Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 9
Initial assessment of the benefits of implementing pharmacogenetics into the medical management of patients in a long-term care facility
Authors Saldivar J, Taylor D, Sugarman E, Cullors A, Garces J, Oades K, Centeno J
Received 31 July 2015
Accepted for publication 24 November 2015
Published 19 January 2016 Volume 2016:9 Pages 1—6
DOI https://doi.org/10.2147/PGPM.S93480
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Video abstract presented by Juan-Sebastian Saldivar
Views: 1372
Juan-Sebastian Saldivar, David Taylor, Elaine A Sugarman, Ali Cullors, Jorge A Garces, Kahuku Oades, Joel Centeno
AltheaDx, San Diego, CA, USA
Abstract: The health care costs associated with prescription drugs are enormous, particularly in patients with polypharmacy (taking more than five prescription medications), and they continue to grow annually. The evolution of pharmacogenetics has provided clinicians with a valuable tool that allows for a smarter, more fine-tuned approach to treating patients for a number of clinical conditions. Applying a pharmacogenetics approach to the medical management of patients can provide a significant improvement to their care, result in cost savings by reducing the use of ineffective drugs, and decrease overall health care utilization. AltheaDx has begun a study to look at the benefits associated with incorporating pharmacogenetics into the medical management of patients who are on five or more medications. Applying pharmacogenetic guided PharmD recommendations across this patient population resulted in the elimination and/or replacement of one to three drugs, for 50% of the polypharmacy patient population tested, and an estimated US$621 in annual savings per patient. The initial assessment of this study shows that there is a clear opportunity for concrete health care savings solely from prescription drug management when incorporating pharmacogenetic testing.
Keywords: personalized medicine, pharmacogenetics, pharmacokinetics, pharmacodynamics, adverse drug reaction, polypharmacy
Introduction
Since the mapping of the human genome roughly 12 years ago, many new discoveries and clinical applications have been introduced into health care. Initially, these consisted primarily of confirming a clinical diagnosis for a genetic condition or syndrome, such as cystic fibrosis, hereditary non-polyposis colorectal cancer, or Li–Fraumeni syndrome, where little could be done other than family counseling/testing, increased monitoring, and symptomatic management. Molecular diagnostics then evolved into a tool that could be used for therapeutic decision making in the treatment of specific disease states, such as KRAS for colorectal cancer, EGFR for lung cancer, Her2/Neu and BRAF for breast cancer and melanoma, respectively. In parallel, research has yielded a better understanding of how humans metabolize and respond to common medications used to treat more widespread conditions including cardiovascular disease, depression, and chronic pain. AltheaDx is a clinical pharmacogenetics (PGx) laboratory that has consolidated current knowledge of genetics as it relates to the pharmacokinetics (PK), or how the body alters a drug once administered, and pharmacodynamics (PD), which is how a drug impacts the body, of many commonly prescribed drugs currently marketed in the US. The IDgenetix product uses a proprietary algorithm to interpret genetic findings obtained from the analysis of a patient’s DNA collected using a cheek swab to predict the most favorable drug response for that individual. IDgenetix considers these genetic findings within the context of other factors (ie, environmental and dietary factors, existing drug regimen, herbal and over-the-counter medications) with the goal of not only selecting the most appropriate and effective therapy for patients, but also to look for opportunities to eliminate unnecessary medications and dangerous adverse events.
The US health care system spent approximately US$300 billion dollars on prescription medications in 2014 and estimates an incremental 6.3% spend annually over the next decade.1 This economic problem is exacerbated by a growing and aging population accompanied by relatively disjointed prescribing behaviors and poor patient monitoring. Patients often receive prescriptions from multiple providers with inadequate measures toward medication reconciliation. Polypharmacy has become a huge problem, with approximately 40% of the elderly and nearly 11% of the US population now taking five or more prescription drugs at any one time.2,3 The US Food and Drug Administration (FDA) reports spending US$136 billion annually on adverse drug reactions (ADRs).4 That means for every dollar that is spent on prescription drugs, an additional US$0.50 is spent on ADRs. The pharmacogenetic information that is currently available to guide therapy selection and adjust dose requirements can significantly enhance the utility of prescription drugs and provides an opportunity for improving patient care. Additionally, this new technology can lead to a decrease in the number of ADRs and the enormous costs associated with these iatrogenic hospitalizations and emergency room visits.
