Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Long-Noncoding RNA ANCR Activates the Hedgehog Signaling Pathway to Promote Basal Cell Carcinoma Progression by Binding to PTCH
Authors Wu H, He P, Xie D, Wang J, Wan C
Received 1 November 2021
Accepted for publication 7 April 2022
Published 25 May 2022 Volume 2022:15 Pages 955—965
DOI https://doi.org/10.2147/CCID.S345371
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Hongxuan Wu,* Pingxiu He,* Dong Xie, Jianqiao Wang, Chuan Wan
Department of Dermatology, The First Affiliated Hospital of Nanchang University, Nanchang, 330006, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chuan Wan, Department of Dermatology, The First Affiliated Hospital of Nanchang University, Nanchang, 330006, People’s Republic of China, Tel +86-791-88692799, Email [email protected]
Purpose: The long non-coding RNA (lncRNA) anti-differentiation noncoding RNA (ANCR) is closely related to the occurrence and development of various malignancies. However, its expression and potential role in basal cell carcinoma (BCC) have not been established. In this study, we characterized the effects of ANCR in BCC and its underlying mechanism.
Methods: The expression of ANCR in BCC tissues and cells was detected by qRT-PCR. Proliferation, invasion, migration and apoptosis of ANCR overexpressed or knock down TE354.T and A431 cells were examined by CCK8, transwell assay, wound healing assay and flow cytometry analysis, respectively. Western blot was performed to measure the expression of apoptosis-related proteins (BAX, BCL2 and Cleaved-caspase3), epithelial-mesenchymal transformation-related proteins (E-cadherin, N-cadherin, vimentin and β-catenin), and Hedgehog-pathway-related proteins (PTCH, GLI1 and SMO). RNA pull-down assay was used to analyze the relationship between ANCR and PTCH. The effect of ANCR on BCC growth in vivo was analyzed using xenograft model. TUNEL assay was used to determine the cell apoptosis.
Results: ANCR and Hedgehog pathway were more highly expressed in BCC tissues than in adjacent normal tissues. ANCR overexpression substantially promoted BCC cell proliferation, invasion, and migration, inhibited apoptosis, and up-regulated BCL2 and decreased the expression of BAX and Cleaved-caspase3 proteins. Additionally, the upregulation of N-cadherin, vimentin, β-catenin, PTCH, GLI1, and SMO expression, and downregulation of E-cadherin expression were observed after ANCR overexpression. Moreover, ANCR knockdown had the opposite effects. An RNA pull-down assay further revealed that ANCR is specifically bound to PTCH. In vivo experiments also showed that ANCR overexpression significantly increased tumor growth and decreased apoptosis, which was reversed by cyclopamine, a specific inhibitor of the Hedgehog signaling pathway.
Conclusion: ANCR activates the Hedgehog signaling pathway by binding to PTCH, thereby promoting BCC progression; accordingly, ANCR could be a candidate therapeutic target in BCC.
Keywords: anti-differentiation noncoding RNA, basal cell carcinoma, Hedgehog signaling pathway, PTCH, epithelial–mesenchymal transition
Introduction
Basal cell carcinoma (BCC) is the most common skin malignancy worldwide, accounting for approximately 80% of non-melanoma skin cancers.1 Although BCC has a low rate of metastasis, it has strong local invasion ability and can therefore destroy adjacent structures. The high prevalence of BCC imposes a marked burden on healthcare worldwide and has a major impact on patient quality of life. The Hedgehog signaling pathway plays a significant role in normal embryonic development, and its abnormal function is associated with a variety of tumors. In particular, the abnormal activation of the Hedgehog signaling pathway can induce the occurrence, metastasis, and treatment resistance of BCC.2 Approximately 85% of patients with BCC have mutations in the Hedgehog signaling pathway.3 Among these, 67% have Patched (PTCH) deletions and 10% have Smoothened (SMO) mutations, leading to the overexpression of the GLI family, and promotion of cell division and tumorigenesis.3 Although the Hedgehog signaling pathway is closely related to the occurrence of BCC, the decided relationship between them has not been fully elucidated. Further studies on the specific molecular mechanism linking this pathway to BCC are needed to provide a theoretical basis for effective prevention and treatment and to identify candidate therapeutic targets.
