Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA PITPNA-AS1 Accelerates the Progression of Colorectal Cancer Through miR-129-5p/HMGB1 Axis
Received 29 June 2020
Accepted for publication 7 September 2020
Published 3 December 2020 Volume 2020:12 Pages 12497—12507
DOI https://doi.org/10.2147/CMAR.S267844
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Chuanping Yuan, Ling Yang
Department of Oncology, The Xinyu People’s Hospital, Xinyu, Jiangxi 338000, People’s Republic of China
Correspondence: Ling Yang
Department of Oncology, The Xinyu People’s Hospital, No.369 Xinxin North Road, Xinyu, Jiangxi 338000, People’s Republic of China
Email [email protected]
Background: Long non-coding RNAs (lncRNAs) are essential regulators in colorectal cancer (CRC) progression. This work aimed to delve into the characteristics of lncRNA PITPNA-AS1 in CRC.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was adopted to examine PITPNA-AS1, miR-129-5p, high-mobility group box-1 (HMGB1) mRNA expressions in CRC tissues and cell lines. Functionally, cell counting kit-8 (CCK-8) and 5-ethynyl-2-deoxyuridine (EdU) incorporation assays were employed to examine cell proliferation; wound healing assay was utilized to detect cell migration; and flow cytometry was used to detect the cell apoptosis. Luciferase reporter assay, RNA immunoprecipitation assay and Western blot were conducted to detect the regulatory relationships among PITPNA-AS1, miR-129-5p and HMGB1.
Results: PITPNA-AS1 and HMGB1 mRNA expressions were observably elevated in CRC tissues and cell lines. Knocking down PITPNA-AS1 could significantly inhibit cell proliferation and migration, and promote apoptosis of CRC cells. PITPNA-AS1 could serve as a competitive endogenous RNA, and up-regulate HMGB1 expression by adsorbing miR-129-5p.
Conclusion: PITPNA-AS1 can expedite CRC cell proliferation and migration, and inhibit apoptosis through miR-129-5p/HMGB1 axis.
Keywords: colorectal cancer, PITPNA-AS1, miR-129-5p, HMGB1
Introduction
Colorectal cancer (CRC) is a common tumor globally with its incidence still on the rise.1 Although great progress has been made in surgery, chemotherapy, radiotherapy and targeted therapy, CRC patients still suffer from poor prognosis due to relapse, metastasis and drug resistance.2 Hence, it is still essential to investigate the underlying mechanisms of CRC progression to provide novel insights for the diagnosis and treatment of CRC.
Long non-coding RNAs (lncRNAs) are RNA molecules with over 200 nt in length. At present, more and more studies find that lncRNA can be a participant in diverse physiological and pathological processes, and it plays a regulatory role in cancers.3 It is confirmed that some lncRNAs participate in the CRC progression, such as MALAT1,4 SNHG3,5 etc. PITPNA antisense RNA 1 (PITPNA-AS1) is a newly discovered lncRNA, which serves as a cancer-promoting molecule in hepatocellular carcinoma.6 Instead, the role and potential mechanism of PITPNA-AS1 in CRC still require a clearer interpretation.
MicroRNAs (miRs), known as non-coding RNAs with about 20 nt, are implicated in tumor progression by suppressing target gene expressions.7 Accumulating evidence proves that miRs take part in the progression of CRC, such as miR-143 and miR-145.8 As a tumor suppressor, miR-129-5p participates in various tumors, like rectal adenocarcinoma9 and gastric cancer.10 It is reported that miR-129-5p expression is declined in CRC cell lines, and miR-129-5p can impede cancer cell proliferation and apoptosis.4 Nonetheless, the mechanism of miR-129-5p dysregulation in CRC is unclear.
High-mobility group box-1 (HMGB1) partakes in the carcinogenesis and development of cancers, and is regarded as a new diagnostic marker and therapy target.11 Besides, HMGB1 is highly expressed in diverse tumors, CRC included.12 Moreover, lymph node metastasis, distant metastasis and poor prognosis in patients with CRC are related to the high expression of HMGB1.13
Recent evidence authenticates that lncRNAs figure prominently in tumorigenesis and cancer progression by acting as competing endogenous RNAs (ceRNAs), which means that lncRNAs act as molecular sponges to reduce the availability of miRNAs, and modulate the expressions of target genes.14 In this work, we analyzed PITPNA-AS1 expression in CRC tissues and cell lines and its relationship with clinical pathological characteristics. Then, we investigated the role of PITPNA-AS1 in regulating CRC cell proliferation, migration and apoptosis. Subsequently, the regulatory function of PITPNA-AS1 on miR-129-5p/HMGB1 axis was explored. It was demonstrated that PITPNA-AS1 could function as a ceRNA, by repressing miR-129-5p expression to up-regulate HMGB1 expression, to facilitate the progression of CRC.
