Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding RNA DIO3OS Hinders Cell Malignant Behaviors of Hepatocellular Carcinoma Cells Through the microRNA-328/Hhip Axis
Authors Wang Z, Song L, Ye Y, Li W
Received 14 January 2020
Accepted for publication 30 April 2020
Published 25 May 2020 Volume 2020:12 Pages 3903—3914
DOI https://doi.org/10.2147/CMAR.S245990
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Zhanpeng Wang,1 Lina Song,2 Yanshuo Ye,1 Wei Li1
1Department of Hepatobiliary-Pancreatic Surgery, China-Japan Union Hospital of Jilin University, Changchun 130000, People’s Republic of China; 2Department of Laboratory Medicine Center, China-Japan Union Hospital of Jilin University, Changchun 130000, People’s Republic of China
Correspondence: Wei Li
Department of Hepatobiliary-Pancreatic Surgery, China-Japan Union Hospital of Jilin University, No. 126 Xiantai Street, Changchun 130000, People’s Republic of China
Tel/ Fax +86-431-89876756
Email [email protected]
Background: The decline of a long non-coding RNA (lncRNA) DIO3OS was implicated in a plethora of cancers, while the relevance in hepatocellular carcinoma (HCC) has not been mentioned. Accordingly, we set to determine the functional role of DIO3OS and the molecular mechanism in HCC progression.
Materials and Methods: The differentially expressed lncRNAs, mRNAs, and microRNAs (miRNAs) were obtained through the datasets GSE101728 and GES57555. Afterwards, DIO3OS was enhanced in HCC cells to examine the behavior changes. Subcellular localization of DIO3OS was determined through website prediction and experimental validation. The expression of Hedgehog (Hh) signaling pathway-related genes was detected. The effects of DIO3OS overexpression on tumor growth were evaluated as well.
Results: DIO3OS was lower in HCC tissues and cells, while upregulation of DIO3OS repressed malignant biological behavior both in vitro and in vivo. DIO3OS, localized in the cytoplasm, inhibited the occurrence of HCC by disrupting the Hh pathway by sponging miR-328 to mediate Hh interacting protein (Hhip).
Conclusion: All in all, the obtained data suggested that DIO3OS interacted with Hhip-dependent Hh signaling pathway to inhibit HCC progression through binding to miR-328, which may be a potent therapeutic target for HCC.
Keywords: hepatocellular carcinoma, DIO3OS, competing endogenous RNA, microRNA-328, Hhip, hedgehog signaling pathway
Introduction
Liver cancer ranked the sixth most common carcinoma and the fourth most frequent consequence of cancer-associated death in 2018, among which hepatocellular carcinoma (HCC) contributed to 75%-85% of all cases diagnosed.1 The most important risk factor for the majority of HCC is the chronic liver disease caused from viral hepatitis, alcohol over-consumption, and/or nonalcoholic steatohepatitis.2 If diagnosed at an early stage, patients may be curable via resection, liver transplantation, or ablation to achieve an over 50% 5-year survival rate.3 Since HCC is recurrently resistant to chemotherapy and radiotherapy, many adjuvant therapies for HCC, such as small molecular targets, monoclonal antibodies and microRNA (miRNAs) have emerged.4 Long non-coding RNAs (lncRNAs) have been implicated in the pathogenesis and progression of various cancers, including HCC, and certain lncRNAs may function as diagnostic and/or prognostic markers for HCC.5
Type 3 deiodinase (DIO3) and DIO3 opposite strand (DIO3OS) genes located at the distal end of the GTL2/DIO3 imprinted cluster are preferentially expressed in the GTL2/DIO3 imprinted region of mouse and humans.6 DIO3OS was remarkably reduced in patients with ulcerative colitis relative to normal controls.7 While its role in cancers, especially HCC remains elusive. Interestingly, lncRNAs may regulate the expression of microRNAs (miRNAs) which are closely related to the progression of HCC and have the potency to bind to lncRNA sponges to suppress gene expression and protein synthesis.8 miR-328 expression was promoted in 48 HCC tissue samples, and downregulation of miR-328 in HepG2 cells attenuated cell invasion and proliferation and promoted cell apoptosis.9 Moreover, circRNA-5692 bound to miR-328-5p which potentiated the malignant behaviors of HCC cells.10 Thus, we proposed that DIO3OS may interact with miR-328 to revert the malignant aggressiveness of HCC cells. More recently, hedgehog-interacting protein antisense RNA 1, a novel lncRNA, was monitored to interact with and stabilize the mRNA of hedgehog-interacting protein (Hhip) to enhance Hhip expression, thereby inhibiting the progression of HCC.11 In addition to facilitating development and regeneration, the Hedgehog (Hh) pathway results in alteration of progenitors when incessantly evoked, which might lead to small-cell lung carcinomas, prostate and digestive tract tumors.12 Considering the aforementioned investigations, in this work, we elaborated the probable roles of DIO3OS and miR-328 in HCC progression with the intention of offering insights for the treatment of HCC.
