Back to Archived Journals » Hypoxia » Volume 3
Induction of long noncoding RNA MALAT1 in hypoxic mice
Authors Lelli A, Nolan K, Santambrogio S, Gonçalves AF, Schönenberger M, Guinot A, Frew I, Marti H, Hoogewijs D, Wenger R
Received 16 June 2015
Accepted for publication 19 August 2015
Published 8 October 2015 Volume 2015:3 Pages 45—52
DOI https://doi.org/10.2147/HP.S90555
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Dörthe Katschinski
Aurelia Lelli,1,2,* Karen A Nolan,1,2,* Sara Santambrogio,1,2 Ana Filipa Gonçalves,1,2 Miriam J Schönenberger,1,2 Anna Guinot,1,2 Ian J Frew,1,2 Hugo H Marti,3 David Hoogewijs,1,2,4 Roland H Wenger1,2
1Institute of Physiology and Zurich Center for Human Physiology (ZIHP), University of Zurich, Zurich, Switzerland; 2National Center of Competence in Research "Kidney.CH", Zurich, Switzerland; 3Institute of Physiology and Pathophysiology, University of Heidelberg, Heidelberg, Germany; 4Institute of Physiology, University of Duisburg-Essen, Essen, Germany
*These authors contributed equally to this work
Abstract: Long thought to be “junk DNA”, in recent years it has become clear that a substantial fraction of intergenic genomic DNA is actually transcribed, forming long noncoding RNA (lncRNA). Like mRNA, lncRNA can also be spliced, capped, and polyadenylated, affecting a multitude of biological processes. While the molecular mechanisms underlying the function of lncRNAs have just begun to be elucidated, the conditional regulation of lncRNAs remains largely unexplored. In genome-wide studies our group and others recently found hypoxic transcriptional induction of a subset of lncRNAs, whereof nuclear-enriched abundant/autosomal transcript 1 (NEAT1) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) appear to be the lncRNAs most ubiquitously and most strongly induced by hypoxia in cultured cells. Hypoxia-inducible factor (HIF)-2 rather than HIF-1 seems to be the preferred transcriptional activator of these lncRNAs. For the first time, we also found strong induction primarily of MALAT1 in organs of mice exposed to inspiratory hypoxia. Most abundant hypoxic levels of MALAT1 lncRNA were found in kidney and testis. In situ hybridization revealed that the hypoxic induction in the kidney was confined to proximal rather than distal tubular epithelial cells. Direct oxygen-dependent regulation of MALAT1 lncRNA was confirmed using isolated primary kidney epithelial cells. In summary, high expression levels and acute, profound hypoxic induction of MALAT1 suggest a hitherto unrecognized role of this lncRNA in renal proximal tubular function.
Keywords: hypoxia-inducible factor, kidney, oxygen, proximal tubule, testis
Introduction
One of the most amazing outcomes of the high-throughput sequencing era is the discovery that approximately 75% of the genome is transcribed and that there might be approximately as many noncoding (nc) RNAs as protein-coding messenger RNAs (mRNAs). While most of our knowledge focuses on small ncRNA functions, much less is known about long noncoding RNA (lncRNA; >200 nucleotides). Several recent reports have shown that lncRNAs are involved in a wide variety of physiological or pathological processes including X-chromosome inactivation, stem cell specification, neurodegenerative disorders, and cancer.1,2 Mechanistically, lncRNAs located at regulatory DNA cis-elements can recruit RNA-binding proteins to trigger gene expression by transcriptional and/or epigenetic gene regulation.3–5 While many of these studies documented a transcriptional dysregulation of lncRNAs in pathological states, the actual mechanisms of conditional regulation of lncRNA expression under physiological conditions have scarcely been investigated.
Conditional regulation of mRNA expression is a major principle in the adaptation of cells and organisms to environmental changes. Conditional regulation of lncRNAs in cancer cell lines suggests that this class of RNA also plays a (largely unknown) role in the adaptation to microenvironmental changes such as tumor hypoxia. However, as it is unlikely that this mechanism evolved in cancer cells, we set out to analyze lncRNA induction also in normal tissues of animals exposed to inspiratory hypoxia.
