Back to Archived Journals » Research and Reports in Biology » Volume 8
Intrinsically disordered proteins: structural and functional dynamics
Authors Wallin S
Received 5 July 2016
Accepted for publication 13 October 2016
Published 17 February 2017 Volume 2017:8 Pages 7—16
DOI https://doi.org/10.2147/RRB.S57282
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Zvi Kelman
Stefan Wallin
Department of Physics and Physical Oceanography, Memorial University of Newfoundland, St. John’s, NL, Canada
Abstract: The classical view holds that proteins fold into essentially unique three-dimensional structures before becoming biologically active. However, studies over the last several years have provided broad and convincing evidence that some proteins do not adopt a single structure and yet are fully functional. These intrinsically disordered proteins (IDPs) have been found to be highly prevalent in many genomes, including human, and play key roles in central cellular processes, such as regulation of transcription and translation, cell cycle, and cell signaling. Moreover, IDPs are overrepresented among proteins implicated in disease, including various cancers and neurodegenerative disorders. Intense efforts, by using both experimental and computational approaches, are consequently under way to uncover the molecular mechanisms that underpin the roles of IDPs in biology and disease. This review provides an introduction to the general biophysical properties of IDPs and discusses some of the recent emerging areas in IDP research, including the roles of IDPs in allosteric regulation, regulatory unfolding, and formation of intracellular membrane-less organelles. In addition, recent attempts at therapeutic targeting of IDPs by small molecules, noting in particular that IDPs represent a potentially important source of new drug targets in light of their central role in protein–protein interaction networks, are also reviewed.
Keywords: natively unfolded proteins, unstructured proteins, protein folding, protein–protein interaction, cell regulation, signaling, drug development, inhibitors
Introduction
Following synthesis on the ribosome, most proteins organize themselves into an essentially unique three-dimensional structure determined by their amino acid sequence. This folding process, whether it occurs spontaneously or with the aid of molecular chaperones, is typically required for biological activity.1 However, an increasing number of proteins investigated at the molecular level are found not to rely on any single structure for their function.2–4 Instead, for this class of so-called intrinsically disordered proteins (IDPs), the native state is best characterized as a dynamic ensemble of interconverting conformations.
The term “disordered” is, in this context, meant to indicate a lack of a specific stable three-dimensional structure. Several other terms, including “unstructured” and “unfolded,” have also been widely used to describe the same phenomenon, but the designation IDP for this class of protein is now the most commonly used term. The structural disorder in IDPs can be located to one or more separate regions along the chain, or it can span the entire length of the protein.
The notion of disorder in protein structures is in fact not new. For instance, it has been known for a long time that substantial structural heterogeneity can exist in peptide hormones,5 in “linkers” that connect domains in multidomain proteins,6 or in loops connecting secondary structure elements in otherwise ordered protein, which typically appear as gaps in the electron density maps from X-ray crystallography experiments.7
In the past decade, however, advances in techniques for biophysical characterization of proteins, notably nuclear magnetic resonance (NMR) spectroscopy methods8 and the development of bioinformatics tools for disorder prediction directly from the amino acid sequence,9 have led to two key insights with far-reaching implications. First, it has become clear that disorder in proteins is highly prevalent in many organisms, particularly complex organisms. While in bacterial genomes only ~4% of all proteins are predicted to contain disordered regions of at least 30 amino acids in length, the corresponding number for eukaryotes is around one-third,10 and for humans, it is around one half.11 Second, protein disorder has been found to play a crucial role in many central cellular processes. For example, IDPs have been shown to function as signaling hubs in protein–protein interaction networks,12,13 make up components of the nuclear pore,14 transport lipids and cholesterol in blood plasma,15 create membrane-less organelles in cytosol and nucleus for posttranscriptional modification of RNA,16,17 and control the progression of the cell cycle.18
The prevalence of disorder varies substantially across different functional classes of proteins. For example, proteins with catalytic cellular functions are predicted to contain significantly less disorder than proteins that perform regulatory functions.11,12 This skewed distribution is a reflection of the important role played by disorder in specific functional molecular mechanisms. It is also in line with the traditional view of enzymes, as molecules relying on a rigid structural framework (with important but minor dynamical behavior) to correctly position key residues in the catalytic site. However, it should be pointed out that structural disorder in enzymes does occur19 and has been linked to a wider substrate specificity.20
Because of their widespread role in regulation, IDPs are, somewhat counterintuitively, often involved in protein binding and molecular recognition. In order to achieve recognition of a specific partner molecule, IDPs typically undergo a disorder–order transition upon contacting a target. This coupled folding–binding process lies at the center of many functional advantages provided by IDPs over ordered proteins. One such advantage is that IDPs can bind several structurally diverse molecules by adopting different conformations depending on the target.21 For example, the intrinsically disordered GTPase binding domain of the Wiskott–Aldrich syndrome protein (WASP) can bind its own VCA domain, leading to autoinhibition, and bind to the GTPase Cdc42 in a different structure, leading to the activation of WASP and the stimulation of actin polymerization.22
Another example is the intrinsically disordered C terminal region of the tumor suppressor p53, which is capable of binding at least four different ordered proteins, Sirtuin, Cyclin A2, the transcriptional coactivator CBP (CREB binding protein), and S100B, assuming four different structures in the process.23
In this review article, some of the molecular mechanisms underlying the functional abilities of IDPs are highlighted and discussed. The field of IDP research has grown rapidly over the last years and is now rather wide. There are several excellent reviews that discuss the broad roles of IDPs in biology (eg, Dunker et al,2 Wright and Dyson,3 and Habchi et al4). The present review focuses on four emerging areas where IDPs have recently been found to play an important role: allostery, regulated unfolding, formation of membrane-less organelles, and fuzzy complexes. In addition, the potential of IDPs and their interactions as therapeutic targets in drug development are discussed.
