Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding RNA (lncRNA) MT1JP Suppresses Hepatocellular Carcinoma (HCC) in vitro
Authors Mo W , Dai Y, Chen J, Liang L, Xu S, Xu X
Received 11 March 2020
Accepted for publication 22 July 2020
Published 2 September 2020 Volume 2020:12 Pages 7949—7960
DOI https://doi.org/10.2147/CMAR.S253496
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Wenhui Mo,* Ying Dai,* Jianqing Chen, Liwei Liang, Shuqi Xu, Xuanfu Xu
Department of Gastroenterology, Shidong Hospital of Shanghai, Shanghai 200433, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xuanfu Xu
Department of Gastroenterology, Shidong Hospital of Shanghai, Shanghai 200433, People’s Republic of China
Tel +86-21-25066666
Email [email protected]
Introduction: The purpose of this study was to evaluate the effects and mechanisms of the long noncoding RNA (lncRNA) MT1JP on hepatocellular carcinoma (HCC) in vitro.
Patients and Methods: Thirty pairs of tumor and adjacent normal tissues were collected from HCC patients. Tissue pathology and MT1JP expression were evaluated by hematoxylin and eosin staining and in situ hybridization (ISH), respectively. The correlation between MT1JP and HCC prognosis was investigated. MTT assays, cloning, flow cytometry, transwell assays, and wound-healing assays were used to evaluate the effects of MT1JP on HCC cell lines. RT-qPCR and Western blot were used to measure the relative mRNA and protein expression levels.
Results: The expression of MT1JP was downregulated in HCC tumor tissues compared with that in adjacent normal tissues, while the percent survival was significantly greater in the high MT1JP expression group than in the low MT1JP expression group (P=0.0238). In vitro, overexpression of MT1JP suppressed the proliferation, invasion, and migration, reduced colony cell number, increased cell apoptosis, and induced G1-phase cell cycle arrest in Bel-7402 and Huh-7 cells. Meanwhile, the mRNA and protein expression levels of RUNX3 and P21 were significantly upregulated, whereas those of MMP2 and MMP9 were significantly downregulated, in Bel-7402 and Huh-7 cells overexpressing MT1JP (all P< 0.001).
Conclusion: LncRNA MT1JP may function as a tumor suppressor in HCC. Overexpression of MT1JP suppressed HCC cell biological activities through the regulation of RUNX3.
Keywords: MT1JP, HCC, Bel-7402, Huh-7, cell biological activities
Introduction
Hepatocellular carcinoma (HCC) has a high rate of morbidity and mortality in developing countries;1 however, its etiology and underlying molecular mechanism remain poorly understood. The pathogenesis of HCC can be attributed to several factors, including the environment, genetics, hepatitis virus infection, cirrhosis, aflatoxin, and nitrosamines. The early symptoms of HCC are often nonspecific, with no significant positive vital signs or similar signs of cirrhosis, and therefore most patients present with advanced disease that is not amenable to curative therapy. The lack of effective diagnostic and therapeutic targets for HCC remains a problem in clinical practice, and it is important that HCC-related biomarkers are identified.
Long noncoding RNAs (lncRNAs) comprise a group of RNAs that do not encode proteins. LncRNAs have been implicated in the pathology of many diseases, especially the development of malignant tumors, making them an important focus of basic and clinical oncological research.2,3 Numerous studies have shown that specific lncRNAs are abnormally expressed in various tumors, regulating the biological activities of cancer cells and exerting important regulatory effects on tumor development and progression. For instance, lncRNA HAND2-AS1 is significantly downregulated in endometrioid carcinoma tissues and functions as a tumor suppressor,4 while lncRNA GPR65-1 is significantly upregulated in gastric cancer tissues and can promote tumor growth via the PTEN-AKT pathway.5 In addition, many lncRNAs can serve as independent risk factors in predicting the prognosis of patients with malignant tumors. The expression of lncRNA AB073614, for example, is significantly increased in glioma tissues and increases with increasing tumor grade. Elevated expression of lncRNA AB073614 is an independent risk factor for poor prognosis in patients with glioma.6 However, relatively few studies have reported on the role of lncRNA MT1JP in HCC, and the ability to predict the prognosis of this disease remains limited. Therefore, in this study, we determined the expression of MT1JP in HCC tissues and evaluated the clinical significance of MT1JP in the diagnosis and treatment of HCC. Moreover, the ability of lncRNA MT1JP to inhibit HCC and the potential underlying mechanisms were also investigated using cell-based assays.