Materials and Methods
Subjects
We collected 31 tumor tissues and matched paracancerous tissues from patients with HCC diagnosed in China-Japan Union Hospital of Jilin University from April 2018 to July 2019, of which 19 were male and 12 were female, with a median age of 59.7 years and an average age of 56.31 ± 8.3 years. All patients were followed up every three months for a period of five years. All patients with clinical samples were diagnosed through pathological examination and none of them received radiotherapy or chemotherapy before enrollment. Patients complicated with chronic systemic diseases or other malignancies were excluded. All the samples were collected with written informed consent from patients, and the protocol was permitted by the ethics committee of China-Japan Union Hospital of Jilin University.
Bioinformatics Analysis
HCC-related datasets involving mRNAs and lncRNAs (GSE101728) and miRNAs (GSE57555) were found in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Differential expression of microarray expression matrix was conducted using in R-language limma package (http://master.bioconductor.org/packages/release/bioc/html/limma.html). The differentially expressed genes were screened out with the conditions of adj.p.Val < 0.05 and |LogFoldChange| > 1.5, and the R language pheatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html) was applied for the heatmap plotting. The expression of DIO3OS and Hhip in The Cancer Genome Atlas (TCGA)-liver HCC database were then predicted with the help of GEPIA website (http://gepia.cancer-pku.cn/index.html). The binding miRNAs of DIO3OS and Hhip were predicted by StarBase, and screened by the jvenn website (http://jvenn.toulouse.inra.fr/app/example.html) for mapping.
Construction of Overexpression or Silencing Plasmids
The overexpression plasmid of DIO3OS, the corresponding empty vector (EV), two small interfering RNAs (siRNAs) targeting DIO3OS and corresponding scramble siRNA were synthesized by GenePharm Bioengineering Co., Ltd. (Shanghai, China).
Cell Treatment and Grouping
Human immortalized normal stem cell LO2 and HCC cell lines MHCC97, BEL-7402, HepG2, SMMC7721, and BEL-7405 cells were from the Cell Bank of Shanghai Institute of Cells, Chinese Academy of Science (Shanghai, China). All cells were cultivated in Roswell Park Memorial Institute-1640 medium supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA). Subsequently, the well-grown BEL-7405 cells and HepG2 cells were transfected with constructed DIO3OS overexpression plasmid and the corresponding empty plasmid, respectively. The LO2 cells were transfected with siRNAs targeting DIO3OS and corresponding scramble siRNA. Transfection was performed using the lipofectamine 2000 kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) strictly in accordance with the instructions for 48 h. Cells were harvested for following experiments. RT-qPCR was applied to examine DIO3OS expression to evaluate the transfection efficiency.
RT-qPCR
The total RNA in the tissues and cells was obtained using a TRizol kit (Thermo Fisher Scientific). The TaqMan miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) was used for reverse transcription of miRNA, while reverse transcription reagents (Applied Biosystems) was applied for mRNA and lncRNA reverse transcription. SYBR Green (Takara Bio Inc., Japan) was applied for real-time quantitative PCR. To synthesize cDNA, the reaction mixture was maintained at 16°C for 30 min, at 42°C for 30 min, at 85°C for 5 min, and then at 4°C. Next, 0.67 μL cDNA was amplified using 5 μL TaqMan 2× Universal PCR Master Mix (Applied Biosystems) without AmpErase Ung, 0.5 μL gene-specific primers/probes, and 3.83 μL ddH2O. Quantitative PCR was performed on the StepOnePlus PCR system (Applied Biosystems), and the reaction mixture was incubated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, followed by 1 min at 60°C. StepOne software v2.0 (Applied Biosystems) was applied to calculate the cycle threshold (Ct). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as internal reference. The sequences were listed below: DIO3OS forward: AGAGTGGCACCATCATGTACCA, reverse: CCAAGTGCGCAACTCAGACA; GAPDH: forward: CGTGAAAAGATGACCCAGATCA; reverse: CACAGCCTGGATGGCTACGT.