Sequence and conformational characteristics of IDPs
The special biophysical properties of IDPs are directly reflected in the frequencies of different amino acids in their sequences. Relative to globular proteins, IDPs are significantly depressed in aliphatic (val, leu, ile) and aromatic (tyr, phe, trp) amino acids, as well as cys, and enriched in most polar and charged amino acids (met, lys, arg, ser, asn, glu) and structure-breaking amino acids (pro, gly).2 This skewed distribution of amino acid types can been exploited to predict the occurrence of disordered protein regions (comparisons between various disorder prediction methods can be found in Jin and Liu24 and Atkins et al25). Indeed, advances in sequence-based prediction of disorder were the key to establishing the prevalence of disorder within natural protein sequences.9
Despite their designation as “disordered,” IDPs do not generally behave as structureless, random polymer chains. The sequences of globular proteins encode their three-dimensional structures (and how to fold to these structures), whereas the sequences of IDPs encode the stability, location, and character of partial structure and overall chain organization. In order to study the sequence determinants of chain compactness in IDPs, Marsh and Forman-Kay26 compiled literature data on the hydrodynamic radii of 32 different IDPs. They found that a high net charge (usually negative) and high fraction of proline tend to increase the size of the IDPs.26 Interestingly, a high fraction of hydrophobic residues did not correlate with the compaction of IDPs, as one might have suspected given the important role of hydrophobic amino acids in driving the folding of globular proteins.27,28
Residual secondary structure is frequently observed and quantified in IDPs by using various biophysical techniques, including NMR29 and circular dichroism. However, the dynamic nature of IDPs and the inherent ensemble averaging of in-bulk experiments can make poorly populated conformational states difficult to detect with traditional techniques. In such cases, single-molecule and computational approaches can provide important additional insights into IDP conformations. The protein α-synuclein, for example, which is best known for its implication in Parkinson’s disease, contains relatively little secondary structure in its monomeric state as judged by circular dichroism.30,31
Yet, single-molecule force spectroscopy experiments, in which individual molecules are mechanically stretched and their mechanical properties are recorded, have detected mechanoresistence forces in a range of values for monomeric α-synuclein, suggesting a heterogeneous conformational population.32 In fact, forces as high as ~400 pN were detected, which is on par with or higher than for many ordered proteins, providing direct evidence for partial structure in IDPs. These results are also in line with electrospray ionization–mass spectrometry experiments, a technique that relies on relating the extent of multiple charging of proteins in solution and the compactness of the conformations, which could discern four subpopulations in α-synuclein (including a compact state).33 Computer simulations of α-synuclein monomers have suggested that the high resistance forces may originate from three-stranded, antiparallel β-sheet structures present in a subset of the α-synuclein conformational ensemble.34
Overall, IDPs can be seen as a diverse class of proteins with a range of conformational properties, from mainly ordered structures with locally disordered segments to highly dynamic conformations with little secondary structure, in line with the proposal of Dyson and Wright that proteins fall on a structural continuum.13 Perhaps the most puzzling basic question in IDP research is why organisms make use of these dynamic proteins at all when they apparently increase the risk for disease, through either protein overexpression35 or aggregation.36,37 The answer lies in the unusual biophysics of IDPs that allows them to function in other ways than ordered proteins, as discussed in the following section.