Long non-coding RNAs (lncRNAs) are a type of RNA with lengths greater than 200 nucleotides that lack protein-coding ability that have important regulatory roles in a variety of complex diseases, including the occurrence and progression of human cancers.4 LncRNA anti-differentiation noncoding RNA (ANCR), also termed differentiation antagonizing nonprotein coding RNA (DANCR), was originally discovered in epidermal stem cells, which is essential for maintaining the undifferentiated state of epidermal precursor cells.5,6 Since its discovery, ANCR has been widely investigated in various human malignancies. ANCR is aberrantly expressed in multiple tumors, including hepatocellular carcinoma, gastric cancer, colorectal cancer, bladder cancer, lung cancer, and glioma and others, in which its primary function is performed as an oncogene, contributing to cancer development and progression.6–10 In contrast, Li et al11 detected the downregulation of ANCR expression in breast cancer tissues and cell lines, and revealing that ANCR could facilitate EZH2 degradation to suppress breast cancer progression. However, the role of ANCR in BCC has not been reported. Furthermore, the regulatory mechanisms of ANCR are complex and involved in multiple factors. ANCR can exert oncogenic effects through the regulation of several signaling pathways by protein binding in cancers. For instance, in bladder cancer, ANCR was found to interact with leucine-rich PPR motif-containing protein to stabilize IL11 mRNA, resulting in the overexpression of IL11 stimulated phosphorylation of the JAK2/STAT3 signaling pathway, ultimately upregulating matrix metalloproteinase 9 expression and promoting tumor progression.9 However, whether ANCR can affect BCC by regulating the Hedgehog signaling pathway remains unexplored.
In this study, we investigated the expression level of ANCR and Hedgehog pathway components (PTCH, GLI1, and SMO) in BCC tissues and adjacent normal tissues. Subsequently, we, respectively, knocked down and overexpressed ANCR in a BCC cell line to clarify its contribution to BCC, and explored the role of the Hedgehog signaling pathway in moderating effects of this lncRNA in vitro and in vivo. Moreover, we identified the interaction between ANCR and PTCH using an RNA pull-down assay. Our results can provide new horizon into the effects of ANCR on BCC and its potential molecular mechanisms, which may offer new theoretical foundation for exploring therapeutic targets of BCC.
Materials and Methods
Tissue Specimens
In total, ten pairs of BCC tissues and corresponding adjacent normal tissues were collected from patients who had undergone surgical resection at the Department of Dermatology, The First Affiliated Hospital of Nanchang University, China. All specimens were immediately frozen in liquid nitrogen and stored at –80°C for subsequent use. None of the patients had been treated with chemotherapy, radiotherapy, immunotherapy, or targeted therapy before surgery. BCC diagnosis was histopathologically confirmed. This study was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (NO. (2022)1-024) and were performed in accordance with the Declaration of Helsinki, and written informed consent was obtained from all patients.
Cell Culture
The human BCC cell line TE354.T and A431 were purchased from FuHeng Biology (FH1066 and FH0188, Shanghai, China) and were incubated in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 100 U/mL penicillin, and 100 µg/mL streptomycin at 37°C in a 5% CO2 atmosphere.
Western Blotting
Total proteins were extracted from the tissues and cells using RIPA lysis buffer (Beyotime, Shanghai, China) and measured the protein concentration with BCA Protein Assay Kit (Beyotime). The protein samples were then separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). The membranes were incubated overnight at 4°C with the following primary antibodies (all from Abcam, Cambridge, MA, USA): anti-PTCH (1:1000, ab53715), anti-GLI1 (1:1000, ab217326), anti-SMO (1:1000, ab236465), anti-E-cadherin (1:1000, ab231303), anti-N-cadherin (1:1000, ab76011), anti-vimentin (1:1000, ab92547), anti-β-catenin (1:1000, ab32572), anti-BAX (1:1000, ab32503), anti-BCL2 (1:1000, ab32124), anti-Cleaved caspase 3 (1:500, ab2302) and anti-GAPDH (1:5000, ab8245). After washing with Tris-buffered saline with Tween (Beyotime), the membranes were incubated with goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (1:20,000, ab6721; Abcam) for 1 h at room temperature. Protein bands were visualized using an electrochemiluminescence detection system (Tanon, Shanghai, China) and analyzed using ImageJ (National Institutes of Health, Bethesda, MD, USA).