Hypoxia represents a major microenvironmental condition that profoundly impacts upon the regulation of several hundred protein-coding genes involved in the adaptation to limited oxygen supply. Hypoxia-inducible factor (HIF) is a heterodimeric DNA-binding protein usually composed of a constitutively expressed β subunit and either a HIF-1α or a HIF-2α subunit whose stability and activity are regulated by oxygen-dependent protein hydroxylation.6,7 HIF-1α and HIF-2α show distinct expression kinetics with HIF-1α protein levels decreasing under prolonged hypoxia, while HIF-2α levels are increasing, suggesting a HIFα isoform-specific kinetics of target gene expression.8–10
During a microarray study using a prostate cancer cell line, we previously found that among 608 genes that were induced by at least twofold in hypoxic conditions, more than 20 encoded lncRNAs.11 These data were confirmed by a recent genome-wide study which focused on the hypoxia-inducible expression of ncRNA species in a breast cancer cell line.12 This study systematically demonstrated for the first time that not only the transcription of mRNA but also the transcription of virtually all RNA classes, including lncRNA, miRNA, piwiRNA, snRNA, and tRNA is conditionally regulated by hypoxia in cultured cells.
Based on their abundant and widespread expression, we focused on two highly conserved, mostly nuclear lncRNAs that were robustly induced by hypoxia: NEAT1 (nuclear- enriched abundant/autosomal transcript 1; also termed MENµ/β) and MALAT1 (metastasis-associated lung adenocarcinoma transcript 1; also termed NEAT2). NEAT1 is a 4 kb polyadenylated, unspliced lncRNA known to associate with proteins that demark paraspeckles (nuclear domains involved in mRNA retention) and to play a central role in paraspeckle formation.13 MALAT1 is a >6.5 kb lncRNA that has been shown to regulate RNA-splicing factors in nuclear speckles that serve the assembly, modification, and storage of splice factors.14 However, at least in one report, MALAT1 has been shown to translocate to the cytoplasm during the G2/M phase, which was associated with the cytoplasmic translocation of hnRNP C required for cell cycle progression.15
Materials and methods
Cell culture and RNA interference
Human Hep3B hepatoma, HeLa cervical carcinoma, and MCF-7 breast cancer cells were cultured in high-glucose Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St Louis, MO, USA). The authors advise that their study did not require ethics approval from the Cantonal Ethics Commission Zurich, as only established, commercially available cancer cell lines were used. Primary mouse kidney epithelial cells were isolated and cultured as reported previously.16 Hypoxic experiments were performed for 24 hours at 0.2% oxygen and 5% CO2 in a gas-controlled glove box (InvivO2 400; Ruskinn Technology Ltd, Bridgend, UK) as described previously.17 Stable knockdown MCF-7 cell lines were established by transduction with lentiviral particles containing short-hairpin RNA expression constructs followed by selection with puromycin as described before.10
Animal experimentation
Animal housing, hypoxic exposure (inspiratory O2 concentration of 8% for 0–108 hours), and organ excision for RNA isolation of male C57BL/6 mice have been described previously.18 For in situ hybridization experiments, male C57BL/6 mice were exposed to 8% oxygen for 24 hours in a hypoxia tent (Coy Laboratory Products, Grass Lake, MI, USA). Control animals were maintained at ambient oxygen concentration. Animal experiments were conducted at the Zurich Integrative Rodent Physiology (ZIRP) facility with the appropriate consent from the Veterinary Office of the Canton Zurich (license number 126/13) following consent by the Animal Experimentation Commission of the Canton Zurich which ensures that the guidelines of the Animal Welfare Act will be followed.
lncRNA quantification
Total cellular RNA was extracted as previously described.19 Total RNA (2 μg) was reverse transcribed (RT) using AffinityScript reverse transcriptase (Agilent, Santa Clara, CA, USA) and the cDNA levels were estimated by quantitative polymerase chain reaction (qPCR) using a SybrGreen qPCR reagent kit (Sigma-Aldrich) in a MX3000P light cycler (Agilent). Transcript levels were calculated by comparison with a calibrated standard and expressed as ratios relative to ribosomal protein L28 or β-actin mRNA levels for MCF-7 and mouse tissues, respectively.
