Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA PVT1 Regulates BAMBI To Promote Tumor Progression In Non-Small Cell Lung Cancer By Sponging miR-17-5p
Authors Wang Z, Zhang Q, Sun Y, Shao F
Received 27 May 2019
Accepted for publication 14 September 2019
Published 7 January 2020 Volume 2020:13 Pages 131—142
DOI https://doi.org/10.2147/OTT.S217335
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Federico Perche
Zhao Wang,* Qiang Zhang,* Yungang Sun, Feng Shao
Department of Thoracic Surgery, The Nanjing Chest Hospital, Nanjing, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Feng Shao
Department of Thoracic Surgery, The Nanjing Chest Hospital, No. 215, Guangzhou Road, Nanjing, Jiangsu 210029, People’s Republic of China
Tel +86 25 5861 9746
Email [email protected]
Background: Non-small cell lung cancer (NSCLC) is a common malignancy over the world. Previous report indicated that the plasmacytoma variant translocation 1 (PVT1) has been documented to function as an oncogene in various types of human cancers. However, the biological mechanism of PVT1 was still rarely reported in NSCLC.
Methods: The levels of PVT1, miR-17-5p, and bone morphogenetic protein and activin membrane-bound inhibitor (BAMBI) in NSCLC tissues (n=64) and cells (H1299 and A549) were detected by qRT-PCR and immunohistochemistry (IHC). The protein level of BAMBI was measured by Western blot assay. Cell viability and apoptotic rate were evaluated by MTT assay and flow cytometry, respectively. The migrated and invaded abilities were assessed by Transwell assay and Wound healing assay. The interactions between miR-17-5p and PVT1 or BAMBI were predicted by starBase v2.0 and TargetScan, respectively, and then dual-luciferase reporter assay and RNA pull-down assay were performed to verify these interactions. The mice model experiments were constructed to further validate the roles of PVT1 in vivo.
Results: The levels of PVT1 and BAMBI were both apparently increased, and miR-17-5p was declined in NSCLC tissues and cells. The depletion of PVT1 or BAMBI blocked cell viability, migrated and invaded abilities but impelled apoptotic rate in A549 and H1299 cells. PVT1 was validated as a sponge to miR-17-5p and BAMBI was a direct target of miR-17-5p. PVT1 promoted cell viability, migrated and invaded abilities but repressed apoptotic rate by targeting BAMBI. MiR-17-5p regulated cell behaviors mediated by PVT1. PVT1 silencing decreased BAMBI expression by sponging miR-17-5p. In addition, PVT1 knockdown blocked the xenograft tumor growth in vivo.
Conclusion: These results manifested that PVT1 modulated BAMBI to promote tumor progression in NSCLC by sponging miR-17-5p. Thus, the novel regulatory pathway may provide a new therapeutic target for NSCLC patients.
Keywords: PVT1, miR-17-5p, BAMBI, tumor progression, non-small cell lung cancer
Corrigendum for this paper has been published
Introduction
Non-small cell lung cancer (NSCLC) is a common cancer worldwide. More than 85% of the lung cancer patients are diagnosed as NSCLC.1 Despite surgical operations and drugs were improved in recent decades, the 5‑year survival rate is as low as 15.9%.2 Therefore, it is imperative to find novel therapeutic targets in the early stage of NSCLC.
Long non-coding RNAs (lncRNAs), a class of non-coding RNAs with more than 200 nucleotides (nts), have been documented to regulate gene expression at the transcriptional, posttranscriptional stage.3 The plasmacytoma variant translocation 1 (PVT1) located in the human chromosome 8q24 gene desert and PVT1 has been reported to function as an oncogene and highly expressed in devious human tumors.4 For example, Fang et al reported that PVT1 was strikingly increased in glioma tissues, and its expression was higher in high grade (III-IV) than that in low grade (I-II) tumors.5 Another study in gallbladder cancer demonstrated that the level of PVT1 was remarkably elevated in gallbladder cancer tissues and cells, and its knockdown retarded cell proliferation, migration and invasion in vitro and also suppressed xenograft tumor growth in vivo.6 In fact, emerging documents in NSCLC revealed that PVT1 was notably upregulated, and its depletion significantly blocked cell proliferation, invasion but induced cell apoptosis in NSCLC.7 Although PVT1 has been reported to be an oncogene in NSCLC, the biological mechanism of PVT1 was rarely defined.