Materials and Methods
Ethics Approval
This project complied with the principles of the Declaration of Helsinki and was endorsed by the Research Ethics Committee of the Xinyu People’s Hospital (No.201508005). All subjects involved signed written informed consent.
Patient Tissues Collection
From 2015 to 2018, the tumor and paracancerous tissues from 66 patients with CRC who had been surgically resected were randomly collected from the Xinyu People’s Hospital. After being verified by two pathologists, all samples were frozen in liquid nitrogen and then stored at −196°C.
Cell Culture
The American Type Culture Collection (ATCC, Rockville, MD, USA) was the provider of human CRC cancer cell lines, including DLD-1, HCT116, SW480, HT-29 cells, and the normal human intestinal epithelial NCM460 cells. The above cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, HyClone, South Logan, UT, USA) with 10% fetal bovine serum (FBS, Gibco, Rockville, MD, USA), 100 U/mL penicillin, 100 μg/mL streptomycin (Hyclone, Logan, UT, USA) in 5% CO2 at 37°C.
Cell Transfection
The small interfering RNA (siRNA) against PITPNA-AS1 (si-PITPNA-AS1) and the negative control (si-NC) were available from GeneChem (Shanghai, China). MiR-129-5p mimic (miR-129-5p), miR-129-5p inhibitor (miR-129-5p in) and corresponding negative control oligonucleotides (miR-NC) were synthesized by GenePharma (Shanghai, China). They were transfected into the cells with LipofectamineTM2000 (Invitrogen, Carlsbad, CA, USA). Cells were harvested for subsequent experiments 48 h after transfection.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The TRIzol reagent (Invitrogen, Carlsbad, CA) was used to extract RNA from tissues and cells. AMV reverse transcriptase (Promega, Madison, WI, USA) was adopted to reversely transcribe 2 μg total RNA into cDNA, and then qRT-PCR was performed with SYBR Green RealTime PCR Master Mix kit (Roche, Basel, Switzerland) in ABI 7900 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA), and finally, relative RNA expression was estimated by the 2–ΔΔCt method. The primer sequences are listed in Table 1.
 |
Table 1 The Primers Used in This Study |
Cell Counting Kit-8 (CCK-8) Assay
Cell counting kit-8 (Dojindo, Kumamoto, Japan) was applied to evaluate CRC cell proliferation. Transfected cells were inoculated into 96-well plates (2 × 103/well). Next, 10 μL CCK-8 solution (Dojindo Molecular Technologies, Japan) was dripped into each well at 0 h, 24 h, 48 h, 72 h, respectively, with which the cells were incubated at 37°C for 2 h. Ultimately, a microplate reader (BioRad, Hercules, CA, USA) was used to record the absorbance of each well at 450 nm.
5-Ethynyl-2-Deoxyuridine (EdU) Incorporation Assay
Cell proliferation was examined by EdU assay kit (RiboBio, Guangzhou, China). To be specific, transfected cells were inoculated into 96-well plates. Then, 50 µM EdU labeling medium was loaded into the wells and incubated for 2 h. Subsequently, 4% paraformaldehyde was dripped into each well for 30 min to fix the cells, followed by the addition of 1 × Apollo® reaction cocktail (100 µL). Thirty-minute later, 100 µL of Hoechst 33,342 staining solution was added to stain the nuclei, and then a fluorescent microscopy (Nikon, Japan) was adopted to observe the cells in 5 random wells.
Wound Healing Assay
Transfected cells were inoculated into 6-well plates. After the wells were fully covered by the cells, a 200 μL pipette tip was employed to draw a straight line on the surface of the monolayer cells to form a clear cell-free zone, and then the cells were cultured in serum-free medium for 24 h. Wound closure was observed at 0 h and 24 h, respectively, under the microscope.
Flow Cytometry
Annexin V-FITC/PI Apoptosis Detection Kit (KeyGEN BioTECH, Jiangsu, China) was adopted to measure the apoptosis rate. Transfected cells were stained with Annexin V-FITC staining solution and propidium iodide (PI) solution to detect the early and late apoptotic cells, thus calculating the total apoptotic rate.
