Back to Journals » OncoTargets and Therapy » Volume 13
lncRNA CRNDE is Upregulated in Glioblastoma Multiforme and Facilitates Cancer Progression Through Targeting miR-337-3p and ELMOD2 Axis
Authors Gao J, Chen Q, Zhao Y, Hou R
Received 15 February 2020
Accepted for publication 15 July 2020
Published 16 September 2020 Volume 2020:13 Pages 9225—9234
DOI https://doi.org/10.2147/OTT.S249887
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
This paper has been retracted.
Jian Gao, 1,* Qunbang Chen, 1,* Yingjia Zhao, 2 Ruizhe Hou 1
1Department of Neurosurgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin 130033, People’s Republic of China; 2Department of Gynecology and Obstetrics, The Second Hospital of Jilin University, Changchun, Jilin 130000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ruizhe Hou Department of Neurosurgery
China-Japan Union Hospital of Jilin University, No. 126 Xiantai Street, Changchun, Jilin 130033, People’s Republic of China
Email [email protected]
Introduction: Colorectal neoplasia differentially expressed (CRNDE) was reported to promote carcinogenesis in several cancers. However, the role of CRNDE in glioblastoma multiforme (GBM) needs to be further explored.
Methods: CRNDE expression levels in GBM tissues and cells were explored using real-time quantitative PCR at first. Effects of CRNDE on GBM cell behaviors were detected by conducting in vitro experiments. Interactions of CRNDE, microRNA-337-3p (miR-337-3p), and ELMO domain containing 2 (ELMOD2) were verified by bioinformatics analysis tools and dual-luciferase reporter assay. Expression correlations of CRNDE and ELMOD2 in GBM tissues were analyzed at GEPIA website.
Results: CRNDE expression was upregulated in GBM tissues and cells compared with normal counterparts. CRDNE knockdown inhibits proliferation and migration, but promotes apoptosis in GBM cell, while CRNDE overexpression caused opposite effects. Mechanisms exploration indicated CRNDE serves as sponge of miR-337-3p to upregulate ELMOD2 expression. Furthermore, we showed CRNDE and ELMOD2 were positively correlated in GBM tissues.
Discussion: In conclusion, our study highlighted the importance of CRNDE/miR-337-3p/ELMOD2 axis in GBM progression and offered novel strategies for GBM treatment.
Keywords: CRNDE, miR-337-3p, ELMOD2, glioblastoma multiforme
Introduction
Although the newly occurred glioblastoma multiforme (GBM) cases each year is not large, its overall survival is only around 15 months.1,2 Hence, comprehensive understanding genetic basis behind GBM progression is necessary with the aim to seek novel treatment targets.
Recent studies indicated most of genome transcripts were belonged to non-coding RNAs (ncRNAs). ncRNAs can be classified into two major types based on their length: long ncRNAs (lncRNAs) and short ncRNAs.3 Increasing evidence indicated lncRNAs play crucial roles in affecting cancer progression through mediating messenger RNA translation, chromatin modification, or microRNA mediated gene expression regulation.4,5 In GBM, there are many lncRNAs been identified closely involved in cancer development, and thus could serve as prognostic prediction markers.3
Colorectal neoplasia differentially expressed (CRNDE) is a lncRNA located at chromosome 16 and elevated expression in a bunch of human cancers including non-small cell lung cancer, cervical cancer, melanoma, hepatocellular carcinoma, tongue squamous cell carcinoma, and so on.6–10 For instance, CRNDE was found increased expression in non-small cell lung cancer and correlated with the advanced tumor stages and poorer overall survival.6 Also, CRNDE knockdown could suppress non-small cell lung cancer cell growth and metastasis.6 Another work indicated CRNDE was increased expression in cervical cancer and its knockdown could inhibit tumor growth via p53 upregulated modulator of apoptosis.7 There is a study showed CRNDE stimulates glioma growth and metastasis through serving as competing endogenous RNA for miR-136-5p.11 However, the functions and mechanisms of CRNDE in regulating GBM initiation and progression remain to be further explored.
Here, we aimed to further validate the roles of CRNDE in GBM and to identify the miRNA and mRNA involved in the roles of CRNDE, which will advance our understanding of mechanisms behind GBM and even provide novel targets for the therapy of GBM.