MicroRNAs (miRNAs) is a type of small non-coding RNAs about 22 nt in length and could function as mRNA inhibitor by blocking mRNA 3ʹUTR or mediating the degradation pathway.8 The aberrant expression of miR-17-5p often associates with cancer progression. For example, the level of miR-17-5p was significantly decreased in triple-negative breast cancer tissues and cells.9 Actually, previous study in NSCLC showed that miR-17-5p was conspicuously reduced in NSCLC tissues and cell lines (SK-MES-1, A549, SPCA-1, H460, H1229 and HCC827) and miR-17-5p overexpression distinctly constrained cell proliferation while induced cell apoptosis in vitro.10 Bone morphogenetic protein and activin membrane-bound inhibitor (BAMBI) is a transmembrane glycoprotein and upregulated in gastric cancer,11 gliomas.12 However, the mechanisms of miR-17-5p and BAMBI have scarcely reported in NSCLC. In this study, we mainly explored the effects of PVT1 in NSCLC.
Materials And Methods
Patients And Specimens
This project was approved by the Ethics Committee of the Nanjing Chest Hospital and performed in accordance with the Declaration of Helsinki Principles. Sixty-four tumor tissue samples and its corresponding adjacent normal tissue samples were collected from NSCLC patients in the Nanjing Chest Hospital and then stored in −80°C refrigerator until further use. Written informed consents were provided for all patients in this project.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA in ESCLC tissues or cells was extracted using miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) and the reverse transcription was carried out using miScript RT Kit (TaKaRa, Dalian, China). The quantitative PCR was performed using FastStart Universal SYBR Green Master (Roche, Germany) on C1000 thermal cycler (Bio-Rad, Hercules, CA, USA). The levels of PTV1, BAMBI and miR-17-5p were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or small nuclear RNA U6, respectively, and calculated by the 2−ΔΔCt method. The primers used in this program were synthesized in Beijing Genomics Institute (BGI, Shenzhen, China) and listed as follows: PVT1: (Forward, 5ʹ-TGAGAACTGTCCTTACGTGACC-3ʹ, Reverse, 5ʹ-AGAGCACCAAGACTGGCTCT-3ʹ), miR-17-5p: (Forward, 5ʹ-CCAGGATCCTTTATAGTTGTTAGAGTTT-3ʹ, Reverse, 5ʹ-CGGAATTCTAATCTACTTCACTATCTGCAC-3ʹ), BAMBI: (Forward, 5ʹ-CTCAAATTCCCCACTCACCCA-3ʹ, Reverse, 5ʹ-GCTGATACCTGTTTCCTTGTCCTG-3ʹ), GAPDH: (Forward, 5ʹ-TGTTCGTCATGGGTGTGAAC-3ʹ, Reverse, 5ʹ-ATGGCATGGACTGTGGTCAT-3ʹ) and U6: (Forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, Reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
Western Blot Assay
Protein in ESCLC tissues and cells was extracted using a protein extraction kit (Beyotime, Shanghai, China) and the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockville, MD, USA) was used to detect the protein concentration. The protein samples were separated by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Subsequently, the membrane was blocked in non-fat milk for 4 hrs and incubated with primary antibody at 4°C overnight. Then, the membrane was incubated with the secondary antibody for another 2 hrs at 37°C. RapidStep ECL Reagent (Millipore) was used to detect the chemiluminescence intensity. BAMBI (1:200; ab203070) primary antibody and goat anti-rabbit IgG (1:5000; ab6721) secondary antibody were purchased from Abcam (Cambridge, MA, USA).