Cell Viability Assays
The viability of HCC cells was detected by a cell counting kit-8 (CCK-8, Roche, Sweden) following the manufacturer’s instructions. Then, HCC cells were subjected to EdU staining with an EdU proliferation kit (ab219801, Abcam, Shanghai, China) for the proliferation activity measurement strictly according to a previous literature.13 Briefly, 1 × 106 cells were cultured with EdU for 4 h, followed by harvesting and fixation with 4% formaldehyde. Afterwards, EdU labeled cells were analyzed using a MoFlo Astrios (Beckman Coulter, Brea, CA, USA).
Detection of Apoptosis
Firstly, the propidium iodide (PI)/Annexin-V kit was used to label the cells in each group, and then the number of apoptotic cells was detected by a flow cytometer (Thermo Fisher). In short, the medium was discarded from each well, and cells were carefully washed with phosphate buffered saline (PBS) and preserved on ice. Cells were then detached with trypsin and isolated with 0.5 mL 0.5× trypsin. After that, they were resuspended in the original medium to insert isolated cells into the suspension for further examination. After exposure of 1.0 × 106 cells to 10 mL binding buffer and 1.25 mL Annexin V-fluorescein isothiocyanate, the cells were incubated at room temperature for 15 min in the dark. Then, the cell suspension was centrifuged at 1000 ×g for 5 min to remove the supernatant, and the cell was resuspended in 0.5 mL ice-cold 1× binding buffer. Finally, the cells were incubated with 10 mL propidium iodide and transferred to a fluorescent activated cell sorter tube (Fahrenheit, Munich, Germany). Fluorescence data analysis was performed via WinMDI 2.8 software.
Apoptotic cell levels were detected using a Hoechst 33258 staining kit (Roche) in strict accordance with the instructions. In short, Hoechst 33258 could bind to DNA molecules as fluorescent probes. The intake of Hoechst 33258 in apoptotic cells increased, and apoptotic cells showed strong blue fluorescence. After cell transfection, cells were fixed at 4°C for 10 min with 4% paraformaldehyde in 0.1 M PBS (pH = 7.4). Apoptotic cells were examined using a fluorescence microscope after staining with 5 mg/mL Hoechst 33258 for 10 min.
Transwell Assay for Cell Migration and Invasion
Transwell assay was fulfilled following a previous report.14 Serum-free medium was used to dilute Matrigel (BD Biosciences, San Jose, CA, USA) at a ratio of 6:1, and 100 μL diluted Matrigel was placed in the apical chamber at 37°C for 4 h for solidification. The apical chamber was filled with 100 μL cell suspension (1 × 105 cells), while the basolateral chamber was filled with 500 μL serum-free medium. After a night, noninvasive cells were removed with cotton swabs, and invaded cells on the surface of basolateral chamber were fixed in 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 20 min. Finally, the invasive cells were visualized and calculated under a microscope. In terms of migration capacity, the steps were consistent except for Matrigel processing.
Sublocalization of DIO3OS
First, we used Lncatlas (http://lncatlas.crg.eu/) to predict the cellular sublocalization of DIO3OS. The localization of DIO3OS in HepG2 and BEL-7405 cells was then detected using a nuclear separation kit (PARIS kit; cat. no. AM1921; Invitrogen Inc., Carlsbad, CA, USA) in accordance with the instructions. Subsequently, the positioning of DIO3OS in HepG2 and BEL7405 cells was further determined by fluorescent in situ hybridization (FISH) experiments, all of which were carried out as previously illustrated.15 In short, nucleic acid-modified oligonucleotide probes targeting DIO3OS (Exiqon, Vedbaek, Denmark) was used. 4ʹ,6-diamidino-2-phenylindole (DAPI) was used to label nuclear DNA (in blue). The RNA signal was detected by incubation with biotinylated coupled antibody against digoxigenin, and amplified using SABC-Alexa Fluro 448 (in red).
