Back to Journals » Infection and Drug Resistance » Volume 13
Metagenomic Next-Generation Sequencing versus Traditional Pathogen Detection in the Diagnosis of Peripheral Pulmonary Infectious Lesions
Authors Huang J, Jiang E, Yang D , Wei J, Zhao M, Feng J, Cao J
Received 18 October 2019
Accepted for publication 24 January 2020
Published 19 February 2020 Volume 2020:13 Pages 567—576
DOI https://doi.org/10.2147/IDR.S235182
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Jie Huang,1,2 Erlie Jiang,3 Donglin Yang,3 Jialin Wei,3 Mingfeng Zhao,4 Jing Feng,1 Jie Cao1
1Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, Tianjin, People’s Republic of China; 2Graduate School, Tianjin Medical University, Tianjin, People’s Republic of China; 3Hematopoietic Stem Cell Transplantation Center, Institute of Hematology and Blood Disease Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin, People’s Republic of China; 4Department of Hematology, Tianjin First Central Hospital, Tianjin, People’s Republic of China
Correspondence: Jing Feng; Jie Cao
Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, Anshan Road No. 154, Heping District, Tianjin 300052, People’s Republic of China
Tel +86 22 6036 2255
Email [email protected]; [email protected]
Purpose: The aim of this study was to evaluate the value of metagenomic next-generation sequencing (mNGS) in peripheral pulmonary infection management by comparing the diagnostic yield of mNGS and traditional pathogen detection methods on interventional specimens obtained by bronchoscopy.
Patients and Methods: This study enrolled patients suspected with pulmonary infection who were admitted to Tianjin Medical University General Hospital from June 2018 to August 2019. Specimens were obtained from bronchoscopy for mNGS analysis and traditional pathogen detection (including bronchoalveolar lavage fluid microbial culture, smear microscopy, and lung biopsy histopathology), and the diagnostic yields were compared between mNGS and traditional methods to evaluate the diagnostic value of mNGS in peripheral pulmonary infection diagnosis.
Results: In this study, by comparing mNGS with traditional pathogen detection, the results indicated that, first, mNGS identified at least one microbial species in almost 89% of the patients with pulmonary infection; second, mNGS detected microbes related to human diseases in 94.49% of samples from pulmonary infection patients who had received negative results from traditional pathogen detection; third, the accuracy and sensitivity of mNGS are higher than those of traditional pathogen detection; and, finally, mNGS could simultaneously detect and identify a large variety of pathogens.
Conclusion: Metagenomic NGS analysis provided fast and precise pathogen detection and identification, contributing to prompt and accurate treatment of peripheral pulmonary infection.
Keywords: pulmonary infection, metagenomic next-generation sequencing, bronchoscopy, bronchoalveolar lavage, smear microscopy, transbronchial lung biopsy
Introduction
Pulmonary infection is prevalent worldwide with high morbidity and mortality, especially in elder and immunocompromised populations.1,2 While fast diagnosis of pulmonary infection is important for prompt disease management and better outcome, accurate detection and identification of pathogens is challenging. Particularly in immunocompromised hosts, almost all bacteria or fungi may be considered potential pathogens responsible for pulmonary infections.3,4 Early detection of pathogens can optimize antibiotic management, shorten hospital stays, and improve survival rates. Delaying diagnosis may impede targeted therapies and lead to poor prognosis.5
Traditional pathogen detection methods of bronchoscopy specimens include bronchoalveolar lavage fluid (BALF) microbial culture, smear microscopy, and histopathology. However, due to the current limitation in microbial cultivating techniques,6 the impact of lesions surrounded by fibrous tissue and antibiotic application history, the positive rate of culture is lower. On the other hand, histopathology analysis is favorable for fungi and cryptococcus detection,7 but has no advantage in the diagnosis of other pathogens. Patients with negative traditional pathogen detection results often undergo empirical antibiotic treatments, which may not cover the real pathogen and lead to reinfection.8,9 This widespread use of antibiotics is likely contributing to the accumulation and dissemination of antibiotic resistance and multidrug-resistant pathogens.10 The traditional pathogen detection methods have low yield and are time-consuming. As these methods are unable to satisfy the current need for fast and precise infection diagnosis, as a result, metagenomic sequencing has emerged as needed.11,12
Metagenomic next-generation sequencing (mNGS), also known as shotgun deep-sequencing, is a high-throughput sequencing approach with high efficiency and short turnaround time. Nowadays mNGS is widely used in but not limited to whole genome sequencing and resequencing, transcriptome sequencing, small ribonucleic acid (RNA) sequencing, deoxyribonucleic acid (DNA) and protein interactions, and DNA methylation analysis.13,15 Pathogen detection and identification by mNGS is an emerging clinical practice, which only needs small amounts of DNA extracted from the sample and simultaneously detects and identifies pathogens. Owing to its high positive rate in pathogen detection, mNGS has already been successfully applied in several clinical trials for infection diagnosis.16,18 In addition, It has been reported that mNGS improved the detection efficiency in culture negative samples.19 However, reports on mNGS application in diagnosis respiratory tract pulmonary infections, particularly by using bronchoscopy samples, remains rare. The aim of this study was to evaluate the value of mNGS in peripheral pulmonary infection diagnosis by comparing the efficiency of mNGS and traditional pathogen detection methods (BALF microbial culture, smear microscopy, and lung biopsy histopathology) in specimens obtained by bronchoscopy.
