Back to Journals » International Medical Case Reports Journal » Volume 14
Machine Learning in an Elderly Man with Heart Failure
Authors Koops J
Received 5 June 2021
Accepted for publication 21 July 2021
Published 28 July 2021 Volume 2021:14 Pages 497—502
DOI https://doi.org/10.2147/IMCRJ.S322827
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ronald Prineas
Joel Koops
Memorial University of Newfoundland and Labrador, Discipline of Family Medicine, Health Sciences Centre, St. John’s, NL, A1B 3V6, Canada
Correspondence: Joel Koops
Ross Family Medicine Clinic, Dr. Leonard A. Miller Center, 6th Floor Southcott Hall, 100 Forest Road, St. John’s, NL, A1A 1E5, Canada
Tel +1 709 777 6301
Fax +1 709 777 8323
Email [email protected]
Abstract: Machine learning is a branch of artificial intelligence and can be used to predict important outcomes in a wide variety of medical conditions. With the widespread use of electronic medical records, the vast amount of data required for this process is now readily available. The following case demonstrates the application of machine learning to an elderly man with heart failure. The algorithms used, namely, decision tree and random forest, both correctly differentiated heart failure with preserved ejection fraction from heart failure with reduced ejection fraction. This has important treatment and prognostic ramifications and can be completed at the point of care while awaiting confirmation via echocardiogram. Viewing the machine learning process through a patient-centered lens, as in this case, highlights the key role we as physicians have in the implementation and supervision of machine learning.
Keywords: artificial intelligence, decision tree, random forest, prediction, heart failure
Introduction
Machine learning (ML) uses large data sets and algorithms to learn as well as predict outcomes without following explicit instructions. It is a branch of artificial intelligence (AI) and while mainstream now, it is not a new concept. The idea of AI was first introduced over 70 years ago by Alan Turing who envisioned making machines that could pass his so-called imitation test and be viewed as intelligent.1 The digitization of electronic health records (EHRs) has provided the tremendous amount of data required for AI to learn. Already, ML has been used to detect skin cancers and lung nodules as well as predict important outcomes such as opioid misuse and emergency department visits.2–5 Physicians are uniquely situated to strengthen the performance of AI tools as we not only collect and interpret data across anatomy but also delivery systems, such as mental and public health.6
Heart failure (HF) is a complex syndrome with multiple variables as well as processes at play and therefore lends itself well to the machine learning process. It is a common condition encountered in primary care with a lifetime risk of around 20%.7 Diagnosis requires clinical suspicion based on an extensive list of varied signs and symptoms that may include dyspnea, orthopnea, paroxysmal nocturnal dyspnea, fatigue, cough, dependent edema, elevated jugular venous pressure, rales, wheezing and a third heart sound (S3), to name a few.8 Ancillary testing often includes chest radiography, an electrocardiogram, lab work, and brain natriuretic peptides (BNP). The gold standard for confirming the diagnosis of HF is echocardiography, which can assess systolic and diastolic ventricular function, chamber sizes, wall thickness, pericardial disease as well as valvular function.8
HF can be further subdivided into HF with preserved ejection fraction (HFrEF) and HF with reduced ejection fraction (HFpEF).8 The former is defined as an ejection fraction (EF) <40% with signs and symptoms of HF.8 The diagnostic criteria for HFpEF vary with no clear consensus or single guideline.9 It is typically defined as an EF >50% with signs and symptoms of HF as well as other criteria.9 The H2FPEF score can be used to estimate the probability of HFpEF versus noncardiac causes of symptoms.10 While there has been no independent external validation of this rule, it does provide some insight into the risk factors for HFpEF. Its variables are composed of heavy: body mass index >30 kg/m2 (2 points), hypertensive: hypertensive and treated with two or more antihypertensives (1 point), atrial fibrillation: paroxysmal or persistent (3 points), pulmonary hypertension: pulmonary artery systolic pressure >35 mmHg using Doppler echocardiography (1 point), elder: age >60 (1 point), and filling pressure: Doppler echocardiographic E/e’ >9 (1 point).10 The score is the sum of points with 0–1 signifying a low probability of HFpEF, ≥2 an intermediate probability, and 6–9 a high probability.10 A limitation of this score is the requirement for parameters measured by echocardiography.