Patients and Methods
General Information
Thirty patients (17 men and 13 women) with HCC who underwent hepatic resection in Shidong Hospital, Shanghai, between January 2013 and March 2014 were enrolled. The mean age of the patients was 54.6 years (range: 37–70 years). Thirteen of the patients presented with TNM stage I–II and 17 with stage III–IV disease, and pathological grading based on Edmondson criteria suggested that 15 of the patients presented with grade I–II and 15 with grade III–IV disease. HCC tissue samples were obtained from patients with pathologically confirmed HCC according to the diagnostic criteria recommended by the World Health Organization (WHO). Adjacent liver tissue was taken 3 cm from the margin of the corresponding tumor. Some of the tissue was immediately stored in liquid nitrogen at −80° C for RNA extraction. None of the patients had previously received radiotherapy, chemotherapy, intervention, or radiofrequency ablation and were regularly followed up after surgery until March 2018. This study was approved by the ethics committee of Shidong Hospital, Shanghai. All patients provided written informed consent, and that this was conducted in accordance with the Declaration of Helsinki.
Extraction and Measurement of lncRNA MT1JP
Total RNA was extracted from HCC tissue and paracancerous tissue samples, as well as from cultured cells, using Trizol reagent (Invitrogen, CA, US), and cDNA was synthesized by reverse transcription depending on instructions. The expression of MT1JP in HCC tissues and paracancerous tissues was determined by qPCR using the cDNA as a template and GAPDH as an internal reference. The sequences of the primers used are shown in Table 1.
 |
Table 1 Experimental Primer Sequence |
In situ Hybridization (ISH) of MT1JP
Tissue slices were dewaxed and digested with proteinase K (200 mg/L) at 37 °C for 15 min. The sections were first rinsed with 0.1 mol/L glycine/PBS for 1 min, then with twice with DEPC–PBS for 5 min each wash, and dehydrated using an ethanol gradient. The slices were then incubated with prehybridization solution at 55°C for 30 min. Simultaneously, probe hybridization working solution was heated at 90°C for 4 min and placed on ice for 10 min. The slices were incubated in this probe hybridization working solution at 55°C for 1 h and then washed with 5×, 1×, and 0.2× saline sodium citrate (SSC) buffer at 55°C, 5 min each wash. The slices were incubated with 1× wash buffer at room temperature (RT) for 15 min and then with anti-DIG-AP solution (1:800, diluted in 1× blocking buffer) at RT for 1 h. After washing three times with 1× washing buffer at RT, 10 min each wash, the slices were equilibrated in 1× detection buffer at 30°C for 5 min, and then developed with BCIP/NBT solution in 1× detection buffer (1:50) at 30°C for 30 min in the dark. After washing twice with KTBT buffer at RT, 5 min each wash, and twice with H2O, 1 min each wash, the slices were counterstained with nuclear fast red for 30 min. Slices were mounted and observed under a microscopic, and MT1JP expression was evaluated using ImageJ software.
Cell Culture and Transfection
The normal hepatocyte L-02 cell line and the hepatoma cell lines Bel-7402, Huh-7, SMMC-7721, MHCC97, and HepG2 were purchased from the Shanghai Institute of Biochemistry and Cell Biology, CAS (Shanghai, China). All the cells were cultured in RPMI-1680 medium at 37°C in an incubator with 5% CO2 and subcultured after 48 h. MT1JP-containing pcDNA3.1 plasmids were transfected into cells. Bel-7402 and Huh-7 cells were each divided into three groups: a normal control group (Normal), blank control group (pcDNA3.1), and lncRNA MT1JP-overexpression group (MT1JP).
MTT Assay
Treated hepatoma cells were cultured to log phase and 2 × 103 cells were seeded in each well of a 96-well cell culture plate with 6 replicates per group. Wells with culture medium alone were used as the blank control. After culturing for 48 h, 20 μL of a 5 mg/mL thiazolyl blue tetrazolium bromide (MTT) solution was added to each well and the cells were incubated for another 4 h. The culture medium was removed and 150 μL of dimethyl sulfoxide solution was added to each well and mixed for 10 min. When the crystals had dissolved, the optical density (OD value) was measured at a wavelength of 490 nm with a microplate reader and cell viability was calculated.