Materials and Methods
Cell Culture
GBM cells (U87, U251 and LN229) and NHA (normal human astrocyte) cell were bought from Cell Culture Center in Shanghai. Cell culture atmosphere was used as previously report.12
Detection the Prognostic Value of CRNDE in GBM
Effects of CRNDE expression on the overall survival of GBM patients (Tumor number = 163, Normal number = 207) were analyzed at GEPIA website (http://gepia2.cancer-pku.cn/#index).
siRNA, miRNA, and Plasmid Transfection
Full length of CRNDE was inserted into pcDNA3.1 to obtain pCRNDE. miR-337-3p mimic and its scramble (miR-scramble) were synthesized by GeneChem (Shanghai, China). siRNAs targeting CRNDE or ELMO domain containing 2 (ELMOD2) named as si-CRNDE or si-ELMOD2 and scrambles were bought from GeneChem. For transfection, 1 × 107 cells were seeded in 6-well plate and cultured to about 80% confluence. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for oligonucleotides and plasmids transfection for 48 h.
Real-Time Quantitative PCR (RT-qPCR)
RNA samples of cultured cells were extracted with Trizol based on recommended protocols. After concentration quantification, equal amounts of samples were transcribed into cDNA using PrimerScript kit (Qiagen, Hilden, Germany). RT-qPCR was performed using SYBR Green (Takara, Otsu, Japan) at ABI 7900 (Applied Biosystems, Foster City, CA, USA). Relative gene expression level was analyzed with 2−ΔΔCt method.13 Primer sequences are listed in Table 1.
 |
Table 1 Primer Sequences Used in This Work |
Western Blot
Cells were washed with PBS and lysed in RIPA buffer containing protease inhibitors (Beyotime, Haimen, Jiangsu, China). Supernatant was collected after centrifugation, and the measure protein concentration with BCA kit (Beyotime). Equal amounts of protein were separated at 10% SDS-polyacrylamide gel, transferred to PVDF membrane and blocked by fat-free milk. Primary antibodies against GAPDH (ab181602), ELMOD2 (ab224732, Abcam, Cambridge, MA, USA), and secondary antibody (ab6721, Abcam) were used. Blots were visualized by BeyoECLPlus (Beyotime) and analyzed with Image Pro Plus 6.0 software.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 assay was performed according to the instruction provided by the manufacturer. In brief, cells after transfection were seeded into plates and incubated for indicated times. CCK-8 solution was added and incubated with these cells for 4 h. Optical density of each well at 450 nm was measured using a plate reader.
Wound-Healing Assay
Cells were grown to 100% confluence in serum-free medium. Wound at cell surface was created using a pipette tip and then washed with PBS. Images were observed under a microscope at 0 and 48 h and analyzed with Image Pro Plus 6.0 software to measure cell migration percentage.
Transwell Invasion Assay
Cell invasion ability was measured using the Matrigel-coated transwell chamber (Corning, NY, USA). 1 × 104 cells resuspended in serum-free medium were seeded into the upper chamber, while FBS contains medium was filled into the lower chamber. After 24 h of incubation, inserts were removed and non-invaded cells were wiped with a cotton swab. Afterwards, invaded cells were fixed with 4% paraformaldehyde and dyed with crystal violet. Invasive cell numbers were counted under a microscope from five independent fields.
Cell Apoptosis
Annexin V-FITC/propidium iodide (PI) kit (BD Biosciences, Franklin Lakes, NJ, USA) was used to detect cell apoptosis percentage. Cells suspended in binding buffer were incubated with Annexin V-FITC and PI reagent in dark for 15 min. FACS flow cytometry (BD Biosciences) was used to detect cell apoptosis percentage.
In vivo Tumor Growth
Animal study protocol was approved by the ethics committee of China–Japan Union Hospital of Jilin University. Animal experiments were performed in accordance with the Guidelines for Care and Use of Laboratory Animal. 1 × 107 U87 cells with sh-CRNDE (5ʹ- GUGCUCGAGUGGUUUAAAUTT-3ʹ) or shR-scramble (5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ, both obtained from GeneChem) transfection were injected into nude mice purchased from National Laboratory Animal Center (Beijing, China). Tumor volume was measured every 7 days until mice scarified with the formula V = 0.5 × length × width2. Tissues were weighed at the end of 4th week.