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT‑qPCR)
Total RNA was isolated from tissue specimens and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cDNA was synthesized using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen Biotech, Beijing, China) according to the manufacturer’s protocols. The total RNA was analyzed via qPCR (Bio Rad, Martinez, CA, USA) using SYBR green qPCR Master Mix (Thermo Fisher Scientific). The RT‑qPCR cycles were as follows: step 1, preparative denaturation (30 s at 95℃); step 2, 40 cycles of denaturation (5 s at 95℃) and annealing (30 s at 60℃); and step 3, dissociation following the manufacturer’s protocol. Primer sequences were as follows: ANCR forward, 5′-CTTGTAGCAACCACGTGTCC-3′; ANCR reverse, 5′-TTCAGGGTAAGGGTCAGCTG-3′; PTCH forward, 5′-CCGGCTATGGGGAAGGC-3′; PTCH reverse, 5′- CCACAACCAAGAACTTGCCG −3′; GLI1 forward, 5′- GCTGCGCTGCCGTGG −3′; GLI1 reverse, 5′- GGTGTGGGGACACTCTGTCT −3′; SMO forward, 5′- GATTGCCGCCTGTTTCTGTC −3′; SMO reverse, 5′- GGCATTCCGGAGGCCAAG −3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′; GAPDH reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′.
ANCR Knockdown and Overexpression
Lentiviral vectors carrying a short hairpin RNA (shRNA) targeting ANCR (shANCR) and negative control shRNA (shNC) were obtained from Shanghai Yazai Biological Technology Co., Ltd. (Shanghai, China). The target sequences of the ANCR shRNAs were as follows: shANCR-1, 5′-GCCATTGAAGCTGGAATGT-3′; shANCR-2, 5′-GGCCAAATATGCGTACTAA-3′; shANCR-3, 5′-CCAACTATCCCTTCAGTTA-3′; shNC, 5′-TTCTCCGAACGTGTCACGT-3′. The blank control vector (Vector) and ANCR overexpressing vector (oeANCR) were obtained from Shanghai Jierui Biological Technology Co., Ltd. (Shanghai, China). Thereafter, 293T cells (Sigma–Aldrich, Shanghai, China) were transfected with the recombinant vector to obtain the packaging viruses. After 48 hours of transfection, collected the lentivirus and purified via ultra-centrifugation at 3000 × g for 2.5 h at 4℃. TE354.T and A431 cells were infected by the ANCR-overexpressing or knockdown lentivirus. The transfection efficiency of the above described cells was evaluated using RT-qPCR.
Cell Viability Assay
Cell viability was detected via the Cell Counting Kit-8 (CCK-8) assay (Beyotime) according to the manufacturer’s instructions. After transfection, cells were seeded into 96-well plates at a density of 5000 cells/well and cultured for 12, 48, and 72 h. Thereafter, each well was added 10 μL of CCK-8 solution and incubated for 3 h. Absorbance at 450 nm was measured using a microplate reader (Bio Rad).
Cell Apoptosis Assay
Cells were washed with phosphate-buffered saline (PBS) and resuspended at a density of 1×106 cells/mL. Subsequently, apoptosis was examined using the Annexin V Apoptosis Detection Kit (eBioscience, San Diego, CA, USA) according to the manufacturer’s instructions, and the apoptosis rate was finally analyzed using flow cytometry (Guava easyCyte HT, Millipore, USA).
Wound-Healing Assay
Cells were added to 6-well plates at a high density and formed monolayers overnight. A 100 μL sterile plastic tip was used to create a wound line across the surface of plates, washing the cell debris generated by the scratches with PBS. For about 36 h of culture in an incubator, the images were obtained under a microscope (Nikon, Tokyo, Japan).
Cell Invasion Assay
The cell invasion was detected via a Transwell assay. Briefly, the cells were resuspended in serum-free medium and inoculated to the upper chamber of a Matrigel-precoated plate at a density of 1×105 cells/well. The bottom chamber contained DMEM with 10% FBS. After incubation for 48 h, gently wiped the upper chamber with cotton swabs to remove unmigrated cells. Cells that migrated were fixed by 4% paraformaldehyde, stained with 0.1% crystal violet, and counted under a microscope (Nikon).
RNA Pull-Down Assay
An RNA pull-down assay was performed using Pierce Magnetic RNA-Protein Pull-down Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, proteins were extracted from TE354.T cells and mixed with a biotin-labeled ANCR or anti-sense RNA probe. Thereafter, the protein extract was incubated with streptavidin agarose beads at 25°C for 2 h. Western blotting was used to quantify PTCH binding to the ANCR pull-down complex.