Colony Formation Assay
Log-phase cells were digested and counted, and 1000 cells were inoculated in each well of a 6-well plate with 2 mL of culture medium in each well. The culture medium was replaced every 4 days for 2 weeks. The medium was subsequently removed and the cells fixed in 4% paraformaldehyde, followed by staining with 0.05% crystal violet. The cells were observed, imaged, and counted.
Flow Cytometry for Cell Apoptosis
A total of 1.5×106 transfected cells were incubated with 0.25% trypsin, harvested and rinsed twice with pre-chilled PBS. The cells stained with annexin V and PI. Cell apoptosis was determined by a flow cytometer. The experiment was repeated three times.
PI Staining Assay for Cell Cycle Analysis
Cells of each group were collected and adjusted to a density of 1 × 106 cells/mL. A 1-mL volume of the cell suspension was taken and PBS was removed by centrifugation. These cells were fixed in 1 mL of precooled 70% ethanol at 4°C overnight. After washing twice with cold PBS, 150 μL of PI staining solution and 150 μL of RNase A solution were added and the cells were incubated for 30 min at 4°C in the dark. For flow cytometric analysis, 1 × 104 cells were used from each sample. Curve-fitting analysis was performed using FACSort CeU Quest software (BD, San Diego, CA, USA) to calculate DNA content, and the percentage of cells at each stage of the cell cycle (G1, S, and G2 phases) was calculated. The experiment was repeated three times.
Transwell Assay for Cell Invasion Ability
For each group, 3 × 104 cells were inoculated on the surface of a Transwell chamber and cultured at 37°C for 24 h. The remaining cells on the bottom of the basement membrane were fixed in formaldehyde and stained with 0.2% crystal violet solution for 10 min. Ten fields were randomly examined under a microscope (200× magnification), and the number of invading cells was counted. The experiment was repeated three times.
Wound-Healing Assay for Evaluation of Cell Migratory Ability
Cells were seeded at 1 × 104 cells/well in a 24-well plate. When the cells had reached 90% confluency, a 20-μL pipette tip was used to scratch the cell monolayer along the diameter of the 24-well plate, and floating cells were washed off with PBS. Fresh medium containing 2% fetal bovine serum was added and the cells cultured. The width of the wounds was observed and measured at 0, 24, and 48 h, and the migratory ability of these cells was calculated. The experiment was repeated three times.
Western Blot (WB) for Protein Expression Analysis
Cells were collected and lysed by RIPA buffer. The cell lysate was denatured and loaded at 30 μg/well for electrophoresis separation by SDS-PAGE. Then, the proteins were routinely transferred to PVDF membrane, which was incubated with primary antibodies (anti-RUNX3, anti-P21, anti-MMP2, anti-MMP9, and anti-GAPDH, 1:200 dilution) at 4°C overnight. After washing three times with PBST (PBS + Triton), the anti-rabbit horse-radish peroxidase-conjugated secondary antibodies (1:1000) was added followed by incubation at 37°C for 4 h. After washing three times with PBST, the membrane was developed using ECL solution (Thermo Fishes Scientific, Waltham, MA, USA). The gray value of the target protein was analyzed by Quantity One 1-D software and the relative expression of the target protein was calculated as the gray value of the target protein/gray value of GAPDH. The experiment was repeated three times.
Statistical Analysis
All data were processed with SPSS 20.0 statistical software. An independent samples t-test, χ2 test, or Fisher’s exact probability test for categorical data was employed. Kaplan–Meier survival curves were plotted for each of the patient groups with differing lncRNA MT1JP expression levels and the difference in survival rate between the two groups was compared by the Log rank test. Measurement data were expressed as means ± SD and compared by t-test between two groups. For comparison among three groups, data were initially subjected to analysis of variance followed by the LSD t-test when the difference was statistically significant. A statistical significance level of α = 0.05 was used.
Results
Relationship of the MT1JP Expression Level in HCC Tissue or Adjacent Liver Tissue with Pathology and Prognosis
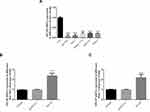
Hematoxylin and eosin (H&E) histological examination (Figure 1A) revealed blurred boundaries between cells in HCC tissue with obvious infiltration, whereas the cell boundaries in normal liver tissue were clear and no obvious infiltration was observed. The ISH assay showed that the expression level of MT1JP in HCC tissue was significantly lower than that in corresponding adjacent liver tissue (P<0.001, Figure 1B). The results of RT-qPCR analysis also confirmed that the expression level of MT1JP in HCC tissue was significantly decreased compared with that in adjacent tissue (P<0.001, Figure 1C). HCC patients were divided into low- and high-expression groups according to the median level of MT1JP lncRNA and survival curve analysis indicated that the postoperative survival rate of patients in the low-expression group was significantly reduced compared with that of the high-expression group (P=0.0238, Figure 1D).
