Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Interactive Association Between Intronic Polymorphism (rs10506151) of the LRRK2 Gene and Type 2 Diabetes on Neurodegenerative Diseases
Authors Huang MH, Liu YF, Nfor ON, Hsu SY, Lin WY, Chang YS, Liaw YP
Received 16 April 2021
Accepted for publication 16 June 2021
Published 13 July 2021 Volume 2021:14 Pages 839—847
DOI https://doi.org/10.2147/PGPM.S316158
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Martin H Bluth
Mei-Hsuen Huang,1 Yu-Fan Liu,2,3 Oswald Ndi Nfor,4 Shu-Yi Hsu,4 Wei-Yong Lin,5– 7 Yuan-Shiun Chang,1 Yung-Po Liaw4,8
1Department of Chinese Pharmaceutical Sciences and Chinese Medicine Resources, College of Chinese Medicine, China Medical University, Taichung, 40402, Taiwan; 2Department of Biomedical Sciences, Chung Shan Medical University, Taichung, Taiwan; 3Division of Allergy, Department of Pediatrics, Chung Shan Medical University Hospital, Taichung, Taiwan; 4Department of Public Health and Institute of Public Health, Chung Shan Medical University, Taichung, 40201, Taiwan; 5Graduate Institute of Integrated Medicine, College of Chinese Medicine, China Medical University, Taichung, 40402, Taiwan; 6Department of Medical Research, China Medical University Hospital, Taichung, 40447, Taiwan; 7Brain Diseases Research Center, China Medical University, Taichung, 40402, Taiwan; 8Department of Medical Imaging, Chung Shan Medical University Hospital, Taichung City, 40201, Taiwan
Correspondence: Yung-Po Liaw
Department of Public Health and Institute of Public Health, Chung Shan Medical University, No. 110, Sec. 1 Jianguo N. Road, Taichung City, 40201, Taiwan
Tel +886-4-24730022 ext.11838
Fax +886-4-23248179
Email [email protected]
Yuan-Shiun Chang
Department of Chinese Pharmaceutical Sciences and Chinese Medicine Resources, College of Chinese Medicine, China Medical University, Taichung, 40402, Taiwan
Tel +886-4-22053366 ext. 5502
Fax +886-4-22083362
Email [email protected]
Purpose: We investigated the interactive effect of rs10506151 polymorphism of the Leucine-rich repeat kinase 2 (LRRK2) gene and type 2 diabetes (T2D) on neurodegenerative disease (ND) risk.
Materials and Methods: Data of 17, 927 participants in the Taiwan Biobank (TWB) assessed between 2008 and 2015 were linked to healthcare records in the National Health Insurance Research Database (NHIRD). The odd ratios (ORs) and 95% confidence intervals (CIs) for NDs were determined using logistic regression analysis.
Results: There were 145 cases with NDs, and 28.28% (n = 41) of these individuals had T2D. Associations of neurodegenerative disorders with LRRK2 rs10506151 variant and T2D were not significant. The corresponding ORs (95% CI) for NDs were 1.06 (0.75– 1.49) in CA/AA compared to CC individuals and 0.93 (0.63– 1.39) in those with T2D compared to non-diabetic participants. However, we found evidence of a significant interaction between rs10506151 and T2D (p = 0.0073). After stratification by genotypes of rs10506151, the OR for NDs was 0.37 (CI, 0.17– 0.82) in CA/AA individuals with T2D and 1.41 (0.88– 2.27) in their CC counterparts. When CA/AA individuals with T2D represented the reference group, the OR (95% CI) was 1.74 (0.81– 3.73) in CC individuals with no T2D, 2.47 (CI, 1.14– 5.38) in CA/AA individuals with no T2D, and 2.34 (CI, 1.07– 5.11) in CC individuals with T2D.
Conclusion: Our data indicated that the risk of NDs was significantly lower among diabetic individuals with combined CA/AA of the LRRK2 rs10506151 variant in Taiwan.
Keywords: polymorphism, neurodegenerative disorders, diabetes, variation
Introduction
Neurodegenerative diseases (NDs) such as Parkinson’s disease (PD), Alzheimer’s disease (AD), and multiple sclerosis, are significant life-threatening diseases in adults.1 They are characterized by degeneration of specific neural clusters.2 Some of the factors associated with neurological diseases include proteinopathy, hypovitaminosis D, and oxidative stress,3,4 even though they express separate pathophysiological features.5
Genetic factors are essential for providing insights into the pathogenesis of NDs. Attempts have been made to investigate mutations in specific ND-related genes.6 The LRRK2 gene is one of the genes that contribute to neurodegeneration, especially in PD.7 Several mutations of this gene have been investigated as risk factors for PD, though the mechanisms remain to be investigated.8,9 Among them, the rs10506151 polymorphism had been associated with a dramatic increase in PD risk among Chinese individuals who were homozygous for the minor allele (rs10506151 A) in both additive and recessive models.10 Despite this, a prior study in Norway found no evidence of an association between the variant and PD.11 It is not yet known whether rs10506151 could increase the risk of NDs.