Animal Xenografts
A total of 15 female BALB/c nude mice (6 weeks old) were obtained from Shanghai Model Organisms and housed in well-ventilated rooms under specific pathogen-free conditions. The mice were randomly divided into three groups (n = 5 per group): control (Ctrl), ANCR overexpression (oeANCR), and oeANCR + cyclopamine (cyc) groups. The BCC model was established by subcutaneously injecting TE354.T cells that stably transfected with the ANCR-overexpression plasmid, whereas the control group was subcutaneously injected with TE354.T cells stably transfected with the control vector plasmid. After 6 d, the mice were intraperitoneally injected with normal saline or cyclopamine (1.5 mg/kg per day) for one week. The tumor volume was recorded every 3 days. After 27 days, euthanized all the mice and weighed the xenografts. The xenograft volume was calculated as follows: (V) = a2 × b × 0.5, in which a is the short diameter and b is the long diameter. The xenografts were embedded in paraffin and subjected to TdT‑mediated dUTP nick end labeling (TUNEL) staining as described below. The animal experiments were approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (No.2021–57).
TUNEL Staining
A TUNEL assay was performed using the colorimetric TUNEL Apoptosis Assay Kit (Beyotime) as per the manufacturer’s instructions. Getting three randomly chosen fields of images per slide via microscopy (Nikon).
Statistical Analysis
All data are presented as the means ± standard deviations (SD) and analyzed using SPSS 20.0 software (SPSS, Chicago, IL, USA). Group means were compared using paired Student’s t-tests or one-way analysis of variance with Tukey post-hoc tests. P < 0.05 was considered statistically significant.
Results
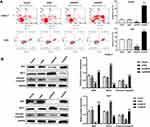
ANCR and Hedgehog Pathway Components are Highly Expressed in BCC Tissues
The expression levels of lncRNA ANCR and Hedgehog pathway components (PTCH, GLI1, and SMO) in BCC and its adjacent normal tissues were detected via RT-qPCR and Western blotting, respectively. As shown in Figure 1A, compared to that in the adjacent normal tissues, ANCR expression in BCC tissues was significantly upregulated (P<0.0001). Additionally, the expression levels of PTCH, GLI1, and SMO in BCC tissues were notably higher than those in adjacent normal tissues (P<0.0001, Figure 1B and C), suggesting that Hedgehog signaling is activated in BCC.
ANCR Promotes Proliferation and Represses Apoptosis in BCC Cells
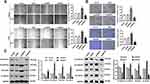
To clarify the effect of ANCR on the biological functions of BCC cells, we modulated the expression of ANCR in TE354.T and A431 cells. Transfection with the overexpression plasmid or shRNAs resulted in significant elevation or suppression of ANCR levels in TE354.T cells relative to those of the Vector group (P < 0.05), indicating that the overexpression vectors and shRNAs had been successfully transfected into TE354.T and A431 cells (Figure 2A–D). Moreover, shANCR-1 showed the greatest effect on the inhibition of ANCR expression. Therefore, shANCR-1 was used for subsequent experiments. Furthermore, the results of CCK-8 and flow cytometry assays showed that ANCR overexpression markedly promoted proliferation and inhibited apoptosis in TE354.T and A431 cells (P < 0.01, Figures 2E, F and 3A). Meanwhile, ANCR overexpression upregulated the pro-apoptosis-related protein BCL2 and downregulated the protein expression of the apoptosis-related proteins BAX and Cleaved-caspase3 (Figure 3B). However, ANCR knockdown had the opposite effects on the above-mentioned cellular biological functions (P < 0.01, Figures 2E, F and 3). These results indicated that ANCR possesses potent proliferation-promoting and apoptosis-inhibiting abilities in BCC cells.
ANCR Promotes Migration and Invasion in BCC Cells
The effects of ANCR on the migration and invasion abilities of TE354.T cells were detected via a wound-healing assay and Transwell assay. Compared with Vector group, ANCR overexpression significantly promoted the migration and invasion of TE354.T and A431 cells (P < 0.01), whereas ANCR knockdown inhibited the migration and invasion compared with shNC group (P < 0.05, Figure 4A and B). Epithelial-mesenchymal transition (EMT) is known to be closely related to cancer cell metastasis.12 Therefore, we further measured the expression of proteins related to the EMT via Western blotting. As shown in Figure 4C, ANCR overexpression significantly increased the expression level of the mesenchymal markers N-cadherin, vimentin, and β-catenin, and decreased the expression level of the epithelial marker E-cadherin in TE354.T and A431 cells, whereas the opposite effect was observed with ANCR knockdown. Collectively, these results demonstrated that ANCR could enhance the migration and invasion ability of BCC cells.