Functional dynamics: protein–protein interactions and regulation
Coupled folding–binding and allosteric regulation
The classical view of allostery is that two distant sites on a protein structure are coupled via a network of energetically linked residues that exists throughout the structure, such that ligand binding at one site affects the affinity for a second ligand at the other site. However, the coupled folding–binding process of IDPs provides a different mechanism to achieve allosteric coupling between distant sites, which relies on thermodynamic rather than energetic considerations alone.38 Allosteric effects mediated by coupled folding–binding have been demonstrated in several regulatory and viral proteins, including the adenovirus early regions 1A protein,39 the protein Phd of the bacterial toxin/antitoxin operon network,40 and the KIX domain of the transcriptional coactivator CBP.41
The precise mechanism underlying the observed allosteric coupling in KIX, in particular, has been heavily discussed in the literature, from both experimental41–44 and theoretical45,46 perspectives. One of the binding partners of KIX, c-Myb, is a transcription factor involved in the differentiation and proliferation of hematopoeitic cells. The centrally located transactivation domain of c-Myb is disordered on its own but folds into a single, kinked α-helix upon binding to KIX (Figure 1). Another disordered KIX binding partner is the activation domain of the mixed lineage leukemia (MLL) protein. Similar to c-Myb, MLL is triggered to fold into a single α-helical structure upon binding to KIX; however, it uses a different binding site on KIX. Therefore, c-Myb and MLL are able to bind KIX simultaneously, and moreover, the loading of KIX with either c-Myb or MLL enhances the affinity for the other ligand by a factor of ~2.47
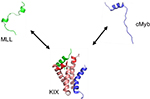  | Figure 1 Coupled folding–binding and allosteric coupling. Notes: The KIX domain (salmon and red) of CBP can simultaneously bind the two ligands c-Myb (blue) and MLL (green). In isolation, both ligands lack a stable structure. Circular dichroism measurements show c-Myb to be ~30% α-helical.100 Upon association with KIX, both cMyb and MLL fold into stable α-helical structures, but binding occurs to different binding sites on KIX. A disorder–order process has been suggested to take place in the C-terminal part of α3 and the L12–G2 loop of KIX (red) upon binding either MLL or c-Myb.45 The binding affinity of MLL for KIX/c-Myb complex is higher by a factor of 1.6 compared to KIX alone, whereas the binding affinity of c-Myb increases by approximately a factor of 2 when KIX associates with MLL. The structure of the KIX complex was reprinted with permission from Brüschweiler S, Konrat R, Tollinger M. Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core. ACS Chem Biol. 2013;8:1600–1610, available from: http://pubs.acs.org/doi/full/10.1021/cb4002188. Copyright © 2013 American Chemical Society.41 Abbreviations: CBP, CREB binding protein; MLL, mixed lineage leukemia. |
Although KIX is mostly a well-ordered protein, it includes significant conformational disorder.48 Although different studies have arrived at different precise allosteric mechanisms, as will be discussed, this conformational heterogeneity likely underpins allosteric coupling in KIX. On the basis of NMR relaxation dispersion techniques, Brüschweiler et al42 found that the binding of MLL induces a redistribution of the conformational ensemble of KIX, including repacking of the hydrophobic core,41 toward a higher energy state that exhibits a higher affinity for the second ligand, c-Myb. Molecular dynamic simulations of the KIX–MLL complex connected this higher-energy state to an “up” state of the L12–G2 loop of the KIX structure.46
More recently, simulations by Law et al45 using a coarse-grained protein model suggested instead a narrowing of the KIX conformational ensemble, that is, a disorder–order transition, upon binding of either MLL or c-Myb. These computational results were also found to be consistent with measurements of the binding kinetics of MLL, c-Myb, and other similar KIX ligands.44 The simulations from Law et al observed, more precisely, that the disorder–order transition involved the C terminal part of the α3 helix as well as the L12–G2 loop (Figure 1). Because this ordering in KIX takes place upon binding either c-Myb or MLL, the view emerging from these studies44,45 is that the first ligand that binds KIX, whether it is MLL or c-Myb, effectively “prepays” an entropic cost that allows the second ligand to bind with an enhanced affinity.
The discovery that IDPs, and indeed all dynamic proteins,49 can exhibit allosteric coupling has had a major impact on the current view of allostery in proteins.50 In particular, it has led to the formulation of new theoretical frameworks,51,52 based on free energy landscape and ensemble views of proteins, which are applicable to IDPs and ordered proteins alike. Common to these frameworks is a focus on the statistical nature of the allosteric process, such that the relative population among different conformational (sub)states of an allosteric protein can be shifted by an effector molecule, thereby altering the properties of a protein including its ligand-binding capabilities. In the case of KIX, for example, both proposed molecular mechanisms can be interpreted as resulting from a shift in the conformational ensemble of monomeric KIX upon binding of the first ligand (albeit in different ways), such that the affinity for the second ligand is enhanced. Much recent work is focused on applying the recently improved understanding of allostery in practical ways, for example, in the rational design of proteins with switch-like behaviors53,54 or drugs that target allosteric sites (see Nussinov and Tsai55 and Dokholyan56 for recent reviews on “allosteric drugs”).