Materials and Methods
Ethics Statement
This study was carried out in accord with the principles of the Declaration of Helsinki and approved by the Ethics Review Committee of Tianjin Medical University General Hospital. Patients’ approval and informed consent were waived because the study involved a retrospective review of patient records.
Specimen Collection
Our study retrospectively reviewed 240 patients suspected of pulmonary infections, who were admitted to Tianjin Medical University General Hospital between June 2018 and August 2019. Eleven patients were excluded from the study due to missing critical data. Among the patients enrolled, 213 patients had underlying disease, including 155 cases of hematological malignancies and 19 cases of rheumatic immune diseases (Figure 1). After confirming no risk of bleeding (platelets (PLT)<50×109/L, activated partial thromboplastin time (APTT)≥50 s, fibrinogen (FIB) ≤1.5 g/L),20 bronchoscopy was performed by the chief physician of the Respiratory Endoscopy Center to obtain specimens including lung tissue, BALF, and brush. The endobronchial ultrasound system (K201/K203, Olympus, Tokyo, Japan) and the virtual bronchoscope navigation system (DirectPath V1.0, Cybernet systems, Olympus) were applied in all patients to precisely locate the lesion, then an ultrathin bronchoscope (BF-P-260F, Olympus) was used to reach the region of the lesion and obtain the specimens. The categories of interventional specimens include lung tissue, BALF, and protected-specimen brush. The average number of tissue samples was 6–10 pieces for lung biopsy, 20 mL for BALF, and two protected-specimen brushes. Each specimen was equally divided into two parts for mNGS analysis and traditional pathogen detection. Although all three different categories of sample were collected in most patients, some patients only went through one or two sampling strategies due to long-term intolerance to bronchoscopy or difficulty in obtaining samples.
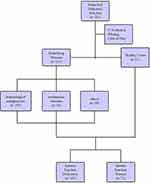 |
Figure 1 Detailing the characteristic of patient samples. |
Specimen Processing
All samples were promptly stored in sterile containers and transported to the laboratory for mNGS, ensuring that the samples were placed at 4°C before analysis. The lung biopsies were fixed with 4% formaldehyde (Fengchuan, Tianjin, China) and were sent within 2 hours of collection to the histopathology laboratory, where they were dehydrated, embedded in paraffin (Haotian Craft Wax, Suzhou, China), sectioned, and stained with hematoxylin and eosin (H&E, Solarbio, Beijing, China) to examine inflammatory cell infiltration and the presence of visible pathogens (fungal hyphae). In addition, all samples underwent special pathological staining, including Ziehl-Neelsen acid-fast staining (after initial dyeing, decolorization and counterstaining, red dyed, unreflective, slender, and slightly curved acid-fast positive Mycobacterium tuberculosis observed under high magnification) and hexamine silver staining (through periodate oxidation, silver solution, and gold chloride staining, sodium thiosulfate washing, bright green solution staining, and the brown and dark brown fungal hyphae can be observed after sealing). The protected-specimen brush (Jingrui, Hangzhou, China) was smeared evenly on a slide (FeiZhou, Jiangsu, China), which was sent to the histopathology laboratory for acid-fast staining after transbronchial brushing. BALF was sent to the microbiology laboratory for microbial culture and microscopic examination smeared with centrifugal sediment. After BALF was centrifuged at 3,000 rpm for 10 minutes, the sediment was smeared on a slide for gram staining (Baso, Zhuhai, China), Ziehl-Neelsen acid-fast staining (Baso), and hexamine silver staining (the reagents needed for the dye were purchased from Yingda Rare, Sinopharm, and Kemiou from China) to distinguish bacteria, fungi, Mycobacterium tuberculosis, and Pneumocystis carinii. BALF culture was performed in a sterile environment, where the collected lavage fluid was centrifuged and the precipitate was inoculated on blood agar plates (bioMérieux, Marcy l’Etoile, France) and sabouraud agar plates (bioMérieux) for culture of bacteria and fungi, respectively, and incubated at 35°C for 24–48 hours for bacteria and 7 days for fungi according to conventional culture method for respiratory specimens. If a typical colony grew, microscopy analysis was performed after colony staining.21 Gram stain, hexamine silver staining, and Ziehl-Neelsen acid-fast stain were used to identify bacteria, fungi, Pneumocystis carinii, and Mycobacterium tuberculosis under a microscope.21
Metagenomic Next-Generation Sequencing