Luciferase Reporter Assay
The binding relationships of miR-328 to DIO3OS and Hhip were detected using a luciferase reporter assay. DIO3OS or Hhip wild-type (WT) and 3ʹuntranslated region (3ʹUTR) binding sequence mutant (MT) was synthesized by GenePharm Bioengineering Co., Ltd. (Shanghai, China) and inserted into pMIR-REPORTTM (Thermo Fisher Scientific).16 Luciferase activity was evaluated by a dual-luciferase reporter assay system (Promega Corporation).17
Immunoblotting
Immunoblotting was exercised to examine the expression of Hh signaling pathway-related markers in HCC cells according to standard protocols as described previously.18 Protein was harvested using radio immunoprecipitation assay lysis buffer, and a bicinchoninic acid assay was applied to determine the protein concentration. Next, we separated equal amount of protein using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred the protein to a polyvinylidene difluoride membrane. After blocking with 1.5% skimmed milk for 1 h at room temperature, membranes were incubated with the primary antibodies at 4°C overnight and with horseradish peroxidase-conjugated secondary antibodies. Membranes were developed using a chemiluminescent substrate. The antibodies were used as follows: Gli1 (1:200, ab49314, Abcam), Gli2 antibodies (1:500, ab26056) and Gli3 antibodies (1:500, ab6050) and the secondary antibodies (1:50,000, ab7090).
Tumorigenesis Assay in vivo
We subcutaneously injected 2 × 106 stably overexpressed HepG2 and BEL-7405 cells (suspended in 200 μL PBS) into the right axils of nude mice. We recorded tumor growth weekly and used the formula (tumor length × tumor width × tumor width × 1/2) to calculate tumor volume. After euthanasia (injection of pentobarbital sodium at 100 mg/kg), we weighed the tumor. We handled the experimental animals according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. The protocol was reviewed and approved by the Ethics Committee of China-Japan Union Hospital of Jilin University.
Immunohistochemistry
Tumor tissues were fixed with formalin, embedded in paraffin and cut into 5 μm thickness sections. The samples underwent strict processes of deparaffinization, rehydration, antigen recovery and endogenous peroxidase inhibition. Next, sections were immunoblotted overnight with the KI67 antibody (ab15580, Abcam) at 4°C and with the appropriate secondary antibody. As a chromogen, diaminobenzidine solution was applied for visualization. At last, nuclei were stained with hematoxylin. A microscope (Leica DM 4000B, Leica, Bannockburn, IL, USA) was used to acquire images.
Quantification and Statistical Analysis
SPSS 21.0 software (SPSS, Chicago, IL, USA) was used for statistical analyses. Normal distribution was tested by Kolmogorov-SmiRnov. Results were displayed as mean ± standard deviation (SD). One-way or two-way analysis of variance (ANOVA) following Tukey’s post hoc test were performed to exhibit the differences among experimental groups. A level of p < 0.05 was considered significant.
Results
Poor Expression of DIO3OS Is Identified in HCC Patients and HCC Cell Lines
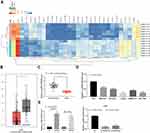
In the GEO database, we analyzed the GSE101728 dataset which included the cancer tissues of seven HCC patients as well as the paired adjacent tissues. Differential analysis of GSE101728 dataset was performed to screen out 30 significantly differentially expressed lncRNAs and to plot the heatmap, as shown in Figure 1A. Additionally, the expression of DIO3OS in HCC patients was analyzed in the TCGA database through the GEPIA bioinformatics website, which revealed the downregulation of DIO3OS in HCC patients (Figure 1B). Later, we tested DIO3OS in tumor tissues and paracancerous tissues of 31 HCC patients by RT-qPCR. The expression of DIO3OS in HCC tissues was found to be significantly reduced (Figure 1C). DIO3OS expression in HCC cells and LO2 cells was examined afterwards. As expected, DIO3OS was diminished in HCC cells (Figure 1D). With the aim to further verify the effect of DIO3OS on HCC, we transfected the DIO3OS overexpression plasmid into HepG2 as well as BEL-7405 cells, whereas two siRNAs targeting DIO3OS into LO2 cells. RT-qPCR was then used to verify the transfection efficiency, and the expression of DIO3OS was significantly enhanced after overexpression plasmid delivery, while downregulated in LO2 cells following introduction of siRNAs (Figure 1E).