Immunohistochemistry (IHC)
The NSCLC tissue samples and normal tissue samples were firstly fixed in 4% neutral formaldehyde and then embedded in paraffin. After being sliced into 4 μm, the slides were blocked with 3% H2O2 for 10 mins to avoid the endogenous peroxidase. Then, the slides were blocked with goat serum and incubated with rabbit primary antibody against BAMBI (Abcam) at 4°C overnight. The slides were probed with second antibody at 37°C for 30 mins. To visualize staining, slides were firstly stained with 3, 30-diaminobenzidine in Tris–HCl buffer and subsequently counterstained with hematoxylin.
Cell Culture And Transfection
Two NSCLC cell lines (H1299 and A549) and human bronchial epithelial cells (HBE) were purchased from China center for type culture collection (Wuhan, China). All cells were cultivated in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific) in an incubator with the parameters of 37°C, 5% CO2. Small interfering RNA (siRNA) targeting PTV1 (si-PTV1, 5ʹ-GCUUGGAGGCUGAGGAGUUTT-3ʹ) and its mock (control), miR-17-5p mimics (miR-17-5p) and its matched negative control (miR-control), miR-17-5p inhibitor and its matched negative control (inhibitor- control), siRNA against BAMBI (si-BAMBI) and its mock (si-control) were obtained from GenePharma (Shanghai, China). The sequences of BAMBI were inserted into pcDNA (Hanbio Biotechnology, Shanghai, China) to construct the overexpression vector of BAMBI (pcDNA-BAMBI). Cell transfection was conducted using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s manual.
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
MTT (Solarbio, Beijing, China) was used for the detection of cell viability. The A549 and H1299 cells (3×104 per well) were injected into a 96-well plate and cultured for 24 hrs. After the transfection, the A549 and H1299 cells were cultivated for another 0, 24, 48, and 72 hrs. Then, MTT was added in the well and incubated for 4 hrs. The dimethyl sulfoxide (DMSO) was then added into each well to dissolve the formazan. The absorbance at 490 nm was detected using a spectrophotometer (Thermo Fisher Scientific).
Cell Apoptosis Assay
Annexin V‑fluorescein isothiocyanate (FITC) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) was used to detect apoptotic rate in accordance with its manual. The resuspended H1299 and A549 cells were incubated with Annexin V‑FITC and propidium iodide (PI) solution for 15 mins in the dark. The apoptotic rate of H1299 and A549 cells was detected using flow cytometry (FACScan, BD Biosciences).
Transwell Assay
The Transwell chambers (Corning, Tewksbury, MA, USA) were used to detect the metastasis ability of H1299 and A549 cells. For migration, the lower chamber was added with RPMI-1640 medium containing 10% FBS while the upper chamber was filled with serum-free medium. The transfected H1299 and A549 cells were injected into the upper chamber and incubated for 48 hrs. The cells on the lower side of upper chamber were stained with 20% methanol and dyed with 0.1% crystal violet for 30 mins. The number of migrated cells was photographed using a microscope (Olympus, Tokyo, Japan) and calculated by Image-Pro Plus (Media Cybernetics, Inc., Rockville, MD, USA). The protocols in the invasion assay were similar with that in migration assay. The difference was that the upper chamber in the invasion assay was coated with Matrigel (BD Biosciences).
Wound Healing Assay
The H1299 and A549 cells were cultivated in 6-well plate until the cell adhesion to 80–90%. Then, the wound was generated by scratching with the 200 μL tip. Then, the cells were treated with 1 μg/mL mitomycin for 1 hr and washed with serum-free medium. The H1299 and A549 cells were maintained at 37°C for another 24 hrs. The wounded areas were photographed at 0 and 24 hrs and then calculated the Wound healing rate with the formula of [1 - (empty area at 48 hrs/empty area at 0 hr)] ×100%.