Diabetes had also been associated with a higher risk of PD and other NDs.12–16 However, a case-control study in Japan reported inverse associations between T2D and PD risk.17 Diabetes mellitus and neurodegenerative conditions seem to share pathophysiological mechanisms. Importantly, Zhu and colleagues previously identified LRRK2 as one of the hub genes for T2D and AD.18
While the LRRK2 rs10506151 polymorphism dramatically increased PD risk in Chinese populations, such associations have not been reported in Taiwan. Therefore, this study investigated the interactive effects of rs10506151 polymorphism and T2D on neurodegenerative disease development in Taiwan Biobank.
Materials and Methods
Study Participants and Setting
We collected basic demographic, anthropometric, lifestyle, and genetic data of participants enrolled in TWB between 2008 and 2015. The disease information was collected from the NHIRD. Data from TWB were linked to electronic healthcare records in the NHIRD at the Health and Welfare Data Science Center (HWDC). The biobank data are not freely available due to restrictions. The Institutional Review Board on Biomedical Science Research in Academia Sinica and the Governance Council of Taiwan Biobank had approved the TWB project.19 Biobank participants had provided written informed consent before enrollment. This study was approved by the Institutional Review Board of Chung Shan Medical University (CS1-20009).
Initial recruitment included 17,985 biobank participants. After excluding persons with incomplete questionnaires (N=37) and genotyping information (N=21), the final analysis included 145 participants with NDs and 17,927 control individuals. Among participants with NDs, 41 were identified with T2D.
Disease Assessment and Covariates
From the linked datasets, we identified participants with NDs including PD (ICD-9-CM: 332.0), other degenerative diseases of the basal ganglia (ICD-9-CM: 333.0), AD (ICD-9-CM: 331.0), dementia subtypes (ICD-9-CM: 290, 331.1, 331.2, 331.7, 331.8, and 331.9), hyperlipidemia (ICD-9-CM: 272), diabetes mellitus (ICD-9-CM: 250), and hypertension (ICD-9-CM: 401–405) based on at least two outpatient visits or inpatient hospitalization.
Cigarette smokers were defined as people who smoked for at least six months and continued to smoke during assessment visits. Alcohol drinkers were defined as persons who reported drinking more than 150 mL of alcohol per week for at least six months and were still drinking during assessment visits. Physical activity included any exercise activity at least three times a week and lasting for at least 30 minutes each time. Body mass index (BMI) was calculated as weight in kilograms divided by height in meters squared (kg/m2).
Genetic Variant, Genotyping and Quality Control
In TWB, genotyping is performed on participants by the National Center for Genome Medicine in Academia Sinica. This is done using the custom Taiwan Biobank Axiom™ Genome-Wide Array Plate (Affymetrix, Santa Clara, CA, USA). In addition to the socio-demographic and lifestyle data, biological samples (including blood and urine samples) are collected from all participants during assessment visits for different assays. Details of the genotyping procedures have been described elsewhere.19 In this study, we chose the LRRK2 rs10506151 variant based on its previous associations with neurodegenerative diseases. This variant passed the quality control test: call rate (>95%), Hardy-Weinberg equilibrium test (p>1.0 × 10−3), and minor allele frequency (>0.05).
Statistical Analysis
Our study outcome was NDs, while rs10506151 and T2D were the exposure variables. We performed descriptive analysis for these variables and covariates based on the presence or absence of T2D. The inclusion and exclusion of study participants are shown in Figure 1. We used the t-test and Chi-square test for the continuous and categorical variables. We then used logistic regression analysis to determine the ORs and 95% CIs for the relationship between diabetes, LRRK2 (rs10506151), and NDs. We also tested for the presence of an interaction between the variant and T2D, followed by stratified analyses based on these variables. Our statistical tools included the statistical analysis system (SAS) 9.4 software (SAS Institute, Cary, NC, USA) and PLINK 1.09 beta.