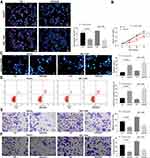
DIO3OS Inhibits Malignant Behaviors in HepG2 and BEL-7405 Cells
We found that after overexpression of DIO3OS, the number of EdU-positive cells was significantly reduced (Figure 2A), and the activity of HepG2 and BEL-7405 cells was significantly inhibited (as revealed by CCK-8) (Figure 2B). We further observed that restoration of DIO3OS resulted in promotions in HepG2 and BEL-7405 cell apoptosis (Figure 2C and D). On top of that, transwell assay unveiled the identical tendency as findings from CCK-8 and EdU assays. Resumption of DIO3OS hampered HepG2 and BEL-7405 cell invasion and migration (Figure 2E and F).
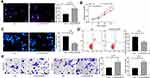
DIO3OS Knockdown Promotes the Malignant Behaviors in LO2 Cells
EdU staining and CCK-8 assays were then used to detect cell activity, we found that the proliferation of LO2 cells increased significantly after DIO3OS knockdown (Figure 3A and B). Moreover, the results of flow cytometry and Hoechst 33258 staining demonstrated that the apoptotic LO2 cells decreased remarkably after DIO3OS knockdown (Figure 3C and D). Finally, we showed through Transwell assays that DIO3OS knockdown contributed to increased number of migrated and invaded cells (Figure 3E).
DIO3OS Is Localized in Cytoplasm
To this end, we predicted the main localization of DIO3OS with cytoplasm (Figure 4A) through the Lncatlas bioinformatics prediction website. Furthermore, FISH experiments and nuclear separation experiments exhibited that DIO3OS mainly expressed in the cytoplasm in HepG2 and BEL-7405 cells (Figure 4B and C).
DIO3OS Serves as a ceRNA for miR-328 and Elevates Hhip Expression
Since DIO3OS was localized in the cytoplasm, it can be preliminarily demonstrated that DIO3OS played a role in stimulating the malignant aggressiveness of HCC cells through the ceRNA mechanism. Consequently, by analyzing the HCC-related miRNA dataset GSE57555, we found that there were 14 miRNAs with significant high expression in HCC. In addition, we used RNA22 to predict the miRNAs that might bound to DIO3OS, and compared the miRNA prediction results with the up-regulated miRNAs in the GSE57555 dataset. Only one miRNA was observed in the intersection: hsa-miR-328 (Figure 5A). It has been reported that downregulation of miR-328 inhibited metastasis and growth of HCC.9 Therefore, we used a luciferase activity assay to authenticate the relationship between miR-328 and DIO3OS (Figure 5B). We next examined miR-328 expression in HepG2 and BEL-7405 cells in the presence of DIO3OS, and found that DIO3OS diminished miR-328 expression (Figure 5C). Furthermore, we examined the miR-328 expression in tumor tissues and paracancerous tissues of 31 HCC patients, and found that miR-328 was higher in tumor tissues. The relationship between the expression of miR-328 and DIO3OS was analyzed by Pearson’s correlation. miR-328 was revealed to be negatively correlated with DIO3OS (Figure 5D and E).
Subsequently, we predicted the direct targets of miR-328 in RNA22, mirDIP, miRDB, DIANA and Targetscan, and compared the predicted results of target genes with those with poor expression in GSE101728 dataset. There were 2 intersection genes Hhip and DLG5 (Figure 5F). We analyzed the expression of Hhip in HCC patients in TCGA database through the GEPIA bioinformatics website. Hhip was significantly reduced in HCC patients (Figure 5G). In addition, Hhip was hypermethylated and transcriptionally reduced in HCC tissues and cells.19 Therefore, we used dual luciferase reporter gene assay to verify the binding relationship between miR-328 and Hhip (Figure 5H). We then examined the expression of Hhip in HepG2 and BEL-7405 cells overexpressing DIO3OS and found that overexpression of DIO3OS simultaneously increased Hhip expression (Figure 5I). Furthermore, we tested Hhip expression in 31 patients with HCC and in paracancerous tissues, and noted that Hhip expression was significantly lower in tumor tissues (Figure 5J). We lastly analyzed the relationship between Hhip and miR-328 or DIO3OS expression by Pearson’s correlation. Hhip was positively correlated with DIO3OS and negatively correlated with miR-328 (Figure 5K and L).