Regulated unfolding
Recent studies have revealed a novel cell regulatory mechanism that relies on the unfolding of ordered protein structures, triggered by signals such as ligand binding, mechanical stress, posttranslational modifications, and changes in pH. Various extents of unfolding have been observed, ranging from local unfolding of a secondary structure element to the global unfolding of an entire protein.57
One example of ligand-controlled regulatory unfolding can be found in the mitochondrial pathway of programmed cell death.58 The tumor suppressor p53 causes apoptosis by associating with various proapoptotic proteins, such as BAX, which leads to mitochondrial outer membrane permeabilization and subsequent cell death. However, the apoptotic function of p53 is usually inhibited by the sequestering of cytosolic p53 in inactive complexes formed with the anti-apoptotic protein Bcl-xL. This sequestering is controlled by the intrinsically disordered BH3 domain of PUMA (p53 upregulated modulator of apoptosis), which binds to Bcl-xL and, in the process, folds into a single α-helix. Intriguingly, the association of PUMA with Bcl-xL also triggers a local unfolding of two of the α-helices of Bcl-xL, α2 and α3, which disrupts the p53/Bcl-xL binding interface. This unfolding thereby releases p53 from Bcl-xL, freeing p53 to carry out its apoptotic function.
An example of global regulatory unfolding is provided by the K-homology splicing regulator protein (KSRP),59 which controls gene expression at the posttranscriptional level. KSRP contains four consecutive KH domains,60 which are regulatory modules that can bind RNA or single-stranded DNA. The third and fourth KH domains (KH3 and KH4) of KSRP bind to adenylate–uridylate-rich elements of mRNAs, whereas the N-terminal KH domain (KH1) recruits the exosome complex, thereby promoting the degradation of the targeted mRNA. Impairment of the exosome recruitment can be achieved by phosphorylation of Ser193 of KSRP, as it causes relocalization of KSRP to the nucleus through an interaction with the 14-3-3ζ protein. Like other 14-3-3 domains, 14-3-3ζ binds short, phosphorylated linear peptide motifs typically found in intrinsically disordered regions.61 Interestingly, Ser193 lies within the domain boundaries of the KH1 domain and is therefore not accessible for recognition by 14-3-3ζ, which requires an extended peptide motif for binding. This apparent contradiction is explained by the complete unfolding of KH1 triggered by the Ser193 phosphorylation, such that the 14-3-3 binding motif is both created and made fully exposed. Biophysical characterization of KH1 confirms that the phosphorylated KH1 domain is ~80% unfolded at 37°C. A dephoshporylation of Ser193 leads to a refolding of the KH domain,59 such that the regulatory mechanism is reversible.
The picture that emerges is that these regulatory proteins are folded on their own but balance on the brink of local or global unfolding. Thermal unfolding curves on KH1 based on circular dichroism show that this domain is less stable than KH2-KH4, and NMR spectroscopy reveals that the N-terminal β strand of KH1 exhibits substantial structural fluctuations, providing access to Ser193 to be phosporylated.59 Similarly, molecular dynamic simulations of free Bcl-xL reveal that the α2 and α3 helices are highly dynamic.62 Therefore, regulatory unfolding appears to be linked to lower stability and increased dynamic behavior of the native state. The situation is reminiscent of the decreased stability and increased dynamics exhibited by so-called “metamorphic” proteins,63 which have an ability to reversibly switch between different ordered (folded) states. Many of the signals that trigger regulatory unfolding, including ligand binding, changes in solution conditions, and chemical modifications, are also those that trigger fold switching in metamorphic proteins.64 Future experimental and theoretical studies will no doubt seek to determine the principles that allow these regulatory proteins to find the delicate balance between native and unfolded (local or global) states and that underpin their unique ability for conformational switching.
IDPs promote the formation of membrane-less organelles
The cytoplasm and nucleoplasm of many cell types contain various organelles that can maintain structural integrity without an enclosing membrane.65 These spherical membrane-less organelles include Cajal bodies, nucleoli, processing bodies, and germ line granules. Because they typically contain both RNA and protein, they are also referred to as ribonucleoprotein (RNP) bodies or granules. RNP bodies have emerged as key players in the posttranscriptional regulation of gene transcription.65 Recently, the formation and biophysical properties of these organelles have come under intense study.16,17,66,67 It appears that RNP bodies are formed via spontaneous liquid–liquid phase separation and display classical liquid-like behaviors, such as wetting, dripping, and relaxation to spherical shape following droplet fusion. Properties such as viscosity and molecular diffusivity are tunable through RNA and salt concentration, which may provide a means for cells to control the RNA processing functions.