Dual-Luciferase Reporter Assay
miRNA target of CRNDE was analyzed at ENCORI, while mRNA target of miRNA was also predicted at ENCORI (http://starbase.sysu.edu.cn/index.php). Wild-type and mutant sequences of CRNDE and ELMOD2 were cloned into psiCHECK and named as WT/MT-CRNDE/ELMOD2. Cells were transfected with luciferase constructs and miRNAs to detect relative luciferase activity using a dual-luciferase assay system (Promega, Madison, WI, USA).
Detection of CRNDE and ELMOD2 Expression in GBM
Expression levels of CRNDE and ELMOD2 in GBM and normal tissues were analyzed at GEPIA. Moreover, the expression correlation between CRNDE and ELMOD2 was also analyzed at GEPIA.
Clinical Sample Information from Oncomine
Expression levels of CRNDE and ELMOD2 in GBM tissues and normal tissues were analyzed using the clinical information obtained from Oncomine (https://www.oncomine.org/resource/login.html). In addition, ELMOD2 expression in different grades of GBM was also explored using the clinical data from Oncomine.
Statistical Analysis
Data were presented in the manner of mean ± SD. Statistical analysis was performed at SPSS (Chicago, IL, USA) using Student’s t-test and one-way ANOVA methods. P < 0.05 was regarded as statistically significant.
Results
Increased CRNDE Expression in GBM
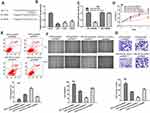
CRNDE level in GBM tissues was firstly analyzed at GEPIA and the results indicated CRNDE was upregulated in GBM tissues compared with normal tissues (Figure 1A). Data from GBM samples from Oncomine indicated that CRNDE was also elevated expression in GBM tissues compared with normal tissues (Figure 1B). High CRNDE level was also found in GBM cells compared with NHA cell (Figure 1C). U87 and U251 cells with first and second highest CRNDE level in the GBM cells investigated were selected for following analyses. In addition, high CRNDE level was found associated with worse overall survival of GBM patients (Figure 1D).
CRNDE Regulates GBM Cell Proliferation and Migration
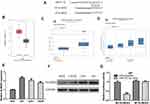
To explore the roles of CRNDE, we knockdown and overexpress CRNDE in U87 and U251 cells. The results showed si-CRNDE decreased, while pCRNDE increased CRNDE levels in U87 and U251 cells (Figure 2A). CCK-8 results showed proliferative ability was increased by pCRNDE and decreased by si-CRNDE in U87 and U251 cells (Figure 2B). Flow cytometry assay showed CRNDE overexpression inhibits cell apoptosis, while CRNDE knockdown caused opposite effects in U87 and U251 cells (Figure 2C). Besides, wound-healing assay and transwell invasion assay were performed and the results showed CRNDE knockdown inhibited cell migration and invasion, while CRNDE overexpression accelerated cell migration and invasion in U87 and U251 cells (Figure 2D and E).
CRNDE Served as miR-337-3p Sponge
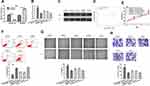
To explore the potential mechanism of CRNDE, we explored the possible target of CRNDE and found miR-337-3p was a potential target for CRNDE (Figure 3A). RT-qPCR showed miR-337-3p level was decreased in GBM cells compared with NHA cell (Figure 3B). Luciferase activity assay indicated luciferase activity of WT-CRNDE was decreased by miR-337-3p mimic, while it was not significantly changed in MT-CRNDE group, which validated the interaction between CRNDE and miR-337-3p (Figure 3C). As shown in Figure 3D and E, miR-337-3p overexpression suppresses cell growth, and reversed the pCRNDE-stimulated cell growth effects. In addition, we showed miR-337-3p mimic suppressed cell migration and invasion abilities, and partially reversed the roles of pCRNDE (Figure 3F and G).
ELMOD2 Serves as Target of miR-337-3p
Then, ENCORI was used to analyze the target for miR-337-3p and the results showed ELMOD2 was a possible target (Figure 4A). We then showed ELMOD2 was increased in GBM tissues and cells (Figure 4B and C). Furthermore, we showed the patients at high tumor grade tend to have higher ELMOD2 expression compared to those at low grade (Figure 4D). RT-qPCR showed ELMOD2 was increased expression in GBM cells in comparison with the normal cell (Figure 4E). Western blot confirmed the results that ELMOD2 was an enhanced expression in GBM cells compared with normal cell (Figure 4F). Luciferase activity reporter assay indicated that WT-ELMOD2 was reduced by miR-337-3p mimic (Figure 4G).