The specimens to be tested were divided into two types. One was used to extract DNA directly from each sample collected from the same patient and sequenced separately. If the patient had poor tolerance to sampling procedure and the volume of BALF was less than 20 mL, DNA from BALF and protected-specimen brush were extracted and sequenced together while lung biopsies were processed separately, at this point the mixed specimen was called a “cocktail specimen”. The sequencing method was performed as previously described.21,22 We added 0.5 mL of BALF sample and 1 g of 0.5 mm glass beads (TUOTAINUO, Shenzhen, China) to a 1.5 mL centrifuge tube (KIRGEN, Shanghai, China), placed on a horizontal plate of the vortex (Kylin-Bell Vortex-5, Haimen, China) at 2,800–3,200 rpm for 30 minutes, and separated 0.3 mL of BALF into a new 1.5 mL centrifugal tube. The tissue was homogenized using a tissue homogenizer (NewZongKe, Shenzhen, China), and the DNA was extracted. The protected-specimen brush was placed on the horizontal plate of the vortex to oscillate for 30 minutes at 2,800–3,200 rpm for subsequent operations. The TIANamp Micro DNA Kit (TIANGEN BIOTECH, Beijing, China) was used to extract DNA from the three samples. The extracted DNA samples were used to construct a DNA library based on the Beijing Genomics Institute sequencer-100; the library construction process included DNA end repairing, adapter connection, polymerase chain reaction (PCR) amplification, and amplification product purification by magnetic beads (MGI, Shenzhen, China). The Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, Canada) and quantitative polymerase chain reaction (qPCR) were used for library quality control. The quality control qualified DNA library was sequenced on the Beijing Genomics Institute sequencer-100 platform (BGI, Shenzhen, China).
Metagenomic Next-Generation Sequencing Analyses
High-quality sequencing data were generated by removing low-quality reads, adapter contamination, duplicated reads, and reads shorter than 35 bp. By Burrows-Wheeler Alignment (BWA), human genome sequence data was deleted. The remaining data was compared with four microbial genome reference sequence databases downloaded from the National Center for Biotechnology Information, which included the whole genome sequence of 2,700 viruses, 1,494 bacteria, 73 fungal and 47 parasite genome sequences associated with human infection.21 The criteria for a positive mNGS test result included: 1) the relative abundance of bacteria (Mycobacterium tuberculosis excluded) and fungi was greater than 30% at the genus level;21 2) Mycobacterium tuberculosis was considered to be positively detected if at least one read was aligned to the reference genome at species or genus level;23 3) positive virus detection was considered when the stringent map read number (SMRN) was no less than 3; and 4) when the pathogen was detected by traditional pathogen detection methods and the mNGS reads number was more than 50, this pathogen can also be considered as positively detected by mNGS.21
Data Analyses
Although histopathology, BALF culture, and smear microscopy were used as traditional pathogen detection methods in our study, the final clinical diagnosis was confirmed by comprehensive evaluation of traditional pathogen detection, mNGS results, other clinical examination results (including X-pert and GM tests of BALF, and relevant tests of blood samples such as Epstein-Barr virus nucleic acid detection, human cytomegalovirus nucleic acid detection, Cryptococcus neoformans capsular polysaccharide detection, Chlamydia pneumoniae, and Mycoplasma pneumoniae serological antibody detection, which was considered viable when a four-fold rise in the paired serum antibody titer was detected between 2–3 weeks, and blood culture), imaging analysis, and prognosis. A mNGS or traditional test result was considered positive only if the pathogen(s) detected was in consistence with the final clinical diagnosis, in that case, even if mNGS or traditional test of only one out of three specimens from one patient was positive, the result of that type of test was considered positive. If the patient’s final clinical diagnosis was non-pulmonary infectious disease, the tests with positive results were considered as false positives. If the patient’s final clinical diagnosis was pulmonary infectious disease, the tests with positive results were considered as true positives, while the tests with negative results were considered as false negatives.
Follow-Up
The patients enrolled in the study were followed up in September 2019, and separated into three groups (Improvement, Non-Change, and Death) according to the outcomes. The Improvement group had significant relief clinical symptoms and absorption of lesion. The Non-Change group had no obvious relief of clinical symptoms and imaging lesions, while the Death group contained all the patients who died due to respiratory events.
Statistical Analyses
With the final clinical diagnosis as the gold standard, the patients were divided into pulmonary infection group and non-pulmonary infection group, and the pulmonary infection group was the positive reference group. The student t-test and χ2 test were used to calculate differences in continuous variables between groups. P-values <0.05 were considered to be statistically significant. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy (ACC) were calculated, and sensitivity and specificity were compared between mNGS and traditional pathogen detection methods using the χ2 test. All statistics were reported as absolute values with their 95% confidence interval (95% CI) and all statistics were calculated by SPSS 19.0 software.