In situ hybridization
Excised organs were embedded in Tissue Tek O.C.T. (Miles Scientific, Naperville, IL, USA), frozen in 2-methylbutane in liquid nitrogen and stored at −80°C before 20 μm cryosections were prepared and mounted on SuperFrost Plus slides (ThermoScientific, Waltham, MA, USA). In situ hybridization was performed according to standard protocols essentially as described previously.20 Briefly, following rehydration in phosphate-buffered saline and acetylation, the slides were prehybridized at room temperature for several hours and then incubated with digoxigenin-labeled cRNA probes (200 ng in 200 μL hybridization buffer) at 60°C overnight. Following sequential washing steps in 5× saline sodium citrate (SSC), 0.2× SSC (20× SSC is 3 M NaCl, 0.3 M Na-citrate, pH 7.0), and Tris-buffered saline (TBS), the slides were blocked with 10% fetal calf serum in TBS for 2 hours and incubated with alkaline phosphatase-labeled anti-digoxigenin antibodies (3.75 U/mL; Roche, Basel, Switzerland) in TBS at 4°C overnight. After washing in TBS, bound antibodies were detected by nitro-blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate staining, and mounted in Mowiol-DABCO medium. For the preparation of a hybridization probe, a 597 nt fragment of MALAT1 was amplified by reverse transcription PCR (RT-PCR) using RNA derived from mouse liver and the primers forward-5′-taagtctcgagggattgggaagccctagttc-3′ and reverse-5′-acttaaagcttcacttgtggggagaccttgt-3′. Following subcloning into pBluescriptKS+, digoxigenin-labeled cRNA sense and antisense probes were synthesized using T3 and T7 RNA polymerases (ThermoScientific).
Results
Hypoxia-inducible expression of NEAT1 and MALAT1 lncRNAs in cultured cells in vitro
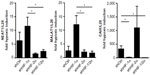
During a microarray study using PC3 prostate carcinoma cells, we detected a previously unrecognized strong oxygen-dependent regulation of lncRNAs, including NEAT1 and MALAT1.11 As shown in Figure 1, robust, albeit somewhat variable, oxygen-dependent regulation of NEAT1 and MALAT1 lncRNA levels was confirmed by RT-qPCR (quantitative RT-PCR) in a number of additional cancer cell lines, including Hep3B hepatocellular carcinoma, HeLa cervical carcinoma, and MCF-7 breast carcinoma, following 24 hours of exposure to 0.2% oxygen.
Preferential HIF-2α isoform-specific regulation of NEAT1 and MALAT1 lncRNAs
Stable shHIF-1α and/or shHIF-2α MCF-7 knockdown breast cancer cell lines were used to assess the role of the HIFα isoforms in NEAT1 and MALAT1 regulation. As shown in Figure 2, both NEAT1 and MALAT1 showed an increased hypoxic induction following knockdown of HIF-1α. Because we previously established that in these (and other) cell lines knockdown of either HIFα isoform leads to an increase in the other isoform,10,21 this result points to a preferential role of HIF-2α in the induction of NEAT1 and MALAT1 gene expression. Indeed, compared with shHIF-1α, shHIF-2α as well as shHIF-1/2α MCF-7 cells showed a significant attenuation of the hypoxic induction of both NEAT1 and MALAT1 lncRNAs. By contrast, the classical HIF-1 target gene carbonic anhydrase 9 (CAIX) displayed an exclusive dependency on HIF-1α in the same experiment (Figure 2).