To explore the impact of the IDgenetix pharmacogenetic testing service on health care costs, therapy selection, and patient outcomes, we began a randomized control study with a large managed care entity focused on improving patient care and reducing pharmaceutical costs in their long-term care settings. Herein, we provide a summary of our initial assessment in this study, which looks at possible polypharmacy reductions and medication cost savings suggested by PGx guided pharmacist regimen reviews.
Materials and methods
Aspire Institutional Review Board-approved clinical research protocol was designed and implemented at several long-term care facilities in Phoenix, Arizona. Male and female patients over the age of 45 experiencing polypharmacy, defined as taking five or more prescription medications per month, were considered for participation. Individuals unwilling or unable to provide informed consent were excluded, as were patients with chronic renal dysfunction, abnormal hepatic function, or a history of malabsorption syndrome. Once written informed patient consent was obtained, a buccal swab sample was obtained and tested. All samples that had passing results were randomized to either a control group, where the results were not provided to the clinician nor the patient, or to the experimental group where the information was provided to determine opportunities to improve the medical management of the patient. While the long-term goal of this study is to measure the difference in patient outcomes, hospital and emergency room usage, and patient and clinician satisfaction, the initial step for the implementation of PGx is to apply the information from the IDgenetix assay to guide clinician recommendations on medication changes, and assess the impact this has on the number of prescription medications being used, as well as the associated costs.
Buccal epithelial samples were collected using cheek swabs at clinical sites and shipped to the lab at ambient temperature. Genomic DNA was extracted using the QIAGEN DSP DNA Midi Kit (Qiagen, Valencia, CA, USA). Extracted genomic DNA concentrations were quantified on a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). After extraction, two platforms were used to characterize pharmacogenomic targets in 17 genes, including CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4/CYP3A5, HTR2A, HTR2C, SLC6A4, SLC6A2, COMT, OPRM1, SLCO1B1, VKORC1, MTHFR, F2, and F5. Methods used for detection were: TaqMan OpenArray Genotyping and CYP2D6 copy number variation determination.5,6
A PGx-assisted assessment was performed on 132 patients by a licensed PharmD, looking for opportunities to improve the medical management of these individuals. The interventions recommended via the IDgenetix tool and standard methods were grouped into three basic categories: 1) a replacement of one medication for another deemed better suited to the patient’s genetic profile, that is, medications negatively impacted by certain genetic mutations were replaced with those metabolized via different pathways or those employing a mechanism of action in which all available evidence indicated little or no effect from the genetic mutations identified; 2) discontinuation of a medication that was determined to be ineffective; and 3) consolidation of two medications into a single prescription. All three recommendations focused on maximizing the safety and efficacy profiles of each individual drug regimen evaluated with an emphasis on chronic medical conditions. A cost analysis associated with these changes was estimated by two models. The first took published average wholesale prices published by Centers for Medicare and Medicaid Services and applied a 15% government discount as estimated for the state of Arizona (Model 1). Assuming that some facilities may negotiate additional discounts, a second, more conservative model (Model 2) was used which estimated a 25% discount from the average wholesale prices.
Results
Metabolizer phenotype frequencies and non-CYP gene allele frequencies were determined for this cohort, and our findings mirror those that have been estimated by others in the field.7 Applying the IDgenetix guided PharmD recommendations across this population would result in the elimination and/or replacement of one to three drugs in about 50% of the polypharmacy patient population tested.
Recommendations were primarily based on gene-drug interactions, with 27.6% comprised of CYP-related gene results (drug metabolism or PK), and 25.0% consisting of non-CYP genetic findings (drug mode of action or PD). Drug-drug interactions accounted for 22.4% of the findings (Figure 1). Among the drug-drug interactions, IDgenetix provided additional data to evaluate the risk of such interactions. For example, clopidogrel’s FDA drug label warns against co-administration with omeprazole because of its capacity to competitively inhibit conversion of clopidogrel to its active thiol metabolite by CYP2C19. This precaution takes on greater significance for CYP2C19 intermediate metabolizers who are more susceptible to this inhibition interaction as a consequence of both lower intrinsic metabolic activity and the resulting greater exposure to omeprazole.
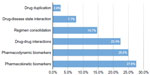  | Figure 1 Basis for therapeutic modification. |
Using standard cost estimates (Model 1), the recommended drug regimen modifications would lead to annualized savings of US$81,972 for the study group, which equates to US$621 in annual savings per patient across the entire population analyzed. When we extrapolate these savings over a 2.5- or 3-year period to account for the average length of stay for these long-term care patients, we estimate a per patient savings of US$1,553 and US$1,863, respectively (Figure 2). The potential savings over the longer time period outweigh the cost of testing by more than two-fold. Figure 2 also breaks down the savings based on drug reduction and replacement/consolidation strategies. Cost savings were broadly analyzed by medical specialty. Psychotropic drug changes accounted for the largest savings by drug class, accounting for 61.3% of total savings. Neurology, cardiovascular, and urology medication changes accounted for 11.0%, 10.5%, and 8.1% of all savings respectively (Figure 3).