CRNDE Modulates GBM Progression by miR-337-3p/ELMOD2
To verify CRNDE regulates GBM via the miR-337-3p/ELMOD2 axis, the following experiments were conducted. Since our previous results indicated that CRNDE and ELMOD2 could interact with miR-337-3p, we introduced pCRNDE into GBM cell to investigate the expression level of miR-337-3p and ELMOD2. RT-qPCR showed miR-337-3p was decreased and ELMOD2 was increased with CRNDE overexpression as the miR-337-3p molecular was mainly bound by CRNDE (Figure 5A). As shown in Figure 5B, miR-337-3p overexpression partially reversed the stimulation effect of CRNDE overexpression on ELMOD2 expression (Figure 5B). Western blot confirmed the results of RT-qPCR (Figure 5C). Moreover, it was found CRNDE and ELMOD2 expression was positively correlated in GBM tissues (Figure 5D). Additionally, the overexpression of miR-337-3p or knockdown of ELMOD2 partially countervailed the functions of CRNDE overexpression on GBM cell growth, migration, and invasion (Figure 5E–H).
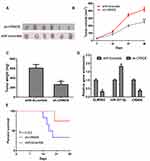
CRNDE Stimulates GBM Progression in vivo
To further understand the oncogenic roles of CRNDE, in vivo experiments on nude mice were conducted. As shown in Figure 6A–C, tumor volume and weight were reduced by sh-CRNDE compared with scramble control. RT-qPCR showed sh-CRNDE significantly decreased CRNDE and ELMOD2 expression, while increased miR-337-3p expression (Figure 6D). Survival analysis showed nude mice with sh-CRNDE tend to have a longer survival rate compared to those without (Figure 6E). These results indicated CRNDE knockdown suppressed GBM carcinogenesis.
Discussion
Numerous lncRNAs have been reported to regulate GBM progression. For example, lncRNA small nucleolar RNA host gene 5 could stimulate GBM cell proliferation via regulating miR-205-5p and zinc finger E-box binding homeobox 2 axis.14 High lncRNA LINC00473 was identified as a predictor for worse survival of glioma patients.15 They also found LINC00473 regulates glioma progression by inhibiting miR-195-5p and activate Yes‑associated protein 1-EA domain family member 1-Hippo signaling pathway.15 In this study, CRNDE was found highly expressed in GBM tissues and cells. Besides, high CRNDE was significantly correlated with poorer overall survival of GBM patients. Through gain and loss-of experiments, we found CRNDE could promote GBM cell growth, migration, and invasion in vitro. Importantly, we showed knockdown of CRNDE hinders GBM tumorigenesis in vivo. These results suggested an oncogenic role of CRNDE in GBM.
Studies have indicated lncRNAs elicit their roles by interacting with miRNAs.8–11 For example, CRNDE was previously reported could interact with miR-205, miR-217, miR-384, and miR-136-5p in cancers.8–11 Here, we explored the potential miRNA target of CRNDE using ENCORI and we found miR-337-3p was a putative target. It could be concluded from previous work that miR-337-3p inhibits epithelial ovarian cancer through regulating the expression of PIK3CA and PIK3CB.16 We validated the miR-337-3p as a functional target of CRNDE using luciferase activity assay and rescue assays. In addition, forcing the expression suppresses GBM cell growth, migration, and invasion in vitro.
ceRNA theory implied mRNA expression could be regulated after the binding of lncRNA and miRNA. Here, we showed ELMOD2 was a possible target of miR-337-3p. ELMOD2 is an ARL2 GTPase-activating protein and promotes the fusion of mitochondrial.17 Also, ELMOD2 is found as a key regulator for the TLR3 pathway and hence to regulate idiopathic pulmonary fibrosis.18 Moreover, ELMOD2 knockdown could affect the adipocyte triglyceride lipase location to regulate cellular lipid metabolism.19 These results indicated that ELMOD2 plays a role in regulating the normal physiological process but its role in cancer is not reported. We showed ELMOD2 was highly expressed in GBM and positively correlated with the expression of CRNDE. A previous work to investigate the roles of CRNDE in glioma indicated that CRNDE serves as ceRNA for miR-136-5p to regulate Bcl-2 and Wnt2 expression and hence to affect glioma cell proliferation, migration, and invasion.11 In our work, we also provided evidence CRNDE affects GBM cell proliferation, migration, invasion, and apoptosis via the miR-337-3p/ELMOD2 axis. Compared with the previous work, we validated their experiment results but provided novel ceRNA triplets for CRNDE, which will help us to further understand the roles of CRNDE in cancers.