Dual-Luciferase Reporter Assay
The putative interactions between miR-17-5p and PVT1 or BAMBI were predicted by starBase v2.0 (http://starbase.sysu.edu.cn) and TargetScan (http://www.targetscan.org) online databases, respectively. The sequences of PTV1 and BAMBI 3ʹUTR or their corresponding mutant were cloned and inserted into the pGL3 vector (Promega, Madison, WI, USA), namely PVT1 WT (PVT1 MUT) and BAMBI 3ʹUTR WT (BAMBI 3ʹUTR MUT). Then, the luciferase reporter and miR-17-5p mimics or miR-NC were co-transfected into H1299 and A549 cells. Dual-luciferase reporter assay kit (Promega) was used to detect the luciferase reporter activity.
RNA Pull-Down Assay
Bio-labeled miR-17-5p (Bio-miR-17-5p) and Bio-labeled negative control (Bio-NC) were obtained from Sangon (Shanghai, China). The lysate samples of H1299 and A549 cells were incubated with Bio-miR-17-5p or Bop-NC. Then, the RNA-RNA complex was conjugated with streptavidin magnetic beads. After elution, the level of PVT1 was measured by qRT-PCR.
Mice Model Experiments
The animal experiment was permitted by the Animal Care Committee of the Nanjing Chest Hospital in accordance with instructions. The five-week-old nude mice were purchased from Shanghai Laboratory Animal Center (Shanghai, China). The H1299 cells (3×106) stably transfected with sh-PVT1 or sh-NC were injected into nude mice (n=6 per group). The volume of tumor was measured every 7 days for 28 days. Following resection, the weight of tumor was detected and the tumor was stored in −80°C for further study.
Statistical Analysis
All data in this study were performed using GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). All quantitative data were repeated for at least three times and presented as mean ± standard deviation (SD). Student’s t-test was used to analyze the differences between two groups, while multigroup was assessed by one-way analysis of variance (ANOVA). P< 0.05 was considered statistically significant difference.
Results
PVT1 And BABMI Were UpRegulated In NSCLC Tissues
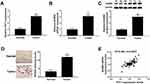
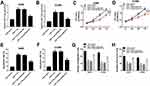
To explore the biological functions of PVT1 and BAMBI in NSCLC, we first detected the level of PVT1 and BAMBI in NACLS tissues. The qRT-PCR results showed that the relative expression of PVT1 and BAMBI was obviously increased in NSCLC tissues compared with that in corresponding adjacent normal tissues (Figure 1A and B). Also, the Western blot exhibited that the protein level of BAMBI was significantly elevated in NSCLC tissues (Figure 1C). The IHC assay exhibited that BAMBI was highly expressed in NSCLC tissues related to that in normal tissues (Figure 1D). In addition, the scatter diagram presented that the level of BAMBI was positively linear correlated with the level of PVT1 (Figure 1E). Taken together, these results manifested that PVT1, BABMI may play vital roles in NSCLC.
The Depletion Of PVT1 Suppressed Cell Proliferation, Migration, And Invasion While Induced Cell Apoptosis In NSCLC Cells
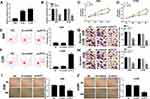
To further detect the biological roles of PVT1 in NSCLC, PVT1 knockdown was conducted in NSCLC cells. First, qRT-PCR results showed that PVT1 was highly expressed in H1299 and A549 cells compared with that in HBE cells (Figure 2A). Then, the knockdown efficiency was confirmed, indicated by the apparent downregulation of PVT1 level in H1299 and A549 cells transfected with si-PVT1 (Figure 2B). Furthermore, the transfection of si-PVT1 retarded cell viability in si-PVT1-transfected H1299 and A549 cells (Figure 2C and D). However, the apoptotic rate was strikingly enhanced in H1299 and A549 cells transfected with si-PVT1 in comparison with that in negative control groups (Figure 2E and F). The transwell assay indicated that the introduction of si-PVT1 contributed to the remarkable decrease of migrated and invaded abilities in H1299 and A549 cells (Figure 2G and H). Also, the wound healing assay presented that the migrated ability was dramatically reduced in H1299 and A549 cells transfected with si-PVT1 (Figure 2I and J). These data demonstrated that PVT1 knockdown blocked cell proliferation, migration, and invasion but promoted cell apoptosis in NSCLC cells.