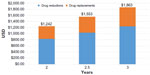  | Figure 2 Average savings by category over 2–3 years. |
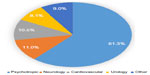  | Figure 3 Savings by drug class. |
The following are representative examples of how PGx was applied:
- Case 1 – AM, a 76 year-old male currently taking: famotidine, gabapentin, levothyroxine, metoclopramide, mirtazapine, morphine, omeprazole, promethazine, sertraline, and tamsulosin. Genetic analysis revealed a CYP2C19 ultra metabolizer phenotype (*1/*17) and an HTR2A diplotype (rs6311 G/G) associated with increased susceptibility to selective serotonin reuptake inhibitor (SSRI)-induced side effects. Increased metabolism of sertraline, a CYP2C19 substrate, suggests standard doses may be sub-therapeutic while the HTR2A variant suggests poor dose escalation tolerability. Given sertraline’s suspect contribution to combination antidepressant therapy with mirtazapine, an attempt at gradual discontinuation seems reasonable. Paring this agent from the regimen would save an estimated US$907 annually.
- Case 2 – WB, a 62 year-old female currently prescribed the following medications: clonidine, duloxetine, hydralazine, insulin glargine, metoprolol, pentoxifylline, quetiapine, and simvastatin. Genetic analysis identified a CYP1A2 ultra metabolizer phenotype (*1/*1F), CYP2D6 intermediate metabolizer phenotype (*1/*5), and an HTR2C diplotype (rs6318 C/C) associated with increased susceptibility to second generation antipsychotic-induced weight gain. Duloxetine, primarily a CYP1A2 substrate and CYP2D6 inhibitor, might be sub-therapeutic at standard doses due to increased metabolism while dose escalation increases drug interaction risk through further reduction of the patient’s already impaired CYP2D6 function. The patient’s vulnerability to second generation antipsychotic cardiometabolic adversity and the potential for exacerbation of comorbid type 2 diabetes suggest an alternative to quetiapine less likely to cause weight gain would be advisable. Venlafaxine offers a therapeutically equivalent alternative to duloxetine that would preserve the patient’s CYP2D6 function and ziprasidone could be considered as a replacement for quetiapine with lower weight gain liability. Implementing these regimen improvements would be accompanied by an estimated US$4,211 reduction in annual prescription costs.
- Case 3 – WF, a 75 year-old male with the following drug regimen: aspirin/dipyridamole, albuterol, amlodipine, baclofen, clonidine, insulin detemir, topical lidocaine patch, lisinopril, nystatin powder, pravastatin, promethazine, sertraline, tamsulosin, and trazodone. Genetic analysis demonstrated an HTR2A diplotype (rs6311 G/G) predisposing increased susceptibility to SSRI-induced side effects. This finding considered with the increased bleeding risk conferred by concomitant SSRI and antiplatelet therapy suggests a non-SSRI alternative to sertraline might be preferable. Mirtazapine presents such an alternative that not only avoids sertraline’s antiplatelet effects and heightened bleeding risk, but offers sedative and antiemetic properties possibly allowing dose reduction or discontinuation of trazodone and promethazine. Although not genetically motivated, replacement of amlodipine and lisinopril as separate agents with the therapeutically equivalent combination product containing amlodipine and benazepril could further reduce pill burden. Assuming discontinuation of trazodone and promethazine on replacement of sertraline with mirtazapine and the combination product substitution, estimated annual drug cost savings reach US$2,868.
Discussion
The use of pharmaceuticals is predicted to grow steadily over the next decade.1 With this increase comes the unwelcomed consequence of more ADRs. While some of the proliferation in prescription drug use can be attributed to our expanding elderly population, we have also seen prescription drug increases in all age groups over the past 15 years.8 Polypharmacy is more prevalent in the elderly population given the increased likelihood of having multiple co-morbidities, and the often fragmented care as a result of multiple specialists making independent prescription recommendations. A more rational approach is needed to provide better care for these complex patients, and to control overall health care costs.