Relationship Between MT1JP and Clinicopathological Features of HCC Patients
The relationship between the clinicopathological features of these HCC patients and the lncRNA MT1JP expression level was analyzed by the χ2 test. The analysis showed that low expression of MT1JP in HCC tissue was correlated with Edmondson grade (P=0.005), TNM stage (P=0.001), degree of differentiation (P=0.025), and tumor size (P=0.013), while no correlation was observed with the age and gender of the patients (P=0.615, P=0.984, Table 2).
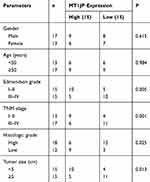 |
Table 2 Correlation of the Expression Level of lncRNA MT1JP with the Clinicopathological Characteristics of Hepatocellular Carcinoma Patients |
Expression of MT1JP in Cell Lines and Groups
The expression of MT1JP in the different cell lines was evaluated by RT-qPCR. The results showed that the expression of MT1JP in hepatoma cells was significantly lower than that in normal liver cells (all P<0.001), with the lowest expression being observed in Bel-7402 and Huh-7 cells (Figure 2A). Following transfection of MT1JP, the expression of MT1JP in the MT1JP-overexpression group was significantly increased compared with that of the Normal group in Bel-7402 and Huh-7 cells (all P<0.001, Figure 2B and C).
Effects of MT1JP on the Proliferative and Clonogenic Ability of Hepatoma Cells
The MTT assay showed that transfection of MT1JP-overexpression plasmids into Bel-7402 and Huh-7 hepatoma cells markedly inhibited their viability (P<0.001, Figure 3A). The colony formation assay also confirmed that clonogenicity was significantly decreased in Bel-7402 (P<0.001, Figure 3B) and Huh-7 (P<0.001, Figure 3C) cells following transfection with MT1JP-expressing plasmids.
Effects of MT1JP A on Apoptosis and Cell Cycle Progression in Hepatoma Cells
Flow cytometry was applied to test the effects of MT1JP on apoptosis and the cell cycle in each group. The results showed that the apoptosis rates of Bel-7402 and Huh-7 cells were significantly increased after transfection with MT1JP-expressing plasmids (all P<0.001, Figure 4A and B). Moreover, the ratio of Bel-7402 and Huh-7 cells at the G1 phase of the cell cycle significantly increased (P<0.001, Figure 4C and D), while the ratio of cells at the G2 phase significantly decreased (P<0.001, Figure 4C and D).
Effect of MT1JP on Invasive Capacity of Hepatoma Cells
Results of the Transwell assay demonstrated that the number of invading Bel-7402 and Huh-7cells in the MT1JP-overexpression group was significantly inhibited compared with that of the Normal group (P<0.001 for both, Figure 5A and B).
Effect of MT1JP on the Migratory Ability of Hepatoma Cells
The wound-healing assay showed that the migratory ability of Bel-7402 and Huh-7 cells transfected with MT1JP for 24 h and 48 h was significantly inhibited compared with that of the Normal group (P<0.01 or P<0.001, respectively; Figure 6A and B).
Effect of MT1JP on the Expression of Relevant Genes
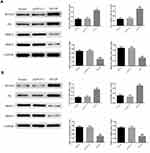
The results of RT-qPCR showed that the expression of the RUNX3 and P21 genes was significantly increased, whereas that of MMP2 and MMP9 was markedly inhibited in Bel-7402 and Huh-7 cells overexpressing MT1JP (P<0.001, Figure 7A and B).
Effect of MT1JP on the Expression of Relevant Proteins
Western blot analysis indicated that the protein expression of RUNX3 and P21 was significantly upregulated, whereas that of MMP2 and MMP9 was markedly downregulated in Bel-7402 and Huh-7 cells overexpressing MT1JP (P<0.001, Figure 8A and B).