DIO3OS Disrupts the Hhip Signaling Pathway
It has been reported that Hhip inhibited proliferation of adipocytes through the Hh signaling pathway.20 Accordingly, the expression patterns of Hh signaling pathway-related proteins Gli1, Gli2 and Gli3 in HepG2 and BEL-7405 cells were evaluated. The expression of Gli1, Gli2 and Gli3 was decreased significantly following overexpression of DIO3OS (Figure 6A and B).
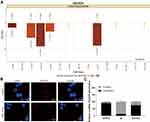
DIO3OS Inhibits HCC Growth in vivo
HepG2 or BEL-7405 cells with stable overexpression of DIO3OS were injected into the armpit of nude mice to evaluate the effect of DIO3OS on the growth of HCC cells in vivo. We monitored that overexpression of DIO3OS inhibited the growth rate of HepG2 or BEL-7405 cells in vivo, and overexpression of DIO3OS significantly reduced tumor weight at the end of the experiment (Figure 7A and B). Subsequently, we performed KI67 immunohistochemical staining on the tumors and found that overexpression of DIO3OS lowered the rate of KI67 positive cells (Figure 7C) in the tumor tissues.
Discussion
China is one of the regions that own the greatest HCC incidence, while the lack of appropriate preemptive or therapeutic treatment regimens reinvigorated wide research against HCC so as to develop novel therapeutic strategies.21 Dysregulation of lncRNAs such as maternally expressed gene 3, metastasis associated in lung adenocarcinoma transcript-1, homeobox transcript antisense RNA as well as H19 have been recognized to be tightly associated with tumorigenesis, metastasis, prognosis and diagnosis of HCC.22 Nevertheless, the relevance of DIO3OS, a newly identified lncRNA, in HCC needs to be excavated. Our results determined that DIO3OS expression was decreased in both HCC tissue and cells. DIO3OS impaired the HCC cell proliferation, migration and invasion as well as tumor growth in vivo via performing as a ceRNA for miR-328 and upregulating Hhip expression through the Hh signaling pathway. Altogether, these observations provided important insights into the cancer-suppressing effect of DIO3OS in HCC cell malignant aggressiveness.
In HCC, we discovered a relatively low expression of DIO3OS in HCC tissue samples by accessing to the TCGA and GEO databases, and RT-qPCR analysis exposed a similar pattern in collected HCC tissues and cell lines. Moreover, upregulation of DIO3OS through delivery of overexpression plasmid of DIO3OS into HepG2 and BEL-7405 resulted in declines in growth, migration and invasion of these two cell lines, while enhancements in cell apoptosis. By contrast, DIO3OS knockdown by siRNAs led to the opposite trends in LO2 cells. Recently, non-coding RNAs, including miRNAs and lncRNAs, has been observed to produce a complex modulatory network through competing with each other to interact with mRNAs, working as a ceRNA.23 Hence, to sightsee the mechanism in which DIO3OS acts in HCC, we investigated candidate miRNAs and established that DIO3OS competed with miR-328 to slow down the progression of HCC. By contrast, DIO3OS promoted cell proliferation and invasion through competing for miR-122 to upregulate ALDOA expression in pancreatic cancer.24 A plethora of researches on miR-328 have paid much attention to its functions on cell growth, invasion and migration in cancers, including gastric cancer,25 invasive breast cancer,26 non-small cell lung cancer,27 and most importantly, HCC.28 Additionally, overexpression of metastasis associated in lung adenocarcinoma transcript-1 sequestered cytoplasmic miR-328-3p, culminating in insulin-like growth factor 1 receptor induction to mediate hypoxia-stimulated proliferation of pulmonary artery smooth muscle cells.29 As a consequence, we considered that DIO3OS may involve in the progression of HCC by competitively binding to miR-328 to regulate the expression of a mRNA.