The distinction between HFrEF and HFpEF is particularly relevant in the clinical setting as it has important prognostic as well as treatment ramifications. The baseline treatment for all patients with symptomatic HFrEF consists of triple therapy, which is comprised of a beta-blocker, an angiotensin converting enzyme (ACE) inhibitor, and a mineralocorticoid receptor antagonist (MRA).11 While these medications have proven benefit in the treatment of HFrEF, beta-blockers and ACE inhibitors are not recommended in HFpEF barring alternative indications and MRAs should be avoided due to concerns regarding adverse effects.11,12 Differentiating between HFrEF and HFpEF requires an echocardiogram, which is often not available at the time of diagnosis and treatment decisions may need to be made prior to obtaining this data. The following case exemplifies the application of ML, namely the decision tree and random forest algorithms, in an elderly man with chronic heart failure. The goal is to determine if it can discriminate between HFrEF and HFpEF based on risk factors and common laboratory tests to better guide treatment as well as discussion with the patient while awaiting confirmation via echocardiography.
Case Description
In early March 2020, an 85-year-old male presented to clinic for follow-up after being diagnosed with congestive heart failure in the emergency department one month prior. His presenting symptoms at that time were shortness of breath on exertion as well as orthopnea. Electrocardiogram demonstrated sinus rhythm with first degree AV block as well as a left bundle branch block (LBBB) that had been documented previously in 2017. The chest x-ray showed mild pulmonary edema as interpreted by the emergency room physician. BNP levels are not available in the region where this patient resides. Medications started in the emergency department included furosemide 20 mg daily as well as ramipril 2.5 mg daily. The patient stated he felt much better and only experienced mild dyspnea with prolonged exertion. Physical exam was unremarkable and vital signs were within normal limits. The patient has a history of benign prostatic hyperplasia and gastroesophageal reflux for which he takes dutasteride 0.5 mg daily, tamsulosin 0.8 mg daily, and rabeprazole 20 mg daily. There is no other past medical history, specifically no diabetes, hypertension, dyslipidemia, kidney disease, sleep apnea or anemia. He quit smoking over 50 years ago and his body mass index is normal. Laboratory investigations immediately prior to starting furosemide and ramipril showed a serum creatinine of 105 µmol/L, sodium of 139 mmol/L, potassium of 4.9 mmol/L, hemoglobin of 140 g/L, and platelets of 212 x 109/L. An echocardiogram had been arranged but would not be completed for several months, possibly longer due to the pandemic restrictions in place. The patient wondered whether he needed to continue the ACE inhibitor and questioned if there were any other medications that would be helpful for his condition.
As the patient had no other medical co-morbities, determining whether this was HFpEF or HFrEF had important implications regarding whether the patient should remain on an ACE inhibitor or if a beta-blocker and MRA would be of benefit. While awaiting the results of the echocardiogram, ML was implemented in the hopes of determining this distinction at the point of care.
The algorithm returned a diagnosis of HFrEF. This result was not entirely unanticipated as a lean male without hypertension or atrial fibrillation is not a classical HFpEF patient. A LBBB is also associated with a decreased ejection fraction, often in the absence of obvious cardiovascular disease.13 While a certain degree of confidence could be placed on this result, it was decided further treatment would await the echocardiogram. He remained stable on these medications with no further exacerbations of his HF. The echocardiogram completed 3 months later demonstrated decreased systolic function with an ejection fraction of 26%. He was subsequently started on metoprolol as well as spironolactone and is currently awaiting cardiac catheterization.
Discussion
ML can primarily be categorized into two fields, supervised learning and unsupervised learning. In supervised learning, the data is first divided into a training and test dataset with the former being used to train the underlying algorithm.14 The test dataset is then fed into the trained algorithm with the goal of predicting an outcome.14 In contrast, the objective of unsupervised learning is not to make any predictions but to find patterns or groupings within a dataset.15 As the aim for this case is to predict an outcome, namely HFrEF or HFpEF, a supervised algorithm was chosen.