Study outcomes related to polypharmacy have demonstrated the benefits of taking fewer prescription medications. Sganga et al,9 classified 480 elderly patients into a non-polypharmacy group (taking <8 drugs,) and a polypharmacy group (taking ≥8 drugs). They found that 26.9% of the non-polypharmacy group, compared to 39.1% of the polypharmacy group were re-hospitalized over a 1-year period (P=0.004). In another study, Rungaga et al,10 looked at 351 patients who were discharged from a transition care program, and found the non-polypharmacy group (taking <5 drugs) had improved outcomes in activities of daily living and fewer falls.
Other studies have examined the clinical impact of deprescribing interventions in the elderly. Garfinkel and Mangin11 applied a deprescribing algorithm based on clinical evidence to a group of 70 community-dwelling elderly patients. Over a 40-month period, only 2% of the discontinued medications were restarted due to a recurrence of symptoms, and no adverse events or deaths were associated with the discontinued medications. Additionally, 88% of the patients reported an improvement in overall health. A similar deprescribing study by Garfinkel et al12 in 119 elderly inpatients showed that the 1-year mortality rate for the control group was 45% versus 21% in the experimental group (P<0.001), and the referral rate to acute care facilities was 30% versus 11.8%, respectively (P<0.002).
Medicare has also recognized the dangers of prescription drug use in the. One of its “star ratings” criteria for measuring the quality and safety of health/drug plans aims at reducing specific high risk drugs in the elderly, as determined by the American Geriatrics Society, known as the Beers Criteria.13 While reducing high risk medication use in the elderly makes a lot of sense, it also makes sense to avoid unnecessary drug use in this population. Deprescribing medications is challenging because clinicians often do not know which medications are helping and which are ineffective. PK and PD research has established significant gene-drug associations, and we now know that genetic factors account for 20%–40% of differences in individual drug metabolism and response.14 Combining this information with the existing evidence based practices gives us an unprecedented opportunity to impact the growing problem of polypharmacy.
In this cohort, patients were taking, on average, 12 prescription medications. Approximately half were found to have an actionable finding based on a PharmD assessment incorporating pharmacogenetic information, resulting in a reduction in the overall medication costs for treating these individuals. The findings were based on either PK (CYP gene), PD (non-CYP gene), or drug-drug interactions. All three categories had a similar impact (Figure 1), indicating that the analysis of any of these factors in isolation is inadequate and does not provide a comprehensive assessment of the individual. Additionally, more than 50% of the therapeutic modifications were based on PK and PD associations, which can only be determined through pharmacogenetic testing.
The pharmacogenetic results from this study influenced regimen changes in a variety of categories including cardiovascular, neurological, and gastrointestinal. However, the majority of the recommendations impacted psychotropics. This could be due to an over-representation of psychiatric co-morbidities in this cohort, but may also reflect the broader knowledge and understanding of the PK and PD associations within the field of psychiatry. Recent studies have demonstrated the clinical utility of incorporating PGx into the treatment of depression and anxiety, as well as the associated cost benefits.15–18 Our findings support the notion that this is an area with significant potential for improvement and cost savings.
The three clinical cases described here demonstrate how pharmacogenetic information was applied in a clinical setting. Given the variability of patient genotypes as well as individual morbidities and drug regimens, it is not possible to provide an example of every scenario that was encountered. Each case is unique and requires a coordinated effort by the clinical care team. Clinicians will have to bring this knowledge into their armamentarium, but it will require the adoption and participation of the entire health care team for successful implementation. Ideally, this type of information will be housed in the electronic health record where contraindicated medications can be flagged in real time. This is especially true for larger hospital settings.
While the clinical acumen of the PharmD providing the recommended changes can introduce some variability, a detailed breakdown of the source for the recommended changes found that about 20% of the proposals could be made purely based on PharmD training, and 80% were made based on IDgenetix test results. This indicates that the large majority of the recommendations are based on objective data. A group of experts from ~80 academic and commercial institutions, called the Clinical Pharmacogenetics Implementation Consortium, with the support of the National Institute of Health, have developed standardized guidelines for the interpretation of pharmacogenetic results on approximately 45 gene-drug associations. This will help provide a more consistent approach to PGx implementation.