Results
Patient Demographics
In our study, the most prevalent hematological malignancies were acute lymphoblastic leukemia (n=47, 30.32%), acute myelocytic leukemia (n=44, 28.39%), myelodysplastic syndromes (n=16, 10.32%), and aplastic anemia (n=11, 7.10%). Vasculitis (n=7, 36.84%), systemic lupus erythematosus (n=5, 26.32%), and rheumatoid arthritis (n=4, 21.05%) accounted for the majority of autoimmune diseases. Among the other underlying diseases, hypertension (n=19, 48.72%) and diabetes (n=13, 33.33%) were most prevalent, and seven patients had hypertension and diabetes simultaneously. The immune function was normal in 73 patients, while the other 167 had immune deficiency. For the final clinical diagnosis, 171 patients were diagnosed with pulmonary infection, and 69 were diagnosed with non-pulmonary infection (Table 1).
 |
Table 1 Demographic Characteristics of Pulmonary Infection and Non-Pulmonary Infection |
Bioinformatics Information
A total of 467 mNGS results were collected, including data from 170 BALF samples, 160 lung tissue samples, 94 bronchoscopic protected-specimen brushes, and 43 cocktail specimens.
Among the patients with pulmonary infections, the percentage with mNGS-positive result was 151/171 (88.30%). Among the patients with non-pulmonary infection, the percentage with mNGS-positive results was 13/69 (18.84%) (Figure 2), the ACC of mNGS was 86.25%. Among the patients diagnosed with fungal pneumonia, the most prevalent pathogenic fungi were Aspergillus (n=32, 45.07%), Pneumocystis carinii (n=22, 30.99%), and Rhizopus (n=7, 9.86%). When analyzed by mNGS, Aspergillus fumigatus was the major species detected in Aspergillus, followed by Aspergillus oryzae, Aspergillus niger. The number of patients co-infected by Aspergillus flavus and Aspergillus oryzae was four, while co-infection of Aspergillus fumigatus and Aspergillus oryzae was one. In patients with a positive mNGS result, the bacteria most frequently detected were Mycobacterium tuberculosis (n=19, 26.39%), Pseudomonas aeruginosa (n=16, 22.22%), Acinetobacter baumannii (n=9, 12.50%), and Klebsiella pneumoniae (n=7, 9.72%). And the viruses most frequently detected were human herpesvirus type 5 (HSV-5, n=26, 43.33%), human herpesvirus type 4 (HSV-4, n=19, 31.67%), human parvovirus B19 (HPV B19, n=4, 6.67%), and Torque teno virus (TTV, n=4, 6.67%), the number of cases of co-infection of HSV-5 and HSV-4 was six. The pathogen detection yield of mNGS analysis and traditional detection methods in patients with pulmonary infection is shown in Table 2. In the specimens analyzed by mNGS, Prevotella, Streptococcus, Porphyromonas, and Veillonella were detected in both groups. Among the 171 patients with pulmonary infection, the number of patients who had positive mNGS results in all three samples was 40, the number for two positive samples was 63, and 48 patients only had positive detection in one sample sent for analysis.
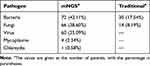 |
Table 2 Detection Rate of Organism of Pulmonary Infection in mNGS Compared with Traditional Detection Method |
Among the patients with pulmonary infections, the positive results of traditional pathogen detection methods were obtained from 44 (25.73%) patients, and the most commonly detected pathogen was mold (the classification of mold could not be clearly identified by traditional culture and histology). Among the patients with non-pulmonary infection, positive results of traditional pathogen detection were achieved in eight (11.59%) patients, and the most commonly detected pathogen was Stenotrophomonas maltophilia. The ACC of traditional pathogen detection was 43.75%.
The most commonly detected pathogens (bacteria, fungi, and viruses) in immunocompromised patients with pulmonary infection were Pseudomonas aeruginosa (n=15, 11.90%), Aspergillus (n=30, 23.81%), and HSV-5 (n=26, 20.63%), respectively. The most commonly detected pathogens (bacteria, fungi, and viruses) in immunocompetent patients with pulmonary infection were Mycobacterium tuberculosis (n=10, 22.22%), Cryptococcus neoformans (n=4, 8.89%), and HSV-4 (n=4, 8.89%), respectively.
Identification of Pathogen in Negative Traditional Pathogen Detection Samples by mNGS
A total of 127 specimens received negative or non-specific detected results from traditional pathogen detection tests, of which 120 had positive results from mNGS, and seven had negative or non-specific detected results from mNGS. In patients with pulmonary infections, the pathogens which traditional pathogen detection generally failed to detect included many fungi (such as the Pneumocystis jirovecii, Aspergillus) and almost all of the viruses, while the common bacteria such as Pseudomonas aeruginosa, Mycobacterium tuberculosis, and Acinetobacter baumannii were easily detected by both traditional methods and mNGS.