BAMBI Knockdown Inhibited Cell Proliferation, Migration, And Invasion And Facilitated Cell Apoptosis In NSCLC Cells
Subsequently, the biological functions of BAMBI were further explored in NSCLC. The mRNA and protein levels of BAMBI were significantly elevated in A549 and H1299 cells related to that in HBE cells (Figure 3A and B). The protein level of BAMBI was obviously decreased in si-BAMBI-transfected H1299 and A549 cells, suggesting the knockdown efficiency (Figure 3C and D). Moreover, the transfection of si-BAMBI resulted in the apparent decrease of cell viability in H1299 and A549 cells (Figure 3E and F), as well as the migrated and invaded abilities (Figure 3I and J). However, the apoptotic rate was drastically elevated in si-BAMBI group compared to that in negative control groups (Figure 3G and H). Summarily, these results revealed that BAMBI silencing repressed NSCLC progression.
BAMBI Overexpression Attenuated The Inhibitory Effects On Cell Proliferation, Migration, And Invasion, As Well As The Facilitated Effect On Cell Apoptosis Mediated By PVT1
Based on the above results, we found that PVT1 or BAMBI silencing can both contribute to the inhibition of cell proliferation, migration and invasion and promotion of cell apoptosis. Subsequently, PVT1 and BAMBI were studied together in NSCLC cells. The protein level of BAMBI was dramatically down-regulated in si-PVT1-transfected A549 and H1299 cells related to that in matched negative control groups (Figure 4A and B). The MTT assay showed that the cell viability was notably decreased in si-PVT1 group, while BAMBI overexpression reversed the inhibitory effect in A549 and H1299 cells (Figure 4C and D). Similarly, the emergence of pcDNA-BAMBI regained migrated and invaded abilities in H1299 and A549 cells constrained by si-PVT1 (Figure 4G and H). Whereas, the flow cytometry results exhibited that the overexpression of BAMBI weakened the accelerated impact on apoptotic rate in H1299 and A549 cells transfected with si-PVT1 (Figure 4E and F). These data illustrated that BAMBI overexpressed alleviated the inhibitory impacts on cell proliferation, migration, and invasion and the promoted effect on cell apoptosis induced by PVT1 silencing.
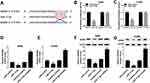
PVT1 Was Sponged To miR-17-5p In A549 And H1299 Cells
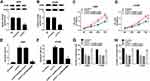
To explore the biological mechanism of PVT1 in NSCLC, the putative target was searched by starBase v2.0. As shown in Figure 5A, miR-17-5p had complementary sequences with PVT1. The dual-luciferase reporter assay implicated that the transfection of miR-17-5p contributed to the prominent downregulation of luciferase activity of PVT1 WT reporter in A549 and H1299 cells, while had no influence on luciferase activity of PVT1 MUT reporter (Figure 5B and C). As presented in Figure 5D and E, the level of PVT1 was much more enriched in Bio-miR-17-5p group related to that in Bio-NC group. Meanwhile, the level of miR-17-5p was markedly reduced in NSCLC tissues and cells (Figure 5F and G). In addition, the level of miR-17-5p was evidently enhanced in A549 and H1299 cells transfected with si-PVT1, while distinctly decreased in pcDNA-PVT1-transfected A549 and H1299 cells (Figure 5H and I). These results unraveled that miR-17-5p negatively interacted with PVT1.
miR-17-5p Blocked Cell Proliferation, Migration, And Invasion But Accelerated Apoptosis In A549 And H1299 Cells Mediated By PVT1
In order to search the effects of miR-17-5p and PVT1 in ESCLC, miR-17-5p and pcDNA-PVT1 were co-transfected into A549 and H1299 cells. The qRT-PCR results indicated that the level of miR-17-5p was conspicuously increased in A549 and H1299 cells transfected with miR-217-5p mimics, while partly declined with the transfection with pcDNA-PVT1 (Figure 6A and B). The cell viability, migrated and invaded abilities were repressed in A549 and H1299 cells transfected with miR-17-5p mimics, while the introduction of PVT1 rescued these inhibitory effects (Figure 6C and D, 6G and H). The transfection of pcDNA-PVT1 contributed to the distinct elevated of apoptotic rate in A549 and H1299 cells, but pvt1 overexpression attenuated the promoted impact (Figure 6E and F). These data disclosed that miR-17-5p suppressed cell proliferation, migration, and invasion but facilitated apoptosis in A549 and H1299 cells regulated by PVT1.