Discussion
The diverse roles of lncRNAs in the development, progression, and metastasis of various cancers, especially malignant tumors, has become a focus of oncological research in recent years.7 Studies have shown that abnormally expressed lncRNAs are widely involved in the regulation of proliferation, differentiation, apoptosis, angiogenesis, migration, and invasion of various tumor cells through a multitude of pathways and molecular mechanisms. LncRNAs can serve as biomarkers for early tumor diagnosis, treatment, and prognosis and targeting lncRNAs provides a new strategy for the diagnosis and treatment of malignant tumors.8 Studies have suggested that the expression of lncRNA MT1JP is downregulated in the tissue of various cancers, including gastric cancer, lung cancer, and tongue squamous cell carcinoma, and that the survival rate of cancer cells is decreased after the transfection of MT1JP-overexpressing vectors.9,10 In the present study, the expression of MT1JP in HCC tissues was initially determined by ISH and RT-qPCR, following which the correlation between MT1JP expression and the pathology and prognosis of HCC was analyzed. The results showed that the survival rate of patients whose HCC tissue exhibited high MT1JP expression levels was significantly better than that of patients whose HCC tissue presented low expression levels of MT1JP. Meanwhile, the size and type of tumor in HCC patients with high MT1JP expression levels improved compared with those of patients exhibiting low MT1JP expression. Accordingly, we hypothesized that MT1JP overexpression may inhibit the biological activity of hepatoma cells. To test this, we selected Bel-7402 and Huh-7 cells with the lowest expression level of MT1JP to conduct the relevant assays. We found that after the transfection of MT1JP-expressing vectors, the proliferative, invasive, and migratory potential of Bel-7402 and Huh-7 cells was significantly inhibited, while cell apoptosis was significantly enhanced.
The RUNX3 gene has critical roles in cell growth, apoptosis, and tumor growth. The RUNX3 protein can regulate cell growth and apoptosis, as well as signal transduction and biological processes, through a variety of complex mechanisms.11,12 The expression of RUNX3 was found to be downregulated or even absent in malignant tumors such as gastric cancer.13,14 This reduced expression of RUNX3 was accompanied by enhanced proliferation and reduced cell apoptosis in gastrointestinal epithelial cells, leading to their abnormal growth and differentiation, and consequently tumorigenesis. Molecular biology studies have confirmed that the mechanisms underlying the reduction of RUNX3 expression are closely related to heterozygous deletions, hypermethylation, and point mutations in the RUNX3 gene.15,16 The results of the present study demonstrated that the expression of RUNX3 in MT1JP cells was significantly upregulated following the transfection of MT1JP-expressing plasmids into hepatoma cells, indicating that MT1JP can inhibit liver cancer by upregulating RUNX3 expression.
Studies have shown that P21 levels are upregulated when RUNX3 is overexpressed, which in turn inhibits cell proliferation by arresting most cells at the G1 phase of the cell cycle, and also enhances cell apoptosis.17,18 Our results showed that the expression level of RUNX3 increased with overexpression of MT1JP, which resulted in a large number of hepatoma cells arresting at the G1 phase as well as increased levels of cell apoptosis. Molecular assays revealed that the main factors contributing to this phenomenon may be related to the upregulation of P21 expression.
The upregulation of MMP2/9 is frequently associated with the invasion and migration of tumor cells.19,20 However, several studies have shown that increased levels of RUNX3 effectively inhibit the expression of MMP2/9 and consequently the invasive and migratory capacity of tumor cells.21,22 Our results suggested that MT1JP may inhibit the invasion and migration of hepatoma cells through the upregulation of RUNX3 and inhibition of MMP2 and MMP9 protein expression.