Performance of mNGS and Compare with Traditional Pathogen Detection
The sensitivity and specificity of mNGS were compared to that of traditional pathogen detection methods (Table 3). mNGS was more sensitive (88.30%) compared to traditional pathogen detection (25.73%), with a difference of 62.57% (P<0.001), However, mNGS was less specific (81.16%) than traditional methods (88.41%), with a difference of 7.25% (P=0.639). The PPV and NPV of mNGS were 92.07% and 73.68%, respectively. The ACC of mNGS was 86.25%, while the ACC of traditional detection methods was 43.75%. The PPV and NPV of traditional pathogen detection were 84.62% and 32.45%, respectively. The follow-up results of all patients after 1 month are shown in Table 4. After the follow-up, 158 patients improved, of which 145 had positive results from mNGS and 42 had positive results from traditional pathogen detection. The number of patients without significant changes or who died from respiratory events were 11 and 2, respectively.
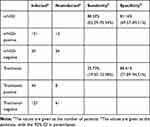 |
Table 3 Comparison of Sensitivity and Specificity Between mNGS and Traditional Detection Method |
 |
Table 4 Infection Status of All Patients at a 1-Month Follow-Up According to the Metagenomic Next-Generation Sequencing Result |
Discussion
Our study retrospectively evaluated the value of mNGS in peripheral pulmonary infection by performing mNGS and traditional pathogen detection in specimens obtained by bronchoscopy. The mNGS analysis was relatively unbiased and highly sensitive for simultaneous detection of hundreds of known pathogens, providing great potential for pathogens analysis in clinical specimens.18 Most of the patients enrolled in this study had hematologic malignancy or rheumatic immune diseases. These patients were chronically immunocompromised due to long-term use of large doses of anti-tumor drugs, cytotoxic drugs, or glucocorticoids, and were prone to complicating diseases, especially pulmonary complications.24,25
When pulmonary lesions occur, the lesions should be judged quickly and accurately to guide the proper follow-up treatment. Since the clinical manifestations of some infectious diseases and non-infectious diseases are very similar, this is more difficult to distinguish by routine laboratory examination and imaging analysis, where there are often overlap characteristics between diseases. In addition, the condition of immunocompromised patients is generally critical, the identification of the pathogens is important for precise diagnosis and necessary for proper treatment,18 while a delay in the diagnosis may impede the targeted treatment and lead to a poor prognosis. So, it is essential to optimize diagnosis efficiency with early and precise management as well as interdisciplinary teamwork.26,27 Although these patients are at high risk with bronchoscopy due to their poor health state and rapid disease progress, studies have pointed out that the risk of complications and the condition of the patients should be evaluated based on the location of the lesion. With the exception of contraindications, an early invasive bronchoscopy examination should be performed to determine the etiology of the lesion.25,28 For patients who are immunocompetent, if the clinical and imaging analysis indicate a suspicion of pulmonary infectious diseases, and the response to initial empirical treatment is poor, timely intervention is also required to obtain lesion samples to identify the pathogen. Currently, the most common detection methods for pathogens in the specimens are microbial culture, histopathology, and smear microscopy.29,30 Microbial culture in bronchoscopy includes tissue culture and BALF culture, but the sensitivity is low and the positive rate is related to many factors, such as sample type and patient’s condition.31 Histopathology and smear microscopy are merely able to identify limited fungal species or Mycobacterium tuberculosis. For virus detection, the above test methods are not applicable. Real-time PCR can be used to detect the virus, but it requires a proper hypothesis of pathogens to synthesize specific primers.18 With the development of diagnostic technology, there has been a rise of some precise detection methods, such as GM test (Aspergillus galactomannan test),32,33 or X-pert (Mycobacterium tuberculosis/rifampicin resistance test).34 These methods are able to improve the diagnostic yield of Aspergillus and Mycobacterium tuberculosis. Real-time PCR also has high sensitivity and specificity for the detection of Pneumocystis jirovecii.35 Although the above detection methods may improve the diagnostic yield of pathogens, their sensitivity is considerably impacted by pathogen load, former antibiotic use, source of samples, and other aspects.21,36,37
Our study obtained interventional specimens from patients with suspected pulmonary infection by bronchoscopy and sent them for mNGS analysis. The results indicated that: first, mNGS was able to identify nearly 89% of patients with pulmonary infection, which was higher than traditional pathogen detection, at 25.73% (P<0.001). However, mNGS was less specific, with a specificity of 81.16%, while the specificity of traditional methods was 88.41% (P=0.639), which may be due to the fact that traditional pathogen detection methods cannot detect more microorganisms, resulting in a higher true negative in non-pulmonary infection; second, and more importantly, mNGS detected microbes related to human diseases in 94.49% of samples from pulmonary infections patients who had received negative results from traditional pathogen detection; third, the ACC of mNGS was 86.25%, while the ACC of traditional pathogen detection was 43.75%; finally, mNGS could simultaneously detect and identify a large variety of pathogens, among which 42.11% were bacteria, 38.60% were fungi, 35.09% were virus, 2.34% were Mycoplasma, and 0.58% were Chlamydia. After a follow-up of 1 month, 158 patients improved after diagnosis and treatment, among whom 145 had positive detection by mNGS analysis and 42 had positive detection by traditional detection methods. However, there were still five patients who had no significant changes after the application of mNGS analysis, which might be due to the high toxicity of the pathogens, the immune deficiency state of patients and drug-resistant. One patient died of massive hemoptysis due to fungal invasion of bronchial arteries in the lungs, another patient died from deteriorative respiratory event. In conclusion, mNGS was superior to traditional pathogen detection in etiological diagnosis of peripheral pulmonary infection.