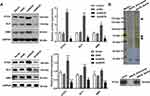
ANCR Acts on the Hedgehog Signaling Pathway
Western blotting analysis of Hedgehog pathway component proteins showed that PTCH, GLI1, and SMO expression levels increased in the ANCR overexpression group compared with Vector group and decreased after ANCR knockdown compared with shNC group (P < 0.05, Figure 5A). The RNA pull-down assay further showed that biotinylated ANCR can bind to PTCH, whereas the anti-sense RNA probe did not bind to PTCH (Figure 5B). Therefore, we speculated that ANCR could interact with PTCH to activate the Hedgehog signaling pathway.
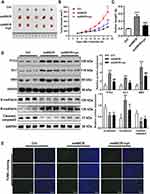
ANCR Promotes BCC Tumor Growth and Inhibits Apoptosis in vivo
TE354.T cells transfected with the ANCR overexpression plasmid were injected into nude mice to explore the pro-oncogenic effect of ANCR in vivo. ANCR overexpression significantly promoted BCC tumor growth (P < 0.001, Figure 6A–C), in the mean time increased the expression levels of PTCH, GLI1, SMO and N-cadherin compared with those in the control group, and decreased the expression level of E-cadherin and Cleaved-caspase3 (P < 0.01, Figure 6D). In addition, TUNEL staining showed that ANCR overexpression repressed cell apoptosis (Figure 6E). However, after the mice were treated with cyclopamine, a specific inhibitor of the Hedgehog signaling pathway, tumor growth and the expression of PTCH, GLI1, SMO and N-cadherin were inhibited compared with oeANCR group (P < 0.01, Figure 6A–D), while apoptosis was promoted (Figure 6E). These results further suggested that ANCR promoted BCC tumor growth and inhibited apoptosis by activating the Hedgehog signaling pathway.
Discussion
BCC is the most common type of skin cancer, with an increasing incidence rate. But yet, the exact molecular mechanism underlying its pathogenesis remains largely unknown. Accumulating evidence supports that lncRNAs, which can regulate gene expression at multiple levels (eg, epigenetic, transcriptional, and post-transcriptional), are widely involved in physiological and pathological processes.13,14 As one type of lncRNA, ANCR is closely associated with the occurrence and development of gastric cancer, hepatocellular carcinoma, glioma, colorectal cancer, and other malignant tumors.6–10 To the best of our knowledge, this is the first study to report the effect of ANCR on BCC. We found that ANCR is more highly expressed in BCC tissues than in adjacent normal tissues. Additionally, ANCR overexpression significantly promoted cell proliferation, migration and invasion, and inhibited the apoptosis of BCC cells, providing insight into the mechanisms by which ANCR contributes to the pathogenesis of BCC.
ANCR plays important roles in the modulation of EMT in many tumours.7,9 EMT is considered to be the main mechanism underlying tumor invasion and metastasis.12 Increasing evidence indicates that this process plays a vital role in the metastasis of multiple malignancies, including BCC, which is accompanied by significant phenotypic changes through the acquisition of mesenchymal marker proteins (N-cadherin, vimentin, and β-catenin) and loss of the epithelial marker protein (E-cadherin).15,16 Moreover, ANCR was reported to be involved in the EMT process.8 Our present results confirmed that ANCR overexpression reduced the expression level of E-cadherin and promoted the expression levels of N-cadherin, vimentin, and β-catenin, indicating that ANCR could promote the metastasis of BCC by inducing the EMT process.
The Hedgehog signaling pathway plays a crucial role in cell differentiation, tissue development, and organ formation during the embryonic period.17–19 Recent studies have suggested that deregulation of the Hedgehog signaling pathway was associated with the initiation and progression of many cancers, including gastric, breast, pancreatic, and prostate cancers, as well as BCC.20–22 In the present study, we also found that the expression levels of PTCH, GLI1, and SMO in BCC tissues were notably elevated, confirming that Hedgehog signaling is activated in these cells. Furthermore, ANCR overexpression elevated the expression levels of proteins related to the Hedgehog signaling pathway (ie, PTCH, GLI1, and SMO), whereas ANCR knockdown resulted in an opposite effect. PTCH and SMO are vital molecules in the Hedgehog signaling pathway, but distinct from which PTCH can directly interact with SMO to inhibit its activity.3,23 An RNA pull-down assay further confirmed that ANCR binds to PTCH, suggesting that ANCR contributes to the regulation of biological processes in BCC via the Hedgehog signaling pathway.