Hypoxic induction of NEAT1 and MALAT1 lncRNAs in mouse organs in vivo
To analyze the oxygen-dependent regulation of lncRNAs in vivo, groups of three mice were exposed to 8% O2 inspiratory normobaric hypoxia for 0–108 hours. Eight percent of O2 has been chosen as a compromise between maximizing the hypoxic response and minimizing the animal burden. Whole organ levels of NEAT1 and MALAT1 lncRNAs were determined by RT-qPCR and normalized to β-actin mRNA levels which did not show any hypoxic alteration. As illustrated in Figure 3, in comparison with β-actin mRNA, NEAT1 (in heart and kidney) and especially MALAT1 lncRNA showed considerably high basal expression levels and a rather high degree of interindividual variability. While NEAT1 lncRNA levels were only slightly induced by chronic (108 vs 0 hours) inspiratory hypoxia in heart, lung, and brain, MALAT1 was induced in most organs measured, with particularly high induction rates in spleen, kidney, testis, lung, and brain. Of note, mainly in kidney, testis, and lung, a marked induction of MALAT1 lncRNA was already detected following acute inspiratory hypoxia of only 4 hours. Because such fast and strong hypoxic changes in expression levels are rather unusual in tissues in vivo, these results suggest a physiological role of MALAT1 in the systemic response to hypoxia.
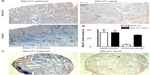
Hypoxic induction of MALAT1 in proximal tubules of the kidney
Because the highest MALAT1 lncRNA levels, relative to β-actin mRNA levels, were found in kidney and testis, we focused on these two organs for further analysis by in situ hybridization. As shown in Figure 4A, MALAT1 was mainly detected in renal tubular epithelial cells. Hypoxic exposure (8% O2 for 24 hours) significantly increased both signal intensity and the number of positive cells. Quantification of the number of MALAT1 positive tubules revealed that proximal but not distal tubules account for the hypoxic induction of MALAT1 lncRNA expression (Figure 4B). In testis, the most outer layer of the seminiferous tubuli was positive for MALAT1 (Figure 4C), suggesting a putative functional role of MALAT1 in spermatogonia. Probably due to the constitutive hypoxic nature of the testis,22 no clear change upon hypoxic exposure could be detected by in situ hybridization (data not shown).
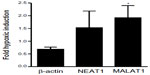
Oxygen-dependent regulation of NEAT1 and MALAT1 lncRNAs in primary mouse kidney epithelial cells
In principle, in vivo regulation of MALAT1 lncRNA by inspiratory hypoxia could result from indirect physiological effects, such as hormonal or sympathetic stimulation, rather than from direct tissue hypoxia. To address this issue, primary epithelial cells were isolated from mouse kidney and exposed to 20% or 0.2% O2 for 24 hours. β-Actin mRNA, NEAT1 and MALAT1 lncRNAs were determined by RT-qPCR and normalized to ribosomal protein S12 mRNA levels. As shown in Figure 5, β-actin mRNA is not induced by hypoxia, confirming its suitability as a constitutive control gene for the in vivo experiments (Figure 3). In contrast to the constitutive expression and low variance of β-actin mRNA, NEAT1 and MALAT1 lncRNAs were induced by hypoxia, albeit with a high variance like found before in the kidney in vivo (Figure 3). Only hypoxic induction of MALAT1 lncRNA reached a statistically significant difference when compared with β-actin mRNA induction (Figure 5), indicating that oxygen is a direct regulator of renal MALAT1 lncRNA expression.
Discussion
During the course of genome-wide studies in PC3 prostate and MCF-7 breast cancer cells, our group and others noticed that there is no principal difference between lncRNA and mRNA expression with regard to the frequency and inducibility of transcriptional regulation by HIFs.11,12 NEAT1 and MALAT1 were among the lncRNAs most strongly induced by hypoxia. In this study, we confirmed this initial observation in several additional cancer cell lines, suggesting that NEAT1 and MALAT1 could serve as general lncRNA biomarkers for tumor hypoxia, which is usually associated with adverse clinical outcome.
While the physiological relevance of oxygen-dependent MALAT1 regulation remains to be elucidated, hypoxia-inducibility of NEAT1 has recently been reported to induce paraspeckle formation, in line with its function as an architectural component of paraspeckles. While the precise function of