Metagenomic NGS can generate tens of thousands of individual sequences,19 while detecting hundreds of common and uncommon pathogens which may evade traditional detection,38 providing comprehensive information on pulmonary microbiota. It has been reported that the core genera of pulmonary microbiota include Prevotella, Pseudomonas, Streptococcus, Fusobacterium, Haemophilus, Veillonella, and Porphyromonas.39,40 In some patients, mNGS could detect Prevotella, Streptococcus, Porphyromonas, and Veillonella in both groups, similar to the core genera of pulmonary microbiota reported previously. In addition to bacterial infection, there was a high percentage of fungal infection in our study. This result might due to the fact that most patients enrolled in our study were immunocompromised or immunodeficient, and fungi such as Aspergillus, Rhizopus, Mucor, and Pneumocystis carinii had become the main opportunistic pathogens in these patients. This was similar to previously research.41,43 The positive rate of mNGS analysis for fungi was 66/71 (92.96%), whereas only 14 cases (19.72%) had positive detection from traditional pathogenic detection methods. According to reports, combined analysis of mNGS and smear can reduce detection time, provide accurate identification of the strain, and serve as a conventional diagnostic tool for invasive fungal infection.19 Since mNGS analysis often detected more than one pathogen in a single test, therefore, clinicians need to have a comprehensive understanding of these infections, in order to diagnose mixed infection and/or distinguish the causative pathogen(s).19
Limitations
Despite the encouraging results of our research, there were still some limitations. First, the interventional specimens obtained by bronchoscopy might not contain valuable pathogens DNA due to the location of the lesion and the tolerance of the patient, which could have contributed to false negative detections. In this case, it is suggested to use a large sum of pooled sample in the future study to improve the detection efficiency. In subsequent studies, standardized procedures should also be developed to further improve detection yield of mNGS. Second, the sample processing procedures used in this study underestimated the possibility of RNA virus infection. As fungal and intracellular bacteria have thicker cell walls, wall-breaking treatments were applied in the DNA extracting procedure, which might affect the detection of RNA viruses. Therefore, DNA extraction procedures presented in this study are not fit for the detection of RNA virus. RNA extraction and reverse transcription procedures should be applied if RNA virus infection is suspected. In addition, although 1/3 positive mNGS samples were interpreted as mNGS positive only if the pathogen(s) detected was in consistence with the final clinical diagnosis, the co-detection of several pathogens inter-sample could have suggested the possibility of false positive detections or the presence of environmental contamination. Despite the increasing use of mNGS, the limitations of this technique need to be noted. Metagenomic NGS analysis is unable to differentiate colonization and pathogenic strains. Also, the precision of mNGS is prone to the impacts of background microbiome and contamination of nucleic acids in reaction kits. Sample processing procedures, bioinformatic analysis, and interpretation of sequencing results are also affecting aspects to the final detection.44 Furthermore, the economy cost is also a problem. The average cost of mNGS is 2,500–3,000 RMB (Ren Min Bi) per specimen, so the costs of 2–3 specimens per patient tend to be a large economic burden compared to traditional pathogenic tests (1,000–1,500 RMB). Last, but not least, our study was a single-center retrospective study, which still requires more prospective and multicenter data to accurately determine the accuracy of mNGS analysis.
Conclusion
At present, there have been few studies on the value of mNGS in etiological detection for pulmonary infection. Our study introduced mNGS into the pathogen detection and identification of specimens obtained by bronchoscopy, and compared its detection yield with the traditional pathogen detection methods, demonstrating that mNGS was able to quickly and accurately detect and identify pathogens. In addition, as most of the patients enrolled in our study had severe underlying disease, obtaining specimens for mNGS testing by bronchoscopy contributed to safe and earlier infection diagnosis, saving precious time for the treatment of critical diseases.