Although the molecular mechanisms underlying the formation and function of RNP bodies remain poorly understood, evidence is pointing to a significant role of IDPs. On a general level, a compilation of proteins in various membrane-less cytoplasmic and nucleoplasmic organelles revealed that intrinsic disorder is highly prevalent within this set of proteins.67 More direct evidence has come from studies of P granules, which are implicated in specifying and maintaining the germ cells of Caenorhabditis elegans.16 P granules are enriched in various RNA-binding proteins, including the RNA helicase LAF-1 that can phase separate into droplets at high protein/low salt concentration in vitro. Elbaum-Garfinkle et al17 showed that the disordered arginine/glycine-rich RGG domain of LAF-1 (∼200 amino acids) is both necessary and sufficient for the formation of P granule–like liquid droplets in vitro. As much remains to be discovered about the biophysics of RNP body formation, one molecular mechanism for P granule assembly has been suggested to involve a loose network of IDP–IDP interactions that drive proteins into dynamic liquid droplets with suitable physical properties.17
IDPs can form “fuzzy” complexes
The coupled folding and binding process provides a useful theoretical framework for understanding the structural basis of recognition in biomolecular interactions involving one or more IDPs. However, there are several examples of interactions in which partial or even full disorder persists in the final complex.68 In other words, the folding of IDPs upon contact with partner molecules can be incomplete or even absent in some cases.
Tompa and Fuxreiter coined the term “fuzziness”69 to describe this phenomenon, inspired by the concept of “fuzzy logic” in mathematics. Further, Tompa and Fuxreiter classified disorder in protein complexes as either “static” or “dynamic”. Static disorder refers to complexes such as Tcf4/β-catenin70 where the IDP adopts a few or a multitude of distinct stable conformations (“polymorphic” model). Dynamic disorder refers to cases where some regions remain unstructured in the complex, either neighboring (“flanking” model) or linking (“clamp” model) ordered binding regions. In an extreme case, formation of the complex does not induce any folding of the IDP, which is therefore left in an entirely disordered but bound state (“random” model).
The above classification can be seen as somewhat arbitrary because there is a continuum between static and dynamic disorder. However, the different interaction models can be useful in distinguishing different functional mechanisms. How dynamic disorder can mediate function is illustrated by the interaction between Sic1, a cyclin-dependent kinase inhibitor, and Cdc4, a ubiquitin ligase subunit. This phosphorylation-dependent interaction regulates the G1/S phase progression in the cell cycle of budding yeast.71
For optimal binding between Sic1 and Cdc4, phosphorylation of at least six of nine possible sites on Sic1 is required. Upon binding Cdc4, Sic1 remains disordered such that the different phosphorylation sites compete for binding to a single binding site on Cdc4.68 This mechanism provides the physical basis for what is essentially a phosphorylation-regulated ultrasensitive threshold for initiation of DNA replication.72
Fuzzy complexes may turn out to be more widespread than currently thought. This possibility is illustrated by computational studies73,74 of the interaction between the calcium-regulated protein S100B and the disordered negative regulatory domain of p53 (p53-NRD), including a study74 from the author’s group. Previous NMR studies have suggested that the p53-NRD folds into a well-organized α-helix upon binding to S100B.75
By characterizing the equilibrium behavior of the p53-NRD/S100B interaction by using an all-atom Monte Carlo-based approach,76 it was found that whereas p53-NRD has a propensity for α-helical structure in the bound state, the peptide exhibits substantial conformational heterogeneity.74 In particular, a centrally located Phe residue was found to interact strongly with the S100B peptide-binding pocket, while flanking segments were substantially disordered. McDowell et al,73 by using two different state-of-the-art explicit water molecular dynamics force fields, confirmed the dynamic nature of this complex. Importantly, these authors also demonstrated that the obtained dynamic ensembles representing the bound state were consistent with the experimentally derived intermolecular NOE nuclear Overhauser effect (NOE) distance restraints for this complex. It should be pointed out that most assigned NOEs are extremely weak for the p53-NRD/S100B complex. Nonetheless, these results73,74 emphasize the importance of considering potential disorder in protein structure calculations and that neglecting this possibility can lead to structural models that are too ordered.
IDPs as potential therapeutic targets
Search for a new class of targets
Over the last several decades, major advances have been made in technologies of key importance for drug discovery efforts, such as high-throughput screening of combinatorial chemistry libraries against protein targets,77 more efficient determination of biomolecular structures via X-ray crystallography,78 and DNA sequencing.79 Despite these and other advances, the efficiency of research and development in the pharmaceutical industry, as measured by the number of new drugs brought to market by per billion US dollars spent, has seen a rather steady decline over the last 60 years.80 This trend, which has been termed Eroom’s law80 (the reverse of Moore’s law,81 ie, the prediction that the number of transistors in dense integrated circuits roughly doubles every 2 years), is prompting a search for new approaches in drug discovery.