Fluorescent Staining of Nuclei
Transfected cells were cultured in 6-well plates. Then, the cell nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI, Sigma-Aldrich, St. Louis, MO) and visualized using a fluorescence microscope (Nikon, Japan).
Luciferase Reporter Assay
The sequences of PITPNA-AS1 and 3ʹUTR of HMGB1 were inserted into the pmirGLO luciferase vector (Promega, Madison, WI, USA), namely, PITPNA-AS1-wild type (WT) or HMGB1-WT reporter vector, and then the PITPNA-AS1-WT or HMGB1-WT binding site was mutated to produce PITPNA-AS1-mutant type (MUT) or HMGB1-MUT reporter vector. Subsequently, the reporter vectors and miR-129-5p/miR-NC were co-transfected into HKE-293T cells employing LipofectamineTM 2000 (Invitrogen, Carlsbad, CA, USA). Forty-eight-hour later, the activity of luciferase was measured by Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
RNA Immunoprecipitation (RIP) Assay
EZ-Magna RIP Kit (Millipore, Bedford, MA, USA) was utilized for RIP experiments. In brief, HEK-293T cells were harvested and then lysed in RIP lysis buffer. The cell extracts were co-immunoprecipitated with RIP buffer containing anti-Ago2 antibody conjugated with magnetic beads or normal mouse IgG. Subsequently, samples were incubated with Proteinase K. Eventually, qRT-PCR was adopted to measure the expressions of PITPNA-AS1 and miR-129-5p, and HMGB1 mRNA and miR-129-5p in immunoprecipitated RNA.
Western Blot Assay
RIPA buffer (Sigma-Aldrich, St. Louis, MO) was adopted to prepare protein samples. After 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to polyvinylidene fluoride membranes (PVDF, Millipore, Billerica, MA, USA) and then blocked by 5% skim milk. Next, the membranes were incubated with primary antibodies at 4°C overnight, and then with horseradish peroxidase-conjugated secondary antibodies at 37°C for 2 h. Ultimately, enhanced chemiluminescence reagents (ECL-Plus; Thermo Fisher Scientific, Waltham, MA) were used to detect protein expressions.
Statistical Analysis
Statistical analysis was completed employing GraphPad Prism 6.0 (GraphPad Software, Inc, CA, USA). Data were expressed as means ± standard deviation, and Student’s t-test or one-way ANOVA was applied for analyzing differences. Chi-square test was performed to evaluate the relationship between PITPNA-AS1 expression and clinical characteristics. Pearson’s correlation coefficient was calculated to analyze the correlations. P < 0.05 indicated statistical significance.
Results
PITPNA-AS1 Expression Was Increased in CRC Tissues and Cell Lines, and PITPNA-AS1 Was Negatively Interrelated with miR-129-5p Expression and Positively Associated with HMGB1 Expression
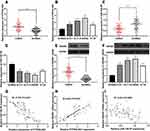
We first detected PITPNA-AS1, miR-129-5p and HMGB1 expressions in CRC tissues and cell lines, and qRT-PCR and Western blot assays denoted that compared with that in paracancerous tissues or NCM460 cell lines, PITPNA-AS1 expression in CRC tissues and cell lines was observably higher (Figure 1A and B), while miR-129-5p expression in CRC tissues and cell lines was lower (Figure 1C and D); HMGB1 expression was remarkably higher in CRC tissues and cell lines (Figure 1E–F). In addition, in CRC tissue, PITPNA-AS1 expression was negatively interrelated with miR-129-5p expression (Figure 1G) and positively associated with HMGB1 mRNA (Figure 1H), and miR-129-5p expression was negatively associated with HMGB1 mRNA expression (Figure 1I). In order to evaluate the clinical significance of PITPNA-AS1 expression in CRC, we divided the CRC patients into a high PITPNA-AS1 expression group and a low PITPNA-AS1 expression group based on the median, and analyzed the relationship between PITPNA-AS1 expression and clinical characteristics of CRC patients, the findings of which depicted that PITPNA-AS1 expression was related to clinical stage and lymph node metastasis (Table 2). These results suggested that the overexpression of PITPNA-AS1 might be associated with CRC progression.