Abbreviations
mNGS, metagenomic next-generation sequencing; BAL, bronchoalveolar lavage; BALF, bronchoalveolar lavage fluid; RNA, ribonucleic acid; DNA, desoxyribonucleic acid; PLT, platelets; APTT, activated partial thromboplastin time; FIB, fibrinogen; MTBC, Mycobacterium tuberculosis complex; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; PPV, positive predictive value; NPV, negative predictive value; ACC, accuracy; CI, confidence interval; BWA, Burrows-Wheeler Alignment; HSV-5, human herpesvirus type 5; HSV-4, human herpesvirus type 4.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgment
We would like to thank Yang Zhou who as a native English speaker helped us in editing the paper and dealing with some language issues.
Author Contributions
All authors contributed to the conception and design, acquisition and analysis of data; drafting and critically revising the paper; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Beigelman-Aubry C, Godet C, Caumes E. Lung infections: the radiologist’s perspective. Diagn Interv Imaging. 2012;93(6):431–440. doi:10.1016/j.diii.2012.04.021
2. Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi:10.1056/NEJMoa1306801
3. De La Cruz O, Silveira FP. Respiratory fungal infections in solid organ and hematopoietic stem cell transplantation. Clin Chest Med. 2017;38(4):727–739. doi:10.1016/j.ccm.2017.07.013
4. Renaud C, Campbell AP. Changing epidemiology of respiratory viral infections in hematopoietic cell transplant recipients and solid organ transplant recipients. Curr Opin Infect Dis. 2011;24(4):333–343. doi:10.1097/QCO.0b013e3283480440
5. Subira M, Martino R, Franquet T, et al. Invasive pulmonary aspergillosis in patients with hematologic malignancies: survival and prognostic factors. Haematologica. 2002;87(5):528–534.
6. Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13(9):601–612. doi:10.1038/nrg3226
7. Guarner J, Brandt ME. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev. 2011;24(2):247–280. doi:10.1128/CMR.00053-10
8. Mortazavi SM, Vegari D, Ho A, Zmistowski B, Parvizi J. Two-stage exchange arthroplasty for infected total knee arthroplasty: predictors of failure. Clin Orthop Relat Res. 2011;469(11):3049–3054. doi:10.1007/s11999-011-2030-8
9. Parvizi J, Erkocak OF, Della Valle CJ. Culture-negative periprosthetic joint infection. J Bone Joint Surg Am. 2014;96(5):430–436. doi:10.2106/JBJS.L.01793
10. Ruppe E, Baud D, Schicklin S, Guigon G, Schrenzel J. Clinical metagenomics for the management of hospital- and healthcare-acquired pneumonia. Future Microbiol. 2016;11(3):427–439. doi:10.2217/fmb.15.144
11. Bragg L, Tyson GW. Metagenomics using next-generation sequencing. Methods Mol Biol. 2014;1096:183–201.
12. Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science (New York, NY). 2006;312(5778):1355–1359. doi:10.1126/science.1124234
13. Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. doi:10.1016/j.tig.2007.12.007
14. Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi:10.1038/nrg2626
15. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi:10.1038/nrg2484
16. Brenner T, Decker SO, Grumaz S, et al. Next-generation sequencing diagnostics of bacteremia in sepsis (Next GeneSiS-Trial): study protocol of a prospective, observational, noninterventional, multicenter, clinical trial. Medicine. 2018;97(6):e9868. doi:10.1097/MD.0000000000009868
17. Joensen KG, Engsbro ALO, Lukjancenko O, et al. Evaluating next-generation sequencing for direct clinical diagnostics in diarrhoeal disease. Eur J Clin Microbiol Infect Dis. 2017;36(7):1325–1338. doi:10.1007/s10096-017-2947-2
18. Kawada J, Okuno Y, Torii Y, et al. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Sci Rep. 2016;6:33452. doi:10.1038/srep33452
19. Tarabichi M, Shohat N, Goswami K, et al. Diagnosis of periprosthetic joint infection: the potential of next-generation sequencing. J Bone Joint Surg Am. 2018;100(2):147–154. doi:10.2106/JBJS.17.00434
20. Bulpa PA, Dive AM, Mertens L, et al. Combined bronchoalveolar lavage and transbronchial lung biopsy: safety and yield in ventilated patients. Eur Respir J. 2003;21(3):489–494. doi:10.1183/09031936.03.00298303
21. Li H, Gao H, Meng H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8:205. doi:10.3389/fcimb.2018.00205
22. Liu N, Kan J, Cao W, et al. Metagenomic next-generation sequencing diagnosis of peripheral pulmonary infectious lesions through virtual navigation, radial EBUS, ultrathin bronchoscopy, and ROSE. J Int Med Res. 2019;47(10):4878–4885. doi:10.1177/0300060519866953
23. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–s240. doi:10.1093/cid/ciy693
24. Nisar MK, Ostor AJ. Pulmonary complications of biological therapies in children and adults with rheumatic diseases. Paediatr Respir Rev. 2013;14(4):236–241. doi:10.1016/j.prrv.2012.12.007
25. Wingard JR, Hiemenz JW, Jantz MA. How I manage pulmonary nodular lesions and nodular infiltrates in patients with hematologic malignancies or undergoing hematopoietic cell transplantation. Blood. 2012;120(9):1791–1800. doi:10.1182/blood-2012-02-378976
26. Hachem RY, Kontoyiannis DP, Boktour MR, et al. Aspergillus terreus: an emerging amphotericin B-resistant opportunistic mold in patients with hematologic malignancies. Cancer. 2004;101(7):1594–1600. doi:10.1002/(ISSN)1097-0142
27. Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. Epidemiology and clinical manifestations of mucormycosis. Clin Infect Dis. 2012;54(Suppl 1):S23–S34. doi:10.1093/cid/cir866
28. Clements PJ, Goldin JG, Kleerup EC, et al. Regional differences in bronchoalveolar lavage and thoracic high-resolution computed tomography results in dyspneic patients with systemic sclerosis. Arthritis Rheum. 2004;50(6):1909–1917. doi:10.1002/(ISSN)1529-0131
29. Menon LR, Divate S, Acharya VN, Mahashur AA, Natrajan G, Almeida AF. Utility of bronchoalveolar lavage in the diagnosis of pulmonary infections in immunosuppressed patients. J Assoc Physicians India. 2002;50:1110–1114.