There are innumerable variations of supervised ML algorithms, all with certain advantages and disadvantages. Logistic regression is a well-known method based on ordinary regression but can only be used to predict the probability of an outcome and not the outcome itself.16 Neural networks are inspired by the human brain and consist of a predetermined number of interconnected nodes.16 These algorithms contain hidden layers and can grow to be complex making them difficult to follow as well as refine.16 The most common and straightforward of the supervised algorithms is decision trees. Decision tree learning builds trees top-down based on a set of hierarchical rules to make predictions for instances.16 The nature of decision trees makes them easy to understand as well as refine if necessary and was therefore the method used for this case. All algorithms were written and executed in Google Colab.
Before making predictions, the first step in the ML process is training the model. In this instance, it involved using a public dataset that is freely available at multiple online repositories and incorporated 299 heart failure patients whose ejection fractions were recorded.17 The included attributes were whether the patient smokes, has diabetes, hypertension and/or anemia, their age as well as serum platelets, sodium and creatinine.17 One attribute available in the dataset, namely creatinine kinase, was not included as the assays for measuring this variable differed. Fifteen percent of the data were then randomly chosen to be a test set and removed. The remaining data were used to train the model. The total computational time from data import to model training took 7.1 seconds. The test dataset was subsequently used to determine the accuracy of the model, which was calculated as 60%. It is important to note that this is the theoretical accuracy as it applies to the model’s ability to predict itself. The model required the patient’s corresponding information prior to making any predictions regarding his diagnosis. The algorithm was asked to make a prediction of whether the patient had HFpEF or HFrEF based on a cutoff of 40%, as this had the most important implications in terms of treatment. The ML process correctly identified the patient as having HFrEF. Total computational time for this prediction was 0.01 seconds.
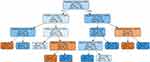
The corresponding decision tree provides a visual representation of the ML process for this case (Figure 1). Each box is termed a leaf or node.16 The root node is the most superior leaf, which contains all the training data to grow the tree.16 The tree grows downward by dividing the data at each level until it ends at a terminal node.16 By default, the number of terminal nodes are typically determined by the algorithm. This usually results in a high number of terminal nodes and a complex tree that is difficult to follow. This may also result in overfitting of the data to the training set, rendering the model useless for real world predictions. For ease of illustration and to reduce complexity, ten terminal nodes were used. Increasing this number had no effect on the outcome or accuracy of the model. Unexpectedly, the decision tree used serum creatinine as the primary rule for differentiating HFrEF from HFpEF. Factors, such as smoking status, age, hypertension and anemia were not included. Physicians understand all too well the importance of these issues in the treatment as well as the prevention of HF. This may have been lost on a software programmer and further exemplifies the partnership we as health care providers need to have in the development and implementation of machine learning in medicine.
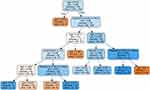
The result seemed suspect given the heavy reliance on creatinine and therefore required further testing to ensure accuracy. The random forest algorithm is a branch of decision tree ML that combines multiple decision trees using different attributes to predict an outcome.16 The default number of trees is one hundred but can be adjusted prior to algorithm implementation. For ease of illustration and to reduce complexity, ten trees were used in conjunction with ten terminal nodes so that each could be directly compared with the decision tree ML approach. Increasing either of these numbers had no effect on the outcome or accuracy of the model. Each tree places a different amount of emphasis on specific features and can be visualized separately (Figure 2). In contrast to the decision tree ML approach, this particular tree utilized age as the primary rule for differentiating HFrEF from HFpEF and is applied in conjunction with other trees to make a final prediction. Accuracy for this method using the same training and test datasets increased to 73% with a computational time of 0.21 seconds. This approach again correctly identified the diagnosis of HFrEF, albeit placing more emphasis on the factors left out in the decision tree process (Figure 3). Specifically, the random forest approach placed particular significance on age, which is relevant in this case. The agreement between these two methods increased confidence in the result, suggesting ML can correctly differentiate between HFrEF and HFpEF in a clinical setting.
Conclusion
ML can effectively be used at the point of care to further quantify and make treatment decisions in patients suspected of having HF. EMRs can offer the massive data required to apply ML to a wide range of medical conditions. Physicians provide patient centered care using an extensive as well as varied knowledge base and are uniquely positioned to be an integral part of the ML revolution in medicine.