 |
Figure 1 A flowchart showing study population. |
Results
Table 1 shows a summary of the demographic characteristics of participants classified by T2D status. The mean age (SD, standard deviation) was 56.86 (8.85) in those with T2D and 47.60 (10.78) in control participants. There were 2443 participants with T2D and 145 with neurodegenerative diseases. Logistic regression analyses showed that the rs10506151 variant and T2D were not independently associated with NDs (Table 2). The OR (95% CI) for NDs was 1.06 (0.75–1.49) in CA/AA compared to CC individuals, and 0.93 (95% CI, 0.63–1.39) in those with T2D (using those with no T2D as reference). Associations with NDs were also seen for age, educational level, hyperlipidemia, and hypertension.
 |
Table 1 Demographic Characteristics of Study Participants with and without T2D |
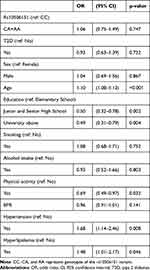 |
Table 2 Odds of Having Neurodegenerative Diseases Among Study Participants |
When we used a multivariate model with the interaction term (T2D X rs10506151), the effect was significant (p=0.0073). When stratified by genotypes (Table 3), the OR (95% CI) was 0.37 (0.17–0.82) in CA/AA individuals who had T2D and 1.41 (0.88–2.27) in their CC counterparts. Compared with the CC genotype (Table 4), CA/AA individuals with and without T2D had ORs (95% CI) of 0.42 (0.19–0.93) and 1.42 (0.96–2.11), respectively. When we used CA/AA and T2D as the reference group (Table 5), the OR (95% CI) was 1.74 (0.81–3.73) for CC and no T2D, 2.47 (1.14–5.38) for CA/AA and no T2D, and 2.34 (1.07–5.11) for CC and T2D.
 |
Table 3 Odds Ratios for Having Neurodegenerative Diseases Based on Genotypes of rs10506151 |
 |
Table 4 Odds Ratios for Neurodegenerative Diseases in Patients with and without T2D |
 |
Table 5 The Odds of Having Neurodegenerative Diseases Based on Rs10506151 Genotype and T2D |
Discussion
As far as we are aware, this is the first work to study the interactive effect of the intronic polymorphism (rs10506151) of the LRRK2 gene and T2D on NDs using large datasets (TWB and the NHIRD). Results indicated that the risk of NDs was significantly lower among individuals having both diabetes and CA/AA genotype in Taiwan. This work provides new insights even though more analyses are needed to better understand the impact of these findings.
As noted earlier, a published study on LRRK2 found no association between rs10506151 and PD among Norwegians.11 On the contrary, a significant relationship was observed between PD and this variant in Chinese populations.10 Both studies differ from the current research in that 1) they focused mainly on PD, whereas we assessed other forms of NDs. 2) We considered diabetes status and confounders, but they did not, and 3) their sample size was relatively smaller.
In our study, the rs10506151 variant was not significantly related to NDs (OR 1.06 (95% CI 0.75–1.49). These results seem to align with those reported by Toft and colleagues.11 However, when we added the interaction terms (rs10506151 and T2D) in our model, we found that the interaction effect was significant. That is to say, the relationship between NDs and T2D was influenced by the rs1050615 variant. Only those with combined CA/AA genotype who had T2D had the lowest risk of developing NDs.
In an attempt to localize susceptibility genes for alterations in the brain structure associated with stroke and dementia risk, Turner and colleagues20 found that the MRI measures of structural brain injury were heritable in non-Hispanic black and white sibships ascertained through hypertensive sibling pairs. They concluded that the susceptibility loci for brain atrophy, ventricular volume, and leukoaraiosis identified by linkage analyses differed among MRI measures and between races. In Framingham,21 investigators noted that the SNP rs10506151 was associated with frontal and temporal brain volumes. That implies this SNP might be related to NDs.
Associations between T2D and the risk of NDs are yet to be clarified.22,23 Prior studies reported associations between diabetes and NDs including but not limited to PD.12–16 However, a case-control study in Japan found that T2D was inversely related to PD risk.17 In this study, we found that the odds ratio for having NDs was not significant among individuals with T2D.