One of the possible new avenues is to target intracellular protein–protein interactions,82 in contrast to the classical drug targets such as the well-defined binding pockets of enzymes and membrane receptors. Protein–protein interfaces are comparatively large and flat and have been seen as challenging targets for disruption by small molecules. However, this problem was alleviated with the discovery that there are “hot spots” in protein–protein interfaces, that is, a few key residues contribute to the bulk of the binding free energy. There have been several recent examples of successful small molecule inhibition of protein–protein interactions that hold promise particularly for cancer,83,84 and some compounds have been taken to clinical trials.85
Inhibiting IDPs with small molecules
Protein–protein interactions involving one or more IDPs are of special interest as drug targets for a few reasons. 1) IDPs are both prevalent and overrepresented in signaling, regulatory, and human disease pathways.86 2) The actions of IDPs are likely sensitive to modulation by small molecules. The reason for this is that IDPs tend to bind multiple targets in the cell with modest affinities, and as a consequence, their actions are likely controlled by cellular concentration. Therefore, even partial inhibition should impact function. In line with this picture, it has been found that the expression of IDPs is tightly regulated in cells,35 and moreover, among the genes that are lethal upon overexpression in yeast, IDPs are overrepresented87 – a too high cellular concentration of IDPs leads to excessive sequestration of IDPs in nonspecific complexes. 3) Although IDPs are significantly enriched in disorder-promoting amino acids, that is, polar and charged amino acids, this is not true for the regions directly involved in target binding. In fact, binding motifs in IDPs are often enriched in hydrophobic amino acids.88 As a result, computationally fast sequence-based methods can be used to predict potential binding sites on IDPs on a genome-wide scale. For example, a set of >35,000 disordered regions with compact hydrophobic clusters, which are likely involved in binding to ordered targets, has already been identified in the human proteome.89
The intrinsically disordered transcription factor c-Myc, which regulates genes involved in various cellular processes, including apoptosis,90 has been studied for inhibition by small molecules. In order to become biologically active, c-Myc must heterodimerize with its partner Max, forming a basic helix-turn-helix leucine zipper with a large protein–protein interface (∼3,200 A˚).91 Because Max is also an IDP, the c-Myc–Max association involves so-called synergistic folding,3,92,93 in which the two partners induce folding in each other upon binding. Several small molecules that inhibit the formation of the c-Myc–Max complex have been obtained through different types of combinatorial library screens.94–96 One of the obtained compounds, 10058-F4, causes reversion of the c-Myc–Max complex into disordered monomers by binding to c-Myc, as shown by a striking shift in the circular dichroism spectra.96
How do small molecules bind c-Myc and how is the conformational ensemble of monomeric c-Myc affected by the binding? In order to better understand the molecular details of this interaction, Michel and Cuchillo97 and Jin et al98 performed molecular dynamic simulations of the interaction of selected regions on c-Myc and a related compound, 10074-A4. Although the c-Myc segments simulated were different in the two computational studies, a similar picture of the interaction was obtained. First, the ligand 10074-A4 does not to induce c-Myc to fold into a specific structure as does Max, consistent with circular dichroism data. Rather, the conformational ensemble of monomeric c-Myc is only perturbed to a small extent by the ligand. The binding of 10074-A4 occurs on multiple points along the c-Myc chain, creating a “ligand cloud” around the IDP conformational ensemble (Figure 2), without a single dominant binding mode. In this sense, the interaction between the small molecule 10074-A4 and c-Myc resembles the fuzzy protein complexes obtained for some IDPs, as discussed above.
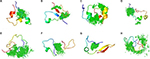  | Figure 2 Ligand cloud around a protein cloud. Notes: (A–H) Interaction between the region 370–409 of c-Myc (cartoon) and the ligand 10074-A4 (green), as represented by the eight most populated structures of c-Myc obtained from a cluster analysis98 and the center-of-mass points of the ligand. Reprinted from Jin F, Yu C, Lai L, Liu Z. Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins. PLOS Comput Biol. 2013;9:e1003249. Creative Commons Attribution (CC BY) license.98 |
In classic structure-based drug design, the aim is to find small molecules that precisely fit a binding pocket, for example, the active site of an enzyme. The difficulties in achieving both sufficient affinity and specificity are complicated by the fact that no protein is entirely static, but includes some structural fluctuations. Consequently, several different strategies to deal with multiple relevant conformations in drug design have been developed.99 However, many IDPs exist as a broad statistical ensemble of conformations with widely fluctuating backbone and side chain torsional angles. How specificity of IDPs for small molecules is achieved is currently unclear. In negative control simulations involving a different region of c-Myc, Jin et al98 unexpectedly found substantial binding with 10058-A4, albeit at a lower affinity. Given this background, future investigations into the molecular mechanisms of specificity in IDP–small molecule binding will be of particular interest.
