194 significantly increased the gelatin-degrading activity of MMP-2. These data suggest that miR-194 is an EMT promoter in CRC cells.
Conclusion
This study indicates that miR-194 is upregulated in CRC specimens and cell lines and that miR-194 promotes cell migration and invasion of CRC cells. These data suggest that miR-194 functions as a tumor promoter in CRC, which may provide new insights for the study of CRC development and metastasis.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
De Rosa M, Pace U, Rega D, et al. Genetics, diagnosis and management of colorectal cancer. Oncol Rep. 2015;34(3):1087–1096. | ||
Nicastri DG, Doucette JT, Godfrey TE, Hughes SJ. Is occult lymph node disease in colorectal cancer patients clinically significant? A review of the relevant literature. J Mol Diagn. 2007;9(5):563–571. | ||
Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs inpluripotency and reprogramming. Nat Cell Biol. 2012;14(11):1114–1121. | ||
Okayama H, Schetter AJ, Harris CC. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig Dis. 2012;30(2):9–15. | ||
Hur K. MicroRNAs: promising biomarkers for diagnosis and therapeutic targets in human colorectal cancer metastasis. BMB Rep. 2015;48(4):217–222. | ||
Amirkhah R, Schmitz U, Linnebacher M, Wolkenhauer O, Farazmand A. MicroRNA – mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer. 2015;54(3):129–141. | ||
Ghanbari R, Mosakhani N, Asadi J, et al. Downregulation of plasma MiR-142-3p and MiR-26a-5p in patients with colorectal carcinoma. Iran J Cancer Prev. 2015;8(3):e2329. | ||
Yazdani Y, Farazmandfar T, Azadeh H, Zekavatian Z. The prognostic effect of PTEN expression status in colorectal cancer development and evaluation of factors affecting it: miR-21 and promoter methylation. J Biomed Sci. 2016;23:9. | ||
Natalwala A, Spychal R, Tselepis C. Epithelial-mesenchymal transition mediated tumourigenesis in the gastrointestinal tract. World J Gastroenterol. 2008;14(24):3792–3797. | ||
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. | ||
Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18. | ||
Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13(9):963–972. | ||
Torzilli PA, Bourne JW, Cigler T, Vincent CT. A new paradigm for mechanobiological mechanisms in tumor metastasis. Semin Cancer Biol. 2012;22(5–6):385–395. | ||
Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis. 2012;29(5):457–469. | ||
Lin QH, Zhang KD, Duan HX, Liu MX, Wei WL, Cao Y. ERGIC3, which is regulated by miR-203a, is a potential biomarker for non-small cell lung cancer. Cancer Sci. 2015;106(10):1463–1473. | ||
Schwickert A, Weghake E, Bruggemann K, et al. microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, integrin alpha V, and additional cytoskeletal elements. PLoS One. 2015;10(12):e0143993. | ||
Passadouro M, Faneca H. Managing pancreatic adenocarcinoma: a special focus in microRNA gene therapy. Int J Mol Sci. 2016;17(5):e718. | ||
Hirata Y, Murai N, Yanaihara N, et al. MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma. BMC Cancer. 2014;14:799. | ||
Chi Y, Zhou D. MicroRNAs in colorectal carcinoma – from pathogenesis to therapy. J Exp Clin Cancer Res. 2016;35:43. | ||
de Krijger I, Mekenkamp LJ, Punt CJ, Nagtegaal ID. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224(4):438–447. | ||
Guo B, Hui Q, Zhang Y, Chang P, Tao K. miR-194 is a negative regulator of GEF-H1 pathway in melanoma. Oncol Rep. 2016;36(4):2412–2420. | ||
Bernuzzi F, Marabita F, Lleo A, et al. Serum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Clin Exp Immunol. 2016;185(1):61–71. | ||
Pecqueux M, Liebetrau I, Werft W, et al. A comprehensive MicroRNA expression profile of liver and lung metastases of colorectal cancer with their corresponding host tissue and its prognostic impact on survival. Int J Mol Sci. 2016;17(10):e1755. | ||
Liang T, Li L, Cheng Y, Ren C, Zhang G. MicroRNA-194 promotes the growth, migration, and invasion of ovarian carcinoma cells by targeting protein tyrosine phosphatase nonreceptor type 12. Onco Targets Ther. 2016;9:4307–4315. | ||
Zhang M, Zhuang Q, Cui L. MiR-194 inhibits cell proliferation and invasion via repression of RAP2B in bladder cancer. Biomed Pharmacother. 2016;80:268–275. | ||
Zhu X, Li D, Yu F, et al. miR-194 inhibits the proliferation, invasion, migration, and enhances the chemosensitivity of non-small cell lung cancer cells by targeting forkhead box A1 protein. Oncotarget. 2016;7(11):13139–13152. | ||
Newell-Litwa KA, Badoual M, Asmussen H, Patel H, Whitmore L, Horwitz AR. ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J Cell Biol. 2015;210(2):225–242. | ||
Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96(7):379–386. | ||
Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. | ||
Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62(9):1315–1326. | ||
Xu X, Wang Y, Chen Z, Sternlicht MD, Hidalgo M, Steffensen B. Matrix metalloproteinase-2 contributes to cancer cell migration on collagen. Cancer Res. 2005;65(1):130–136. | ||
Ricci S, D’Esposito V, Oriente F, Formisano P, Di Carlo A. Substrate-zymography: a still worthwhile method for gelatinases analysis in biological samples. Clin Chem Lab Med. 2016;54(8):1281–1290. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.