BAMBI Was Negatively Interacted With miR-17-5p In A549 And H1299 Cells
To further understanding the biological mechanism of BAMBI in NSCLC, the putative target was searched by TargetScan. The results showed that miR-17-5p also had complementary binding sites with BAMBI 3ʹUTR (Figure 7A). Following dual-luciferase reporter assay indicated that the luciferase activity of BAMBI 3ʹUTR WT reporter was strikingly reduced in A549 and H1299 cells transfected with miR-17-5p mimics compared with that in miR-NC group, while the luciferase activity BAMBI 3ʹUTR MUT reporter had no significant fluctuation in any group (Figure 7B and C). Furthermore, the mRNA and protein levels of BAMBI were dramatically down-regulated in miR-17-5p-transfected A549 and H1299 cells in comparison with that in miR-NC, while exhibited the opposite trend in miR-17-5p inhibitor group (Figure 7D–G). Taken together, BAMBI was a direct candidate of miR-17-5p.
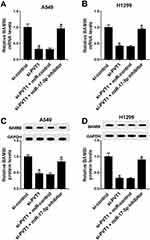
PVT1 Positively Regulated BAMBI Expression By Sponging miR-17-5p
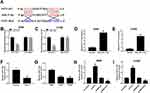
To further elucidate the relationship among PVT1, miR-17-5p and BAMBI in NSCLC, si-PVT1 and miR-17-5p inhibitor were co-transfected into A549 and H1299 cells. The qRT-PCR results showed that the level of BAMBI was distinctly downregulated in si-PVT1-transfected A549 and H1299 cells, while the transfection of miR-17-5p partially regained BAMBI expression (Figure 8A and B). Similarly, the protein level of BAMBI exhibited the same trend in A549 and H1299 cells (Figure 8C and D). These results unraveled that PVT1 regulated BAMBI by sponging miR-17-5p.
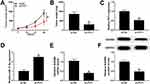
PVT1 Depletion Restrained Xenograft Tumor Growth In Vivo
To further verify the functions of PVT1 in vivo, the mice model was constructed. The measurement results showed that the tumor volume and weight were both markedly reduced in sh-PVT1 group in contrast to that in sh-NC group (Figure 9A and B). Furthermore, the level of PVT1 was obviously declined, while miR-17-5p was significantly upregulated in sh-PVT1 xenograft tumor group (Figure 9C and D). Also, the mRNA and protein levels of BAMBI were evidently reduced in sh-PVT1 group (Figure 9E and F). These data demonstrated that the silencing of PVT1 blocked xenograft tumor growth in vivo.
Discussion
NSCLC is a common malignant cancer. Emerging evidence has manifested that lncRNAs play crucial roles in many types of cancer including NSCLC. In this study, we aimed to explore the biological mechanism of PVT1 in NSCLC. All results revealed that PVT1 promoted NSCLC progression via miR-17-5p/BAMBI axis.
LncRNAs have been reported to serve as oncogenic role in the development of NSCLC. Song et al reported that lncRNA AWPPH was dramatically increased in NSCLC cells, and AWPPH overexpression in NSCLC cells impelled cell proliferation and restrained cell apoptosis.13 Another report showed that CRNDE was upregulated in NSCLC, while its depletion retarded cell proliferation while enhanced apoptosis in NSCLC.14 In the present study, we found that PVT1 was highly expressed in NSCLC tissues and cells, and the depletion of PVT1 suppressed cell proliferation, migration, and invasion but induced cell apoptosis in vitro. PVT1 silencing also curbed xenograft tumor growth in vivo. The results indicated that PVT1 may play a crucial role in NSCLC.