Conclusion
In summary, this work provided novel CRNDE/miR-337-3p/ELMOD2 triplets in regulating GBM progression, which may help to validate novel therapeutic targets for GBM.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi:10.1101/gad.187922.112
2. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-Oncol. 2015;17(Suppl 4):iv1–62. doi:10.1093/neuonc/nov189
3. Paulmurugan R, Malhotra M, Massoud TF. The protean world of non-coding RNAs in glioblastoma. J Mol Med (Berl). 2019;97(7):909–925. doi:10.1007/s00109-019-01798-6
4. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
6. Jing H, Xia H, Qian M, Lv X. Long noncoding RNA CRNDE promotes non-small cell lung cancer progression via sponging microRNA-338-3p. Biomed Pharmacother. 2019;110:825–833. doi:10.1016/j.biopha.2018.12.024
7. Zhang JJ, Fan LP. Long non-coding RNA CRNDE enhances cervical cancer progression by suppressing PUMA expression. Biomed Pharmacother. 2019;117:108726. doi:10.1016/j.biopha.2019.108726
8. Xu L, Zhang Y, Zhao Z, et al. The long non-coding RNA CRNDE competed endogenously with miR-205 to promote proliferation and metastasis of melanoma cells by targeting CCL18. Cell Cycle. 2018;17(18):2296–2308.
9. Wang H, Ke J, Guo Q, Barnabo Nampoukime KP, Yang P, Ma K. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK1 axis. J Cell Mol Med. 2018;22(12):5862–5876. doi:10.1111/jcmm.13856
10. Ren Y, He W, Chen W, et al. CRNDE promotes cell tongue squamous cell carcinoma cell growth and invasion through suppressing miR-384. J Cell Biochem. 2019;120(1):155–163. doi:10.1002/jcb.27206
11. Li DX, Fei XR, Dong YF, et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8(50):88163–88178. doi:10.18632/oncotarget.21513
12. Wu W, Hu Q, Nie E, et al. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci Rep. 2017;7:45029. doi:10.1038/srep45029
13. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‑time quantitative PCR and the 2(‑Δ ΔC(T)) method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
14. Meng X, Deng Y, Lv Z, et al. LncRNA SNHG5 promotes proliferation of glioma by regulating miR-205-5p/ZEB2 axis. Onco Targets Ther. 2019;12:11487–11496. doi:10.2147/OTT.S228439
15. Wang X, Li XD, Fu Z, Zhou Y, Huang X, Jiang X. Long non‑coding RNA LINC00473/miR‑195‑5p promotes glioma progression via YAP1‑TEAD1‑Hippo signaling. Int J Oncol. 2020;56(2):508–521. doi:10.3892/ijo.2019.4946
16. Zhang Z, Zhang L, Wang B, et al. MiR-337-3p suppresses proliferation of epithelial ovarian cancer by targeting PIK3CA and PIK3CB. Cancer Lett. 2020;469:54–67. doi:10.1016/j.canlet.2019.10.021
17. Schiavon CR, Turn RE, Newman LE, Kahn RA, Spang A. ELMOD2 regulates mitochondrial fusion in a mitofusin-dependent manner, downstream of ARL2. Mol Biol Cell. 2019;30(10):1198–1213. doi:10.1091/mbc.E18-12-0804
18. Pulkkinen V, Bruce S, Rintahaka J, et al. ELMOD2, a candidate gene for idiopathic pulmonary fibrosis, regulates antiviral responses. FASEB J. 2010;24(4):1167–1177. doi:10.1096/fj.09-138545
19. Suzuki M, Murakami T, Cheng J, Kano H, Fukata M, Fujimoto T. ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol Biol Cell. 2015;26(12):2333–2342. doi:10.1091/mbc.E14-11-1504
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.