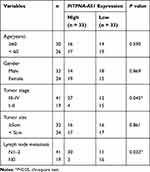 |
Table 2 Correlation Between Clinicopathological Features and PITPNA-AS1 Expression in CRC (n = 66) |
PITPNA-AS1 Facilitated the Proliferation and Metastasis of CRC Cells, and Impeded Apoptosis
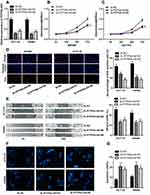
To expound on the function of PITPNA-AS1 in CRC, we used PITPNA-AS1 siRNA#1 and PITPNA-AS1 siRNA#2 to silence PITPNA-AS1 expression in HCT116 and SW480 cell lines (Figure 2A). The cell proliferation changes in CRC cells were analyzed using CCK-8 and EdU experiments. It was revealed that the proliferation of HCT116 and SW480 cells in si-PITPNA-AS1#1 group and si-PITPNA-AS1#2 group was markedly lower than in si-NC group (Figure 2B–D). The migratory ability of CRC cells was determined using wound-healing assay. In comparison to the control group, inhibited cell migration was observed in HCT116 and SW480 with PITPNA-AS1 knockdown (Figure 2E). Additionally, as shown, after the nuclei of CRC cells were stained with DAPI, in PITPNA-AS1 knockdown groups, apoptotic morphological characteristics obviously appeared, such as pyknosis and karyorrhexis (Figure 2F). Consistently, flow cytometry analysis uncovered that the apoptosis rate was dramatically higher than that in PITPNA-AS1 knockdown group (Figure 2G). All findings showed that PITPNA-AS1 promoted cell proliferation and migration, and inhibited cell apoptosis in CRC.
PITPNA-AS1 Could Sponge miR-129-5p and Negatively Regulate Its Expression
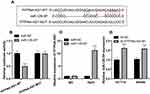
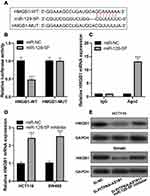
To pinpoint the molecular mechanism by which PITPNA-AS1 promotes CRC progression, we obtained a binding site in the sequence of PITPNA-AS1 for miR-129-5p from LncBase Predicted v.2 database (Figure 3A). Subsequently, dual-luciferase report assay was performed, the findings of which depicted that the luciferase activity in the co-transfection group of PITPNA-AS1-WT and miR-129-5p was markedly lower than that of HEK-293T cells co-transfected with PITPNA-AS1-MUT and miR-129-5p (Figure 3B). RIP experiment found that PITPNA-AS1 and miR-129-5p were rich in Ago2-containing immunoprecipitate, suggesting that PITPNA-AS1 was directly interacted with miR-129-5p (Figure 3C). The dual-luciferase report assay and RIP experiment demonstrated that PITPNA-AS1 could directly bind to miR-129-5p. Moreover, we further confirmed the regulatory relationship between PITPNA-AS1 and miR-129-5p, and miR-129-5p expression in CRC cells with PITPNA-AS1 knockdown was remarkably increased (Figure 3D). Hence, it was verified that PITPNA-AS1 could directly regulate miR-129-5p in CRC cell lines. The results indicated that PITPNA-AS1 could sponge miR-129-5p in CRC.
MiR-129-5p Inhibition Could Reverse the Effect of PITPNA-AS1 Knockdown on CRC Cells
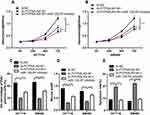
To further elaborate on that PITPNA-AS1 takes part in CRC cells through miR-129-5p, we co-transfected PITPNA-AS1 siRNA#1 and miR-129-5p inhibitors into HCT116 and SW480 cell lines, and CCK-8 and EdU experiments unmasked that miR-129-5p inhibitors could partially counteract the inhibitory effect of PITPNA-AS1 knockdown on CRC cell proliferation (Figure 4A–C). Wound healing assay manifested that miR-129-5p inhibitors could partially reverse the inhibitory effect of PITPNA-AS1 on the migration of CRC cells (Figure 4D). Flow cytometry depicted that miR-129-5p inhibitors could offset the promotion of PITPNA-AS1 knockdown on the apoptosis of CRC cells (Figure 4E). Therefore, we confirmed that PITPNA-AS1 promoted the progression of CRC through regulating miR-129-5p.