Conclusion
Our first snapshot from this randomized controlled trial demonstrates a clear opportunity for concrete health care savings by incorporating pharmacogenetic information into the medication management of patients with polypharmacy. Using a cost-benefit model, on average, patients recognized an annual savings of US$621. Extrapolating over the average length of a nursing home stay of 2–3 years equates to an overall saving ranging from ~US$1,240–US$1,860. At a test cost of roughly US$800, even if only two thirds of the recommendations were implemented, the test would still have a cost benefit over the 2–3 year period. Even using the more conservative cost-benefit model (Model 2), the 2–3 year savings would range from ~US$1,000–US$1,500. In order to provide a more comprehensive economic evaluation of this tool, follow-up studies and clinical outcome observations are needed to confirm the sustainability of the pharmaceutical recommendations discussed here and to explore the potential cost-effectiveness of PGx testing. However, based on previous studies,9–12,18 we would anticipate additional cost savings and benefits from improved patient outcomes and other measures including hospitalizations, adverse drug events, length of stay, cognitive and overall health assessments, patient satisfaction with level of care, physical activity including activities of daily living, fall frequency, and pain management.
The burden on our health care system continues to grow as our population continues to age and live longer. The enormous amount of resources spent on prescription drugs and the adverse events associated with their overutilization also continues to expand. Most clinicians continue to use a “one size fits all” approach to treating their patients. Additionally, the elderly population typically have multiple co-morbidities, requiring multiple physician specialists being involved in their care, but often executed in isolation from one another. We now have advanced genetic tools that provide insight into the individualized drug response for each patient, giving us a better roadmap on how to best treat on a personalized level. This approach increases the likelihood of reaching a therapeutic goal, improves patient adherence to therapy, and decreases the risk for an ADR. All of this translates into decreased health care costs, and it is in the best interest of our patients and the health care system to seek opportunities to administer care in a safer, more efficient and cost-effective manner.
Disclosure
The authors are full time employees of AltheaDx, San Diego, CA, USA. The authors have no other conflicts of interest to disclose in this work.
References
Centers for Medicare and Medicaid Services. Available from: https://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/NationalHealthExpendData/Downloads/proj2014.pdf. CMS; 2013. Accessed November 27, 2015. | |
Gurwitz JH. Polypharmacy – a new paradigm for quality drug therapy in the elderly? Arch Intern Med. 2004;164(18):1957–1959. | |
Centers for Disease Control and Prevention and National Center for Health Statistics. Prescription Drug Use Continues to Increase: US Prescription Drug Data for 2007–2008. CDC; 2010. Available from: http://www.cdc.gov/nchs/data/databriefs/db42.pdf. Accessed November 27, 2015. | |
US Food and Drug Administration [homepage on the Internet. Costs associated with ADRs. Available from: http://www.fda.gov/NewsEvents/Testimony/ucm114949.htm. Accessed November 27, 2015. | |
Lesch KP, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–1531. | |
Pratt VM, Zehnbauer B, Wilson JA, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J Mol Diagn. 2010;12(6):835–846. | |
The Pharmacogenomics Knowledgebase [homepage on the Internet]. Available from: https://www.pharmgkb.org/index.jsp. Accessed November 27, 2015. | |
Centers for Disease Control and Prevention [homepage on the Internet]. Available from: http://www.cdc.gov/nchs/data/hus/hus14.pdf#085. Accessed November 27, 2015. | |
Sganga F, Landi F, Ruggiero C, et al. Polypharmacy and health outcomes among older adults discharged from hospital: Results from the CRIME study. Geratr Gerontol Int. 2015;15(2):141–146. | |
Runganga M, Peel NM, Hubbard RE, et al. Multiple medication use in older patients in post-acute transitional care: a prospective cohort study. Clin Interv Aging. 2014;9:1453–1462. | |
Garfinkel D, Mangin D. Feasibility study of systematic approach for discontinuation of multiple medications in older adults: addressing polypharmacy. Arch Intern Med. 2010;170(18):1648–1654. | |
Garfinkel D, Zur-Gil S, Ben-Israel J. The war against polypharmacy: a new cost-effective geriatric-palliative approach for improving drug therapy in disabled elderly people. Isr Med Assoc J. 2007;9(6):430–434. | |
The American Geriatrics Society [homepage on the Internet]. Available from: http://www.americangeriatrics.org/press/id:5907. Accessed November 27, 2015. | |
Ventola CL. Role of pharmacogenomic biomarkers in predicting and improving drug response: part 1: the clinical significance of pharmacogenetic variants. P T. 2013;38(9):545–560. | |
Hall-Flavin DK, Winner JG, Allen JD, et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Trans Psychiatry. 2012;2:e172. | |
Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo BM. A Prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219–227. | |
Hall-Flavin DK, Winner JG, Allen JD, et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23(10):535–548. | |
Winner J, Allen JD, Altar CA, Spahic-Mihajlovic A. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3:e242. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