Conclusion and prospects
Following the first systematic studies of IDPs in the 1990s, there have been intense computational and experimental efforts to understand the structural and dynamical basis of function for this class of highly flexible proteins. These efforts have revealed that IDPs are highly prevalent in many genomes and involved in a range of cellular processes in which they function through diverse functional dynamics. This review focused in particular on some emerging areas of importance for cell regulation, including allosteric coupling, regulated unfolding, and formation of membrane-less intracellular processing bodies. Given their prevalence and central role in biology, it is not surprising that IDPs are implicated in human disease and that efforts are under way for therapeutic targeting of IDPs. It was noted that mechanisms underlying specificity in small molecule–IDP interactions need to be further elucidated. Although IDP biophysics as a field has reached some level of maturity, future studies are likely to bring additional discoveries about the role of IDPs in biology and disease.
Disclosure
The author reports no conflicts of interest in this work.
References
Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. | ||
Dunker AK, Oldfield CJ, Meng J, et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9(Suppl 2):S1. | ||
Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. | ||
Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114:6561–6588. | ||
Pullen RA, Jenkins JA, Tickle IJ, Wood SP, Blundell TL. The relation of polypeptide hormone structure and flexibility to receptor binding: the relevance of X-ray studies on insulin, glucagon and human placental lactogen. Mol Cell Biochem. 1975;8:5–20. | ||
Radford SE, Laue ED, Perham RN, Martin SR, Appella E. Conformational flexibility and folding of synthetic peptides representing an interdomain segment of polypeptide chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Biol Chem. 1989;264:767–775. | ||
Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. | ||
Kosol S, Contreras-Martos S, Cedeño C, Tompa P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules. 2013;18:10802–10828. | ||
Romero P, Obradovic Z, Kissinger CR, et al. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. 1998;3:437–448. | ||
Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. | ||
Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. | ||
Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. | ||
Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. | ||
Hough LE, Dutta K, Sparks S, et al. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. Elife. 2015;4:e10027. | ||
Gursky O. Structural stability and functional remodeling of high-density lipoproteins. FEBS Lett. 2015;589:2627–2639. | ||
Brangwynne CP, Eckmann CR, Courson DS, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. | ||
Elbaum-Garfinkle S, Kim Y, Szczepaniak K, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. | ||
Mitrea DM, Yoon MK, Ou L, Kriwacki RW. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem. 2012;393:259–274. | ||
Schulenburg C, Hilvert D. Protein conformational disorder and enzyme catalysis. Top Curr Chem. 2013;337:41–67. | ||
Norris AL, Serpersu EH. NMR detected hydrogen-deuterium exchange reveals differential dynamics of antibiotic and nucleotide-bound aminoglycoside phosphotransferase 3’-IIIa. J Am Chem Soc. 2009;131:8587–8594. | ||
Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. | ||
Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. | ||
Hsu WL, Oldfield CJ, Xue B, et al. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22:258–273. | ||
Jin F, Liu Z. Inherent relationships among different biophysical prediction methods for intrinsically disordered proteins. Biophys J. 2013;104:488–495. | ||
Atkins JD, Boateng SY, Sorensen T, McGuffin LJ. Disorder prediction methods, their applicability to different protein targets and their usefulness for guiding experimental studies. Int J Mol Sci. 2015;16:19040–19054. | ||
Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98:2383–2390. | ||
Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. | ||
Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. | ||
Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv Protein Chem. 2002;62:311–340. | ||
Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. | ||
Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. | ||
Sandal M, Valle F, Tessari I, et al. Conformational equilibria in monomeric alpha-synuclein at the single-molecule level. PLoS Biol. 2008;6:e6. | ||
Frimpong AK, Abzalimov RR, Uversky VN, Kaltashov IA. Characterization of intrinsically disordered proteins with electrospray ionization mass spectrometry: conformational heterogeneity of alpha-synuclein. Proteins. 2010;78:714–722. | ||
Jónsson SÆ, Mitternacht S, Irbäck A. Mechanical resistance in unstructured proteins. Biophys J. 2013;104:2725–2732. | ||
Babu MM, Lee R, Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21:432–440. | ||
Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. | ||
Chen Y, Dokholyan NV. Natural selection against protein aggregation on self-interacting and essential proteins in yeast, fly, and worm. Mol Biol Evol. 2008;25:1530–1533. | ||
Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104:8311–8315. | ||
Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. | ||
Garcia-Pino A, Balasubramanian S, Wyns L, et al. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell. 2010;142:101–111. | ||
Brüschweiler S, Konrat R, Tollinger M. Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core. ACS Chem Biol. 2013;8:1600–1610. | ||
Bru¨ schweiler S, Schanda P, Kloiber K, et al. Direct observation of the dynamic process underlying allosteric signal transmission. J Am Chem Soc. 2009;131:3063–3068. | ||