PITPNA-AS1 Could Promote HMGB1 Expressions via Repressing miR-129-5p Expression
To delve into the ceRNA network among PITPNA-AS1, miR-129-5p, and their target gene in CRC, we searched the TargetScan database and found that HMGB1 was a downstream target gene of miR-129-5p (Figure 5A). Dual-luciferase reporter experiments confirmed that miR-129-5p mimics could reduce the luciferin activity of pmirGLO-HMGB1-WT. However, it had no influence on the luciferase activity of pmirGLO-HMGB1-MUT (Figure 5B). The RIP experiment demonstrated that HMGB1 mRNA and miR-129-5p were rich in Ago2-containing immunoprecipitate, which suggested that HMGB1 mRNA and miR-129-5p interacted directly in CRC cells (Figure 5C). In addition, it was found that miR-129-5p inhibitors could significantly improve HMGB1 mRNA expression (Figure 5D). Next, we co-transfected PITPNA-AS1 siRNA#1 and miR-129-5p inhibitors into HCT116 and SW480 cell lines, and observed that as against si-NC group, HMGB1 expression in PITPNA-AS1 siRNA# 1-transfected cells decreased markedly; the expression of HMGB1 in miR-129-5p inhibitors-co-transfected cells was higher than that in si-PITPNA-AS1#1 group (Figure 5E). These data suggested that PITPNA-AS1/miR-129-5p axis modulated the expression of HMGB1 to participate in the progression of CRC.
Discussion
CRC is a common gastrointestinal cancer. Although the current treatments have been greatly enhanced, the 5-year survival rate of patients is still not high.15 Hence, it is fundamental to deeply probe the pathological mechanism of CRC and to find new therapeutic targets. Mounting research shows that lncRNAs are crucial regulators in human cancers, including CRC. For example, ZEB1-AS1 expression is up-regulated in CRC tissues and cell lines, and ZEB1-AS1 can promote cell proliferation and inhibit apoptosis through the miR-205/YAP1 axis.16 SNHG3 facilitates the proliferation, migration and invasion of CRC cells by modulating the miR-539/RUNX2 axis.5 LINC00520 is highly expressed in CRC tissues and cell lines, and can function as a ceRNA for miR-577, thus increasing HSP27 expression and promoting cell proliferation, migration and invasion.17 A recent study implies that PITPNA-AS1 expression is markedly elevated in hepatocellular carcinoma, and PITPNA-AS1 can increase the expression of WNT5A by inhibiting miR-876-5p expression to promote cell proliferation and migration.6 However, its role in CRC is still unknown. Our study found for the first time that PITPNA-AS1 was markedly overexpressed in CRC tissues and cell lines, and that high expression of PITPNA-AS1, interrelated with higher clinical stage of patients and positive lymph node metastasis, could promote cell proliferation and migration, and suppress apoptosis. Therefore, we confirmed that, in CRC PITPNA-AS1 could function as a tumor promoter.
We also explored the underlying mechanism of PITPNA-AS1 in CRC, and luciferase report assay and RIP experiment confirmed that miR-129-5p was one of the targets of PITPNA-AS1. Accumulating studies manifest that miR-129-5p has an anti-tumor effect, and for example, miR-129-5p expression is significantly declined in gastric cancer cell lines and miR-129-5p can suppress the proliferation and invasion of gastric cancer cells through targeting COL1A1.10 MiR-129-5p expression is significantly decreased in rectal adenocarcinoma cell lines and miR-129-5p can impede cell proliferation, migration and invasion via targeting E2F7.9 Another study manifests that miR-129-5p expression is remarkably declined in CRC cell line, and miR-129-5p can impede cell proliferation and promote apoptosis via targeting HMGB1.4 In this study, we unearthed that miR-129-5p expression was dramatically decreased in CRC tissues and cell lines, and mechanistically, it was confirmed that PITPNA-AS1 could adsorb miR-129-5p, which might partly explain the mechanism of miR-129-5p dysregulation in CRC.