Abbreviations
ML, machine learning; AI, artificial intelligence; EHR, electronic health records; HF, heart failure; BNP, brain natriuretic peptide; HFrEF, heart failure with reduced ejection fraction; HFpEF, heart failure with preserved ejection fraction; EF, ejection fraction; ACE, angiotensin converting enzyme; MRA, mineralocorticoid receptor antagonist; LBBB, left bundle branch block.
Informed Consent/Institutional Approval
Informed consent was obtained from the patient to publish their case details. This case did not require any institutional approval as per the HREA of Newfoundland and Labrador.
Disclosure
The author reports no conflicts of interest in this work.
References
1. Turing A. Computing machinery and intelligence. Mind. 1950;49(236):433–460. doi:10.1093/mind/LIX.236.433
2. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi:10.1038/nature21056
3. Henschke CI, Yankelevitz DF, Mateescu I, Brettle DW, Rainy TG, Weingard FS. Neural networks for the analysis of small pulmonary nodules. Clin Imaging. 1997;21(6):390–399. doi:10.1016/S0899-7071(97)81731-7
4. Lo-Ciganic WH, Huang JL, Zhang HH, et al. Evaluation of machine-learning algorithms for predicting opioid overdose risk among medicare beneficiaries with opioid prescriptions. JAMA Netw Open. 2019;2(3):e190968. doi:10.1001/jamanetworkopen.2019.0968
5. Rahimian F, Salimi-Khorshidi G, Payberah AH, et al. Predicting the risk of emergency admission with machine learning: development and validation using linked electronic health records. PLoS Med. 2018;15(11):e1002695. doi:10.1371/journal.pmed.1002695
6. Liaw W, Kakadiaris IA. Artificial intelligence and family medicine: better together. Fam Med. 2020;51(1):8–10. doi:10.22454/FamMed.2020.881454
7. Lloyd-Jones DM, Larson MG, Leip EP, et al.; Framingham Heart Study. Lifetime risk for developing congestive heart failure: the Framingham Heart Study. Circulation. 2002;106(24):3068–3072. doi:10.1161/01.CIR.0000039105.49749.6F
8. Ezekowitz JA, O’Meara E, McDonald MA, et al. 2017 Comprehensive Update of the Canadian Cardiovascular Society Guidelines for the Management of Heart Failure. Can J Cardiol. 2017;33(11):1342–1433.
9. Lekavich CL, Barksdale DJ, Neelon V, Wu JR. Heart failure preserved ejection fraction (HFpEF): an integrated and strategic review. Heart Fail Rev. 2015;20(6):643–653. doi:10.1007/s10741-015-9506-7
10. Reddy YNV, Carter RE, Obokata M, Redfield MM, Borlaug BA. A simple, evidence-based approach to help guide diagnosis of heart failure with preserved ejection fraction. Circulation. 2018;138(9):861–870. doi:10.1161/CIRCULATIONAHA.118.034646
11. Chavey WE, Hogikyan RV, Van Harrison R, Nicklas JM. Heart failure due to reduced ejection fraction: medical management. Am Fam Physician. 2017;95(1):13–20.
12. Gazewood JD, Turner PL. Heart failure with preserved ejection fraction: diagnosis and management. Am Fam Physician. 2017;96(9):582–588.
13. Akhtari S, Chuang ML, Salton CJ, et al. Effect of isolated left bundle-branch block on biventricular volumes and ejection fraction: a cardiovascular magnetic resonance assessment. J Cardiovasc Magn Reson. 2018;20(1):66. doi:10.1186/s12968-018-0457-8
14. Dey A. Machine learning algorithms: a review. Int J Comput Sci Inf Technol. 2016;7(3):1174–1179.
15. Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–1930. doi:10.1161/CIRCULATIONAHA.115.001593
16. Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19(1):1–6. doi:10.1186/s12911-019-1004-8
17. Ahmad T, Munir A, Bhatti SH, Aftab M, Raza MA. Survival analysis of heart failure patients: a case study. PLoS One. 2017;12(7):e0181001. doi:10.1371/journal.pone.0181001
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.