In summary, MT1JP is of potential significance in HCC. Overexpression of MT1JP can effectively inhibit the biological activity of hepatoma cells and the underlying mechanism may be closely associated with the upregulation of RUNX3 expression.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Zhou M, Zhang XY, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother. 2017;85:348–354. doi:10.1016/j.biopha.2016.11.035
2. Gupta SC, Awasthee N, Rai V, Chava S, Gunda V, Challagundla KB. Long non-coding RNAs and nuclear factor- κB crosstalk in cancer and other human diseases. Biochim Biophys Acta Rev Cancer. 2019;19:188316. doi:10.1016/j.bbcan.2019.188316
3. Bermúdez M, Aguilar-Medina M, Lizárraga-Verdugo E, et al. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front Oncol. 2019;9:1008. doi:10.3389/fonc.2019.01008
4. Yang X, Wang CC, Lee WYW, Trovik J, Chung TKH, Kwong J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018;413:23–34. doi:10.1016/j.canlet.2017.10.028
5. Li Y, Shen Z, Wang B, et al. Long non-coding RNA GPR65-1 is up-regulated in gastric cancer and promotes tumor growth through the PTEN-AKT-slug signaling pathway. Cell Cycle. 2018;17(6):759–765. doi:10.1080/15384101.2018.1426414
6. Hu L, Lv QL, Chen SH, et al. Up-regulation of long non-coding RNA AB073614 predicts a poor prognosis in patients with glioma. Int J Environ Res Public Health. 2016;13(4):433. doi:10.3390/ijerph13040433
7. Dianatpour A, Ghafouri-Fard S. Long non coding RNA expression intersecting cancer and spermatogenesis: a systematic review. Asian Pac J Cancer Prev. 2017;18(10):2601–2610. doi:10.22034/APJCP.2017.18.10.2601
8. He JH, Han ZP, Li YG. Association between long non-coding RNA and human rare diseases (Review). Biomed Rep. 2014;2(1):19–23. doi:10.3892/br.2013.191
9. Liu L, Yue H, Liu Q, et al. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget. 2016;7(13):15787–15800. doi:10.18632/oncotarget.7487
10. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi:10.1038/onc.2011.621
11. Yang HD, Kim PJ, Eun JW, et al. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016;7(10):11412–11423. doi:10.18632/oncotarget.7194
12. Xu SQ, Qin Y, Pan DB, et al. Inhibition of WWP2 suppresses proliferation, and induces G1 cell cycle arrest and apoptosis in liver cancer cells. Mol Med Rep. 2016;13(3):2261–2266. doi:10.3892/mmr.2016.4771
13. Li N, Zheng D, Xue J, et al. Cidan inhibits liver cancer cell growth by reducing COX-2 and VEGF expression and cell cycle arrest. Exp Ther Med. 2015;9(5):1709–1718. doi:10.3892/etm.2015.2351
14. Jin G, Peng L, Zhang J, Qu L, Shou C. Cancer and embryo expression protein 65 promotes cancer cell growth and metastasis. Oncol Lett. 2015;9(4):1772–1778. doi:10.3892/ol.2015.2958
15. Emptage RP, Lemmon MA, Ferguson KM. Molecular determinants of KA1 domain-mediated autoinhibition and phospholipid activation of MARK1 kinase. Biochem J. 2017;474(3):385–398. doi:10.1042/BCJ20160792
16. Li X, Zhao Z, Zhang X, et al. Klf4 reduces stemness phenotype, triggers mesenchymal-epithelial transition (MET)-like molecular changes, and prevents tumor progression in nasopharygeal carcinoma. Oncotarget. 2017;8(55):93924–93941. doi:10.18632/oncotarget.21370
17. Deng T, Zhang Y. Possible involvement of activation of P53/P21 and demethylation of RUNX 3 in the cytotoxicity against Lovo cells induced by 5-Aza-2ʹ-deoxycytidine. Life Sci. 2009;84(9–10):311–320. doi:10.1016/j.lfs.2008.12.015
18. El-Aassar MR, Saad EA, Habib SA, Waly HM. Loading of some quinoxaline derivatives in poly (l-lactic) acid/Pluronic® F-127 nanofibers enhances their anticancer efficiency and induces a p53 and p21 apoptotic-signaling pathway. Colloids Surf B Biointerfaces. 2019;183:110444. doi:10.1016/j.colsurfb.2019.110444
19. Abdollahi A, Nozarian Z, Nazar E. Association between expression of tissue inhibitors of metalloproteinases-1, matrix metalloproteinase-2, and matrix metalloproteinase-9 genes and axillary lymph nodes metastasis in patients with breast cancer. Int J Prev Med. 2019;10:127. doi:10.4103/ijpvm.IJPVM_355_16
20. Ciucă FI, Mărăşescu PC, Matei M, et al. The prognostic value of CXCR4, MMP-2 and MMP-9 in tongue squamous carcinoma. Rom J Morphol Embryol. 2019;60(1):59–68.
21. Chen F, Wang M, Bai J, et al. Role of RUNX3 in suppressing metastasis and angiogenesis of human prostate cancer. PLoS One. 2014;9(1):e86917. doi:10.1371/journal.pone.0086917
22. Li Y, Chen F, Chu J, et al. miR-148-3p inhibits growth of glioblastoma targeting DNA methyltransferase-1 (DNMT1). Oncol Res. 2019;27(8):911–921. doi:10.3727/096504019X15516966905337
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.