Next, we searched for downstream genes of miR-129-5p. Bioinformatics analysis, luciferase reporter assay and RIP experiment demonstrated that HMGB1 was a target gene of miR-129-5p. HMGB1 is a versatile regulator that is originally thought to be a nuclear protein involved in the regulation of gene transcription, by which it can regulate inflammation and infection responses.18 In recent years, accumulating studies confirm that HMGB1 features prominently in the tumorigenesis of multiple cancers including liver cancer, lung cancer, breast cancer, prostate cancer, cervical cancer and CRC.19 In CRC, it is demonstrated that HMGB1 overexpression can promote cancer cell proliferation and metastasis, and HMGB1 can also serve as a predictor for the poor prognosis of the patients.20 Another study reports that the HMGB1 expression is associated with lymph node metastasis and distant metastasis of CRC patients, and HMGB1 inhibits the apoptosis of cancer cells by inducing the activation of ERK signaling and c-inhibitor of apoptosis protein 2.12 Besides, HMGB1 promotes CRC cell growth, migration, invasion via JAK/STAT3 pathway.21 Moreover, HMGB1 mediates the proliferation and migration of CRC by increasing the phosphorylation of ERK1/2, ras-related C3 botulinum toxin substrate 1 and Akt, and promoting the expressions of inducible nitric oxide synthase and matrix metalloproteinase 9.22 In this work, we confirmed that HMGB1 expression was markedly raised in CRC tissues and cell lines, and PITPNA-AS1 could modulate its expression via targeting miR-129-5p.
In conclusion, in the present work, we verify the function and downstream mechanism of PITPNA-AS1 in CRC. Specifically, PITPNA-AS1 can adsorb miR-129-5p to modulate HMGB1 expression, and thus exert its indirect cancer-promoting role in CRC. Nonetheless, this work has certain limitations. First of all, these data are only obtained from in vitro experiments and the conclusion needs to be confirmed with animal experiments in the following work. Secondly, whether PITPNA-AS1 can promote other malignant biological behaviors of CRC cells, such as chemoresistance, requires further investigation. Last but not the least, there are other candidate downstream miRNAs of PITPNA-AS1, which remain be screened and verified.
Disclosure
The authors declare no conflicts of interest.
References
1. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):
2. Brenner H, Chen C. The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br J Cancer. 2018;119(7):
3. Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:
4. Wu Q, Meng W-Y, Jie Y, et al. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233(9):
5. Dacheng W, Songhe L, Weidong J, et al. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother. 2020;125:
6. Sun J, Zhang Y, Li B, et al. PITPNA-AS1 abrogates the inhibition of miR-876-5p on WNT5A to facilitate hepatocellular carcinoma progression. Cell Death Dis. 2019;10(11):
7. Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):
8. Balacescu O, Sur D, Cainap C, et al. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int J Mol Sci. 2018;19(12):12. doi:10.3390/ijms19123711
9. Wan P, Bai X, Yang C, et al. miR-129-5p inhibits proliferation, migration, and invasion in rectal adenocarcinoma cells through targeting E2F7. J Cell Physiol. 2020;235(7–8):
10. Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2018;96(1):
11. Hirata Y, Kurobe H, Higashida M, et al. HMGB1 plays a critical role in vascular inflammation and lesion formation via toll-like receptor 9. Atherosclerosis. 2013;231(2):
12. Zhang W, An F, Xia M, et al. Increased HMGB1 expression correlates with higher expression of c-IAP2 and pERK in colorectal cancer. Medicine. 2019;98(3):
13. Tang D, Loze MT, Zeh, III HJ, et al. The redox protein HMGB1 regulates cell death and survival in cancer treatment. Autophagy. 2010;6(8):
14. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):
15. van der Valk M, Hilling DE, Bastiaannet E, et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): an international multicentre registry study. Lancet. 2018;391(10139):
16. Jin Z, Chen B. LncRNA ZEB1-AS1 Regulates Colorectal Cancer Cells by MiR-205/YAP1 Axis. Open Med. 2020;15:
17. Jin XH, Hong Y-G, Li P, et al. Long noncoding RNA LINC00520 accelerates the progression of colorectal cancer by serving as a competing endogenous RNA of microRNA-577 to increase HSP27 expression. Hum Cell. 2020;33(3):
18. Hreggvidsdottir HS, Lundberg AM, Aveberger A-C, et al. High mobility group box protein 1 (HMGB1)-partner molecule complexes enhance cytokine production by signaling through the partner molecule receptor. Mol Med. 2012;18(2):
19. Tripathi A, Shrinet K, Kumar A. HMGB1 protein as a novel target for cancer. Toxicol Rep. 2019;6:
20. He S, Cheng J, Sun L, et al. HMGB1 released by irradiated tumor cells promotes living tumor cell proliferation via paracrine effect. Cell Death Dis. 2018;9(6):
21. Chen X, et al. MiR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. Am J Cancer Res. 2017;7(10):
22. Kuniyasu H, Chihara Y, Kondo H. Differential effects between amphoterin and advanced glycation end products on colon cancer cells. Int J Cancer. 2003;104(6):
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.