Toto A, Giri R, Brunori M, Gianni S. The mechanism of binding of the KIX domain to the mixed lineage leukemia protein and its allosteric role in the recognition of c-Myb. Protein Sci. 2014;23:962–969. | ||
Shammas SL, Travis AJ, Clarke J. Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proc Natl Acad Sci U S A. 2014;111:12055–12060. | ||
Law SM, Gagnon JK, Mapp AK, Brooks CL. Prepaying the entropic cost for allosteric regulation in KIX. Proc Natl Acad Sci U S A. 2014;111:12067–12072. | ||
Palazzesi F, Barducci A, Tollinger M, Parrinello M. The allosteric communication pathways in KIX domain of CBP. Proc Natl Acad Sci U S A. 2013;110:14237–14242. | ||
Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. | ||
De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355:1005–1013. | ||
Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. | ||
Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. | ||
Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. | ||
Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. | ||
Choi JH, Laurent AH, Hilser VJ, Ostermeier M. Design of protein switches based on an ensemble model of allostery. Nat Commun. 2015;6:6968. | ||
Stratton MM, Loh SN. Converting a protein into a switch for biosensing and functional regulation. Protein Sci. 2011;20:19–29. | ||
Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. | ||
Dokholyan NV. Controlling allosteric networks in proteins. Chem Rev. 2016;116:6463–6487. | ||
Mitrea DM, Kriwacki RW. Regulated unfolding of proteins in signaling. FEBS Lett. 2013;587:1081–1088. | ||
Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. | ||
Diaz-Moreno I, Hollingworth D, Frenkiel TA, et al. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nat Struct Mol Biol. 2009;16:238–246. | ||
Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. | ||
Bustos DM, Iglesias AA. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins. 2006;63:35–42. | ||
Liu X, Beugelsdijk A, Chen J. Dynamics of the BH3-only protein binding interface of Bcl-xL. Biophys J. 2015;109:1049–1057. | ||
Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. | ||
Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20:482–488. | ||
Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. | ||
Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nature. 2015;11:899–904. | ||
Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015;589:15–22. | ||
Mittag T, Orlicky S, Choy WY, et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105:17772–17777. | ||
Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. | ||
Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–1052. | ||
Nash P, Tang X, Orlicky S, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. | ||
Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104:9650–9655. | ||
McDowell C, Chen J, Chen J. Potential conformational heterogeneity of p53 bound to S100B(ββ). J Mol Biol. 2013;425:999–1010. | ||
Staneva I, Huang Y, Liu Z, Wallin S. Binding of two intrinsically disordered peptides to a multi-specific protein: a combined Monte Carlo and molecular dynamics study. PLoS Comput Biol. 2012;8:e1002682. | ||
Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta). Nat Struct Biol. 2000;7:570–574. | ||
Staneva I, Wallin S. All-atom Monte Carlo approach to protein-peptide binding. J Mol Biol. 2009;393:1118–1128. | ||
Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. | ||
Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr. 2008;64:88–95. | ||
Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. | ||
Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. | ||
Moore GD. Cramming more components onto integrated circuits. Electronics. 1965;38:114–117. | ||
Mullard A. Protein-protein interaction inhibitors get into the groove. Nat Rev Drug Discov. 2012;11:173–175. | ||
Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. | ||
Fry DC, Vassilev LT. Targeting protein-protein interactions for cancer therapy. J Mol Med. 2005;83:955–963. | ||
Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21:1102–1114. | ||
Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. | ||
Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. | ||
Meszaros B, Tompa P, Simon I, Dosztanyi Z. Molecular principles of the interactions of disordered proteins. J Mol Biol. 2007;372:549–561. | ||
Cheng Y, LeGall T, Oldfield CJ, et al. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. | ||
Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. | ||
Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14:481–488. | ||
Zhang W, Ganguly D, Chen J. Residual structures, conformational fluctuations, and electrostatic interactions in the synergistic folding of two intrinsically disordered proteins. PLoS Comput Biol. 2012;8:e1002353. | ||
Bhattacherjee A, Wallin S. Coupled folding-binding in a hydrophobic/polar protein model: impact of synergistic folding and disordered flanks. Biophys J. 2012;102:569–578. | ||
Berg T, Cohen SB, Desharnais J, et al. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci U S A. 2002;99:3830–3835. | ||
Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg Med Chem Lett. 2009;19:6038–6041. | ||
Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008;15:1149–1155. | ||
Michel J, Cuchillo R. The impact of small molecule binding on the energy landscape of the intrinsically disordered protein C-myc. PLoS One. 2012;7:e41070. | ||
Jin F, Yu C, Lai L, Liu Z. Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins. PLOS Comput Biol. 2013;9:e1003249. | ||
Carlson HA. Protein flexibility and drug design: how to hit a moving target. Curr Opin Struct Biol. 2002;6:447–452. | ||
Shammas SL, Travis AJ, Clarke J. Remarkably fast coupled folding and binding of the intrinsically disordered transactivation domain of cMyb to CBP KIX. J Phys Chem B. 2013;117:13346–13356. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