Certain NDs seem to be influenced by metformin.24 A population-based study in Taiwan showed that metformin was linked to a higher risk for PD in patients with T2D.22 In another observational study, sulfonylurea was associated with a higher risk of PD in Taiwanese patients with T2D.23 Nevertheless, the risk appeared much reduced when combination therapy with metformin was applied. In the current study, we attempted to analyze the effect of metformin use (using ‘no metformin use’ as reference) in those with T2D. We found that the OR for having NDs in metformin users was 1.05 (95% CI 0.55–2.02) as shown in Supplementary Table 1. In Orkaby’s study,25 metformin was associated with a lower risk of subsequent dementia in veterans below the age of 75. Other studies found that metformin may reduce AD risk based on its ability to sensitize neuronal insulin resistance.26,27 According to another study, metformin may be used as a future therapy for NDs.28 However, according to some authors, treatment with metformin might rather stimulate the development of AD.29
One limitation of our study is that we did not focus on a specific disease because the sample size was not adequate. Before performing quality control on samples (n = 19,500), 165 participants were identified with NDs. Of these, 47 had PD, 10 had AD, 9 had other degenerative diseases of the basal ganglia, and 119 had other dementia. Of note, some of these individuals had more than one condition. To benefit from a larger sample size, NDs jointly included PD, other degenerative diseases of the basal ganglia, AD, and other dementia as stated above. Despite the limitation, our results may provide new directions for research in NDs. That is to say, the interactive effect of T2D or its medications and LRRK2 mutation on neurodegenerative conditions is worthy of further investigation.
Conclusion
We found that among the four subpopulations categorized by rs10506151 genotypes and diabetes status, the risk of NDs was significantly lower among individuals having T2D and CA/AA genotype in Taiwan. These preliminary findings expand knowledge about the genetics of NDs and may be relevant to studies investigating disease-modifying therapies for neurodegenerative conditions. Despite these, there is a need for more investigations.
Abbreviations
T2D, type 2 diabetes; OR, odds ratio; CI, 95% confidence interval; BMI, body mass index; LRRK2, Leucine-rich repeat kinase 2; NHIRD, National Health Insurance Research Database; TWB, Taiwan Biobank; PD, Parkinson’s disease; AD, Alzheimer’s disease; SNP, single nucleotide polymorphism.
Funding
This study was supported by the Ministry of Science and Technology (MOST 109-2121-M-040-002). The funder hand no role in the collection, analysis, and interpretation of data and in writing the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wu S, Bekhit AE-DA, Wu Q, et al. Bioactive peptides and gut microbiota: candidates for a novel strategy for reduction and control of neurodegenerative diseases. Trends Food Sci Technol. 2020.
2. Muddapu VR, Dharshini SAP, Chakravarthy VS, Gromiha MM. Neurodegenerative Diseases – is Metabolic Deficiency the Root Cause? Front Neurosci. 2020;14:213. doi:10.3389/fnins.2020.00213
3. Ganguly G, Chakrabarti S, Chatterjee U, Saso L. Proteinopathy, oxidative stress and mitochondrial dysfunction: cross talk in Alzheimer’s disease and Parkinson’s disease. Drug Des Devel Ther. 2017;11:797–810. doi:10.2147/DDDT.S130514
4. Bivona G, Gambino CM, Iacolino G, Ciaccio M. Vitamin D and the nervous system. Neurol Res. 2019;41(9):827–835. doi:10.1080/01616412.2019.1622872
5. Sheikh S, Safia HE, Mir SS. Neurodegenerative Diseases: multifactorial Conformational Diseases and Their Therapeutic Interventions. J Neurodegenerative Dis. 2013;2013:563481. doi:10.1155/2013/563481
6. García J-C, Bustos R-H. The Genetic Diagnosis of Neurodegenerative Diseases and Therapeutic Perspectives. Brain Sci. 2018;8(12):222. doi:10.3390/brainsci8120222
7. Rui Q, Ni H, Li D, Gao R, Chen G. The Role of LRRK2 in Neurodegeneration of Parkinson Disease. Curr Neuropharmacol. 2018;16(9):1348–1357. doi:10.2174/1570159X16666180222165418
8. Chen X, Xie C, Tian W, et al. Parkinson’s disease-related Leucine-rich repeat kinase 2 modulates nuclear morphology and genomic stability in striatal projection neurons during aging. Mol Neurodegener. 2020;15(1):12. doi:10.1186/s13024-020-00360-0
9. Rudenko IN, Kaganovich A, Langston RG, et al. The G2385R risk factor for Parkinson’s disease enhances CHIP-dependent intracellular degradation of LRRK2. Biochem J. 2017;474(9):1547–1558. doi:10.1042/BCJ20160909
10. Skipper L, Li Y, Bonnard C, et al. Comprehensive evaluation of common genetic variation within LRRK2 reveals evidence for association with sporadic Parkinson’s disease. Hum Mol Genet. 2005;14(23):3549–3556. doi:10.1093/hmg/ddi376
11. Toft M, Haugarvoll K, Ross OA, Farrer M, Aasly J. LRRK2 and Parkinson’s disease in Norway. Acta Neurol Scand. 2007;115:72–75. doi:10.1111/j.1600-0404.2007.00852.x
12. Morsi M, Maher A, Aboelmagd O, Johar D, Bernstein L. A shared comparison of diabetes mellitus and neurodegenerative disorders. J Cell Biochem. 2018;119(2):1249–1256. doi:10.1002/jcb.26261
13. Xu Q, Park Y, Huang X, et al. Diabetes and risk of Parkinson’s disease. Diabetes Care. 2011;34(4):910–915. doi:10.2337/dc10-1922
14. Pagano G, Polychronis S, Wilson H, et al. Diabetes mellitus and Parkinson disease. Neurology. 2018;90(19):e1654–e1662. doi:10.1212/WNL.0000000000005475
15. Biosa A, Outeiro TF, Bubacco L, Bisaglia M. Diabetes mellitus as a risk factor for Parkinson’s disease: a molecular point of view. Mol Neurobiol. 2018;55(11):8754–8763. doi:10.1007/s12035-018-1025-9
16. Li Y, Li L, Hölscher C. Incretin-based therapy for type 2 diabetes mellitus is promising for treating neurodegenerative diseases. Rev Neurosci. 2016;27(7):689–711.