Furthermore, in view of the mechanism of lncRNA-miRNA-mRNA network, we screened out one target gene of miR-328, Hhip. The level of Hhip was reduced in adipose tissue development, and recombinant Hhip promoted the differentiation of adipocytes by targeting the canonical Hh pathway.20 More specifically, Hhip, a reverse modulator of the Hh pathway, was downregulated in gastric cancer, and its overexpression hampered cancer cell survival, proliferation, migration as well as invasion.30 Then, we believed that the Hh pathway lies the downstream of the DIO3OS/miR-328/Hhip axis in HCC cells. The regulatory role of Hh pathway in liver cells including hepatic stellate cells, progenitors, and endothelial cells has been increasingly uncovered.31 Hh pathway components (Gli1 and Gli2) were potentiated in both HCC tissues and cells.32 We monitored that introduction of DIO3OS remarkably diminished the Hh pathway-related proteins Gli1, Gli2 and Gli3 expression, while restored Hhip expression. The observations of our study were largely in agreement with a previous study, where the lncRNA-dependent noncanonical Hh pathway in breast cancer sheds some lights on certain patients who would respond optimally to treatment.33 In HCC, resumption of the Hh pathway is accountable for the maintenance of cancer stem cells, and Hh disruption demonstrates an obvious anti-tumor effect.34 Of note, Gli3, a target gene for miR-133, was repressed by miR-133 during the event of embryonic myogenesis.35 However, the correlation between miR-328 and the Hh pathway in HCC cells was not investigated in this work due to the time and fund limitations.
Conclusion
Lastly to generalize our finding, we recapitulate a DIO3OS/miR-328/Hhip regulatory network that might regulate the HCC cell growth, migration and invasion. Meanwhile, DIO3OS and miR-328 procedure a regulatory loop where the release of DIO3OS contributes to the declines in miR-328 expression, eventually enhancing Hhip expression and disrupting the Hh pathway. Thus, this study may provide a new therapeutic maneuver for HCC treatment and offer some insights on the lncRNA-miRNA-mRNA regulatory nexus on the HCC pathogenesis.
Author Contributions
ZPW contributed to the study design and manuscript preparation; All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–399. doi:10.3322/caac.21161
3. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi:10.1016/S0140-6736(11)61347-0
4. Wang Z, Zhang G, Wu J, Jia M. Adjuvant therapy for hepatocellular carcinoma: current situation and prospect. Drug Discov Ther. 2013;7(4):137–143.
5. Niu ZS, Niu XJ, Wang WH. Long non-coding RNAs in hepatocellular carcinoma: potential roles and clinical implications. World J Gastroenterol. 2017;23(32):5860–5874. doi:10.3748/wjg.v23.i32.5860
6. Yang W, Li D, Wang G, et al. Expression and imprinting of DIO3 and DIO3OS genes in Holstein cattle. J Genet. 2017;96(2):333–339.
7. Wang S, Hou Y, Chen W, et al. KIF9AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol Med Rep. 2018;17(2):2195–2202. doi:10.3892/mmr.2017.8118
8. Shi H, Xu Y, Yi X, Fang D, Hou X. Current research progress on long noncoding RNAs associated with hepatocellular carcinoma. Anal Cell Pathol (Amst). 2019;2019:1534607.
9. Liang F, Cui ZJ, Liu JD, Liu KP, Li L, Chen YL. Downregulated miR-328 suppressed cell invasion and growth in hepatocellular carcinoma via targeting PTEN. Eur Rev Med Pharmacol Sci. 2018;22(19):6324–6332. doi:10.26355/eurrev_201810_16043
10. Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi:10.1038/s41419-019-2089-9
11. Bo C, Li X, He L, Zhang S, Li N, An Y. A novel long noncoding RNA HHIP-AS1 suppresses hepatocellular carcinoma progression through stabilizing HHIP mRNA. Biochem Biophys Res Commun. 2019;520(2):333–340. doi:10.1016/j.bbrc.2019.09.137
12. Eichenmuller M, Gruner I, Hagl B, et al. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49(2):482–490. doi:10.1002/hep.22649
13. Chen W, Li K, Zhu K, et al. RP11-81H3.2 acts as an oncogene via microRNA-490-3p inhibition and consequential tankyrase 2 up-regulation in hepatocellular carcinoma. Dig Dis Sci. 2019. doi:10.1007/s10620-019-06007-5
14. Hu WY, Wei HY, Li KM, Wang RB, Xu XQ, Feng R. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed Pharmacother. 2020;121:109642.
15. Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1):143. doi:10.1186/s12943-019-1079-y
16. Jiang C, Zhu W, Xu J, et al. MicroRNA-26a negatively regulates toll-like receptor 3 expression of rat macrophages and ameliorates pristane induced arthritis in rats. Arthritis Res Ther. 2014;16(1):R9. doi:10.1186/ar4435
17. Zhang Y, Guo L, Li Y, et al. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol Cancer. 2018;17(1):1. doi:10.1186/s12943-017-0753-1
18. Wang R, Cheng L, Yang X, et al. Histone methyltransferase SUV39H2 regulates cell growth and chemosensitivity in glioma via regulation of hedgehog signaling. Cancer Cell Int. 2019;19:269. doi:10.1186/s12935-019-0982-z
19. Tada M, Kanai F, Tanaka Y, et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res. 2008;14(12):3768–3776. doi:10.1158/1078-0432.CCR-07-1181
20. Wei H, Li J, Shi S, et al. Hhip inhibits proliferation and promotes differentiation of adipocytes through suppressing hedgehog signaling pathway. Biochem Biophys Res Commun. 2019;514(1):148–156. doi:10.1016/j.bbrc.2019.04.047
21. Jindal A, Thadi A, Shailubhai K. Hepatocellular carcinoma: etiology and current and future drugs. J Clin Exp Hepatol. 2019;9(2):221–232. doi:10.1016/j.jceh.2019.01.004
22. Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018;17:900–913. doi:10.17179/excli2018-1541
23. Roy S, Trautwein C, Luedde T, Roderburg C. A general overview on non-coding RNA-based diagnostic and therapeutic approaches for liver diseases. Front Pharmacol. 2018;9:805. doi:10.3389/fphar.2018.00805
24. Cui K, Jin S, Du Y, et al. Long noncoding RNA DIO3OS interacts with miR-122 to promote proliferation and invasion of pancreatic cancer cells through upregulating ALDOA. Cancer Cell Int. 2019;19:202. doi:10.1186/s12935-019-0922-y
25. Yan BL, Li XL, An JY. MicroRNA-328 acts as an anti-oncogene by targeting ABCG2 in gastric carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(14):6148–6159. doi:10.26355/eurrev_201907_18428
26. Saberi A, Danyaei A, Neisi N, Dastoorpoor M, Tahmasbi Birgani MJ. MiR-328 may be considered as an oncogene in human invasive breast carcinoma. Iran Red Crescent Med J. 2016;18(11):e42360. doi:10.5812/ircmj.42360
27. Arora S, Ranade AR, Tran NL, et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer. 2011;129(11):2621–2631. doi:10.1002/ijc.25939
28. Li JZ, Li J, Liu BZ. MicroRNA-328-3p inhibits malignant progression of hepatocellular carcinoma by regulating MMP-9 level. Eur Rev Med Pharmacol Sci. 2019;23(21):9331–9340. doi:10.26355/eurrev_201911_19426
29. Xing Y, Zheng X, Fu Y, et al. Long Noncoding RNA-maternally expressed gene 3 contributes to hypoxic pulmonary hypertension. Mol Ther. 2019;27(12):2166–2181. doi:10.1016/j.ymthe.2019.07.022
30. Zuo Y, Lv Y, Qian X, et al. Inhibition of HHIP promoter methylation suppresses human gastric cancer cell proliferation and migration. Cell Physiol Biochem. 2018;45(5):1840–1850. doi:10.1159/000487875
31. Magistri P, Leonard SY, Tang CM, Chan JC, Lee TE, Sicklick JK. The glypican 3 hepatocellular carcinoma marker regulates human hepatic stellate cells via Hedgehog signaling. J Surg Res. 2014;187(2):377–385. doi:10.1016/j.jss.2013.12.010
32. Gao L, Zhang Z, Zhang P, Yu M, Yang T. Role of canonical Hedgehog signaling pathway in liver. Int J Biol Sci. 2018;14(12):1636–1644. doi:10.7150/ijbs.28089
33. Xing Z, Lin C, Yang L. Unraveling the therapeutic potential of the LncRNA-dependent noncanonical Hedgehog pathway in cancer. Mol Cell Oncol. 2015;2(4):e998900. doi:10.1080/23723556.2014.998900
34. Della Corte CM, Viscardi G, Papaccio F, et al. Implication of the Hedgehog pathway in hepatocellular carcinoma. World J Gastroenterol. 2017;23(24):4330–4340. doi:10.3748/wjg.v23.i24.4330
35. Mok GF, Lozano-Velasco E, Maniou E, et al. miR-133-mediated regulation of the Hedgehog pathway orchestrates embryo myogenesis. Development. 2018;145(12):12. doi:10.1242/dev.159657
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.