30. Miller SA, van Zante A, Schwartz BS. Cytologic evaluation can predict microbial culture results for infectious causes of pulmonary nodules in patients undergoing fine needle aspiration biopsy. Diagn Microbiol Infect Dis. 2010;68(3):330–333. doi:10.1016/j.diagmicrobio.2010.08.005
31. Al-Nakeeb Z, Gupta V, Bell C, Woodhead M. Are we missing opportunities to confirm the diagnosis of tuberculosis by microbial culture? Respir Med. 2013;107(12):2022–2028. doi:10.1016/j.rmed.2013.09.016
32. Steinbach WJ, Addison RM, McLaughlin L, et al. Prospective Aspergillus galactomannan antigen testing in pediatric hematopoietic stem cell transplant recipients. Pediatr Infect Dis J. 2007;26(7):558–564. doi:10.1097/INF.0b013e3180616cbb
33. Viscoli C, Machetti M, Gazzola P, et al. Aspergillus galactomannan antigen in the cerebrospinal fluid of bone marrow transplant recipients with probable cerebral aspergillosis. J Clin Microbiol. 2002;40(4):1496–1499. doi:10.1128/JCM.40.4.1496-1499.2002
34. Shenai S, Ronacher K, Malherbe S, et al. Bacterial loads measured by the xpert MTB/RIF assay as markers of culture conversion and bacteriological cure in pulmonary TB. PLoS One. 2016;11(8):e0160062. doi:10.1371/journal.pone.0160062
35. Samuel CM, Whitelaw A, Corcoran C, et al. Improved detection of Pneumocystis jirovecii in upper and lower respiratory tract specimens from children with suspected pneumocystis pneumonia using real-time PCR: a prospective study. BMC Infect Dis. 2011;11:329. doi:10.1186/1471-2334-11-329
36. Becker MJ, de Marie S, Fens MH, Verbrugh HA, Bakker-Woudenberg IA. Effect of amphotericin B treatment on kinetics of cytokines and parameters of fungal load in neutropenic rats with invasive pulmonary aspergillosis. J Antimicrob Chemother. 2003;52(3):428–434. doi:10.1093/jac/dkg367
37. Marr KA, Laverdiere M, Gugel A, Leisenring W. Antifungal therapy decreases sensitivity of the Aspergillus galactomannan enzyme immunoassay. Clin Infect Dis. 2005;40(12):1762–1769. doi:10.1086/429921
38. Prachayangprecha S, Schapendonk CM, Koopmans MP, et al. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J Clin Microbiol. 2014;52(10):3722–3730. doi:10.1128/JCM.01641-14
39. Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187(10):1067–1075. doi:10.1164/rccm.201210-1913OC
40. Yu G, Gail MH, Consonni D, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17(1):163. doi:10.1186/s13059-016-1021-1
41. Hu R, Jiang XY, Wu Y. Risk factors for invasive pulmonary fungal infection in patients with hematological malignancies not receiving hematopoietic stem cell transplant. Neoplasma. 2012;59(6):669–675. doi:10.4149/neo_2012_085
42. Kontoyiannis DP, Lewis RE. How I treat mucormycosis. Blood. 2011;118(5):1216–1224. doi:10.1182/blood-2011-03-316430
43. Kurosawa M, Yonezumi M, Hashino S, et al. Epidemiology and treatment outcome of invasive fungal infections in patients with hematological malignancies. Int J Hematol. 2012;96(6):748–757. doi:10.1007/s12185-012-1210-y
44. Simner PJ, Miller S, Carroll KC. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis. 2018;66(5):778–788. doi:10.1093/cid/cix881
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