17. Miyake Y, Tanaka K, Fukushima W, et al. Case–control study of risk of Parkinson’s disease in relation to hypertension, hypercholesterolemia, and diabetes in Japan. J Neurol Sci. 2010;293(1–2):82–86. doi:10.1016/j.jns.2010.03.002
18. Zhu Y, Ding X, She Z, et al. Exploring shared pathogenesis of Alzheimer’s disease and type 2 diabetes mellitus via co-expression networks analysis. Curr Alzheimer Res. 2020;17(6):566–575. doi:10.2174/1567205017666200810164932
19. Lin W-Y, Liu Y-L, Yang AC, Tsai S-J, Kuo P-H. Active cigarette smoking is associated with an exacerbation of genetic susceptibility to diabetes. Diabetes. 2020;69(12):2819–2829. doi:10.2337/db20-0156
20. Turner ST, Fornage M, Jack CR, et al. Genomic susceptibility loci for brain atrophy, ventricular volume, and leukoaraiosis in hypertensive sibships. Arch Neurol. 2009;66(7):847–857. doi:10.1001/archneurol.2009.110
21. Seshadri S, DeStefano AL, Au R, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(S1):S15. doi:10.1186/1471-2350-8-S1-S15
22. Kuan Y-C, Huang K-W, Lin C-L, Hu C-J, Kao C-H. Effects of metformin exposure on neurodegenerative diseases in elderly patients with type 2 diabetes mellitus. Prog Neuropsychopharmacol Biol Psychiatry. 2017;79:77–83. doi:10.1016/j.pnpbp.2017.06.002
23. Wahlqvist ML, Lee M-S, Hsu -C-C, Chuang S-Y, Lee J-T, Tsai H-N. Metformin-inclusive sulfonylurea therapy reduces the risk of Parkinson’s disease occurring with Type 2 diabetes in a Taiwanese population cohort. Parkinsonism Relat Disord. 2012;18(6):753–758. doi:10.1016/j.parkreldis.2012.03.010
24. Rotermund C, Machetanz G, Fitzgerald JC. The Therapeutic Potential of Metformin in Neurodegenerative Diseases. Front Endocrinol (Lausanne). 2018;9:400.
25. Orkaby AR, Cho K, Cormack J, Gagnon DR, Driver JA. Metformin vs sulfonylurea use and risk of dementia in US veterans aged≥ 65 years with diabetes. Neurology. 2017;89(18):1877–1885. doi:10.1212/WNL.0000000000004586
26. Asadbegi M, Yaghmaei P, Salehi I, Ebrahim-Habibi A, Komaki A. Neuroprotective effects of metformin against Aβ-mediated inhibition of long-term potentiation in rats fed a high-fat diet. Brain Res Bull. 2016;121:178–185. doi:10.1016/j.brainresbull.2016.02.005
27. Gupta A, Bisht B, Dey CS. Peripheral insulin-sensitizer drug metformin ameliorates neuronal insulin resistance and Alzheimer’s-like changes. Neuropharmacology. 2011;60(6):910–920. doi:10.1016/j.neuropharm.2011.01.033
28. Markowicz-Piasecka M, Sikora J, Szydłowska A, Skupień A, Mikiciuk-Olasik E, Huttunen KM. Metformin–a future therapy for neurodegenerative diseases. Pharm Res. 2017;34(12):2614–2627. doi:10.1007/s11095-017-2199-y
29. Imfeld P, Bodmer M, Jick SS, Meier CR. Metformin, other antidiabetic drugs, and risk of Alzheimer’s disease: a population‐based case–control study. J Am Geriatr Soc. 2012;60(5):916–921.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