In summary, we have characterized ANCR as a lncRNA that is highly expressed in BCC tissues; effective on promoting cell proliferation, migration, and invasion; and inhibiting apoptosis in BCC by activating the Hedgehog signaling pathway. Therefore, our investigation provides novel insights into the molecular mechanisms underlying BCC pathogenesis, and our findings could serve as the basis for developing a new therapeutic target for the treatment of BCC.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
Animal experiments were performed in accordance with National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals (No.2021-57). All experiments were approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (NO. (2022)1-024). Informed consent was obtained from all patients.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81860484) and the Natural Science Foundation of Jiangxi Province of China (No. 20202BABL206107).
Disclosure
The authors declare that they have no conflict of interest.
References
1. Rogers HW, Weinstock MA, Feldman SR, et al. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012. JAMA Dermatol. 2015;151(10):1081–1086. doi:10.1001/jamadermatol.2015.1187
2. Xin M, Ji X, De La Cruz LK, Thareja S, Wang B. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med Res Rev. 2018;38(3):870–913. doi:10.1002/med.21482
3. Katoh M. Genomic testing, tumor microenvironment and targeted therapy of Hedgehog-related human cancers. Clin Sci. 2019;133(8):953–970. doi:10.1042/CS20180845
4. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
5. Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–343. doi:10.1101/gad.182121.111
6. Pan L, Xiao X, Zhao Y, Yin L, Jiang P. The functional roles of long noncoding RNA DANCR in Human Cancers. J Cancer. 2020;11(23):6970–6981. doi:10.7150/jca.44384
7. Xie C, Guo Y, Lou S. LncRNA ANCR promotes invasion and migration of gastric cancer by regulating FoxO1 expression to inhibit macrophage M1 polarization. Dig Dis Sci. 2020;65(10):2863–2872. doi:10.1007/s10620-019-06019-1
8. Wen Z, Lian L, Ding H, et al. LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 2020;17(3):381–394. doi:10.1080/15476286.2019.1708547
9. Chen Z, Chen X, Xie R, et al. DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing IL-11-STAT3 signaling and CCND1 expression. Mol Ther. 2019;27(2):326–341. doi:10.1016/j.ymthe.2018.12.015
10. Cheng C, Dong Y, Ru X, Xia Y, LncRNA JY. ANCR promotes glioma cells invasion, migration, proliferation and inhibits apoptosis via interacting with EZH2 and repressing PTEN expression. Cancer Gene Ther. 2021;28(9):1025–1034. doi:10.1038/s41417-020-00263-8
11. Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59–71. doi:10.1038/cdd.2016.95
12. Rhim A, Mirek E, Aiello N, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi:10.1016/j.cell.2011.11.025
13. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887
14. Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorg Chem. 2019;92:103214. doi:10.1016/j.bioorg.2019.103214
15. Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): a biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232(12):3261–3272. doi:10.1002/jcp.25797
16. Mochel MC, Liaquat S, Moore JB, Hoang MP. Metastasizing basal cell carcinoma: a clinicopathologic and immunohistochemical study of 22 cases. J Cutan Pathol. 2021;48(3):374–383. doi:10.1111/cup.13888
17. Jia Y, Wang Y, Xie J. The Hedgehog pathway: role in cell differentiation, polarity and proliferation. Arch Toxicol. 2015;89(2):179–191. doi:10.1007/s00204-014-1433-1
18. Kong J, Siebold C, Rohatgi R. Biochemical mechanisms of vertebrate hedgehog signaling. Development. 2019;146(10):dev166892. doi:10.1242/dev.166892
19. Roberts K, Kershner A, Beachy P. The stromal niche for epithelial stem cells: a template for regeneration and a brake on malignancy. Cancer Cell. 2017;32(4):404–410. doi:10.1016/j.ccell.2017.08.007
20. Xu Y, Song S, Wang Z, Ajani JA. The role of hedgehog signaling in gastric cancer: molecular mechanisms, clinical potential, and perspective. Cell Commun Signal. 2019;17(1):157. doi:10.1186/s12964-019-0479-3
21. Riobo-Del Galdo NA, Lara Montero A, Wertheimer EV. Role of Hedgehog signaling in breast cancer: pathogenesis and therapeutics. Cells. 2019;8(4):375. doi:10.3390/cells8040375
22. Thayer SP, Magliano M, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425(6960):851–856. doi:10.1038/nature02009
23. Rohatgi RML, Corcoran RB, Scott MP, Scott MP. Hedgehog signal transduction by smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci U S A. 2009;106(9):3196–3201. doi:10.1073/pnas.0813373106
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.