Emerging evidence indicated that lncRNAs exerted competing endogenous (ceRNAs) function to sponge miRNAs and further affect the expression of mRNA.15 A study in NSCLC reported that lncRNA SNHG1 promoted cell proliferation, migration, and invasion in NSCLC by sponging miR-497.16 Hu et al documented that LINC01296 was upregulated and miR-5095 was downregulated in NSCLC, LINC01296 enhanced cell proliferation and migration via regulating miR-5095.17 In this study, PVT1 was validated as a sponge to miR-17-5p. MiR-17-5p mimics inhibited cell proliferation, migration, and invasion but induced apoptosis in A549 and H1299 cells mediated by PVT1. Taken together, these results demonstrated that PVT1 accelerated NSCLC progression by sponging miR-17-5p.
BAMBI has been known as a transmembrane glycoprotein and is a pseudo-receptor of T-box brain protein 1 (TBR1).18 Previous documents reported that the dysregulation of BAMBI was found in different types of cancers and correlated with tumor progression.19 A study in osteosarcoma demonstrated that BAMBI was highly expressed, and BAMBI overexpression enhanced cell migration and invasion.20 Also, the upregulation of BAMBI was reported in ovarian cancer21 and colorectal cancer.22 In this research, we found that BAMBI was highly expressed in NSCLC cells, and BAMBI knockdown suppressed cell proliferation, migration, and invasion and induced cell apoptosis. The level of BAMBI was a controversy in different types of cancers. For instance, another study demonstrated that BAMBI was significantly reduced in high-grade bladder cancer.23 Also, Marwitz et al reported that BAMBI was strikingly declined in NSCLC tissues and resulted in the NSCLC development.24 These reports were contrary to our results. The controversial conclusions may be attributed to the different TNM stages of tissue samples or microsatellite instability statuses of tumor tissues. Currently, BAMBI was verified as a direct target of miR-17-5p. The restoration experiment exhibited that PVT1 knockdown impeded cell proliferation, migration, and invasion but promoted apoptosis in A549 and H1299 cells by regulating BAMBI. The depletion of PVT1 downregulated BAMBI expression by sponging miR-17-5p. Taken together, these results indicated that PVT1 positively modulated BAMBI expression to regulate cell behaviors in NSCLC by sponging miR-17-5p.
In conclusion, our results demonstrated that PVT1/miR-17-5p/BAMBI regulatory network regulated NSCLC progression. This new regulatory network may provide novel biomarker for NSCLC patients. Since TGF-β as a driver of cancer progression in the middle and advanced stage,25,26 previous study reported that PVT1 facilitated epithelial‑mesenchymal transition (EMT) via TGF‑β/Smad signaling pathway in pancreatic cancer.27 In the future study, we will explore whether the PVT1/miR-17-5p/BAMBI regulatory network on NSCLC progression was mediated by TGF‑β/Smad signaling pathway.
Acknowledgment
Zhao Wang and Qiang Zhang are co-first authors for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535. doi:10.1038/nrc3775
2. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
3. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
4. Cui D, C H Y, Liu M, et al. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol. 2016;37(3):4127–4134. doi:10.1007/s13277-015-4261-x
5. Fang J, Huang J. Clinical significance of the expression of long non-coding RNA PVT1 in glioma. Cancer Biomark. 2019;(Preprint):1–5.
6. Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33. doi:10.1186/s12943-019-1010-6
7. Guo D, Wang Y, Ren K, et al. Knockdown of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer by regulating miR-497 expression. Exp Cell Res. 2018;362(1):172–179. doi:10.1016/j.yexcr.2017.11.014
8. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi:10.1038/nrg3074
9. Li J, Lai Y, Ma J, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer. 2017;17(1):745. doi:10.1186/s12885-017-3674-x
10. Li H, Zhou H, Luo J, et al. MicroRNA-17-5p inhibits proliferation and triggers apoptosis in non-small cell lung cancer by targeting transforming growth factor β receptor 2. Exp Ther Med. 2017;13(6):2715–2722. doi:10.3892/etm.2017.4347
11. Zhang Y, Yu Z, Xiao Q, et al. Expression of BAMBI and its combination with Smad7 correlates with tumor invasion and poor prognosis in gastric cancer. Tumour Biol. 2014;35(7):7047–7056. doi:10.1007/s13277-014-1962-5
12. Wang D, Chen X, Zhang R. BAMBI promotes macrophage proliferation and differentiation in gliomas. Mol Med Rep. 2018;17(3):3960–3966. doi:10.3892/mmr.2017.8320
13. Song Z, Du J, Zhou L, et al. lncRNA AWPPH promotes proliferation and inhibits apoptosis of non‑small cell lung cancer cells by activating the Wnt/β‑catenin signaling pathway. Mol Med Rep. 2019;19(5):4425–4432. doi:10.3892/mmr.2019.10089
14. Fan YF, Yu ZP, Cui XY. lncRNA Colorectal Neoplasia Differentially Expressed (CRNDE) promotes proliferation and inhibits apoptosis in non-small cell lung cancer cells by regulating the miR-641/CDK6 axis. Med Sci Monit. 2019;25:2745. doi:10.12659/MSM.913420
15. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi:10.1016/j.semcdb.2014.05.015
16. Li Z, Lu Q, Zhu D, et al. Lnc-SNHG1 may promote the progression of non-small cell lung cancer by acting as a sponge of miR-497. Biochem Biophys Res Commun. 2018;506(3):632–640. doi:10.1016/j.bbrc.2018.10.086
17. Hu X, Duan L, Liu H, et al. Long noncoding RNA LINC01296 induces non-small cell lung cancer growth and progression through sponging miR-5095. Am J Transl Res. 2019;11(2):895–903.
18. Sekiya T, Adachi S, Kohu K, et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;279(8):6840–6846. doi:10.1074/jbc.M310876200
19. Sekiya T, Oda T, Matsuura K, et al. Transcriptional regulation of the TGF-beta pseudoreceptor BAMBI by TGF-beta signaling. Biochem Biophys Res Commun. 2004;320(3):680–684. doi:10.1016/j.bbrc.2004.06.017
20. Zhou L, Park J, Jang KY, et al. The overexpression of BAMBI and its involvement in the growth and invasion of human osteosarcoma cells. Oncol Rep. 2013;30(3):1315–1322. doi:10.3892/or.2013.2569
21. Pils D, Wittinger M, Petz M, et al. BAMBI is overexpressed in ovarian cancer and co-translocates with Smads into the nucleus upon TGF-β treatment. Gynecol Oncol. 2010;117(2):189–197. doi:10.1016/j.ygyno.2009.12.034
22. Togo N, Ohwada S, Sakurai S, et al. Prognostic significance of BMP and activin membrane-bound inhibitor in colorectal cancer. World J Gastroenterol. 2008;14(31):4880. doi:10.3748/wjg.14.4880
23. Khin SS, Kitazawa R, Win N, et al. BAMBI gene is epigenetically silenced in subset of high-grade bladder cancer. Int J Cancer. 2009;125(2):328–338. doi:10.1002/ijc.24318
24. Marwitz S, Depner S, Dvornikov D, et al. Downregulation of the TGFβ pseudoreceptor BAMBI in non–small cell lung cancer enhances TGFβ signaling and invasion. Cancer Res. 2016;76(13):3785–3801. doi:10.1158/0008-5472.CAN-15-1326
25. Katsuno Y, Lamouille S, Derynck R. TGF-β signaling and epithelial–mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25(1):76–84. doi:10.1097/CCO.0b013e32835b6371
26. Massagué J. TGFβ in cancer. Cell. 2008;134(2):215–230. doi:10.1016/j.cell.2008.07.001
27. Zhang X, Feng W, Zhang J, et al. Long non‑coding RNA PVT1 promotes epithelial‑mesenchymal transition via the TGF‑β/Smad pathway in pancreatic cancer cells. Oncol Rep. 2018;40(2):1093–1102. doi:10.3892/or.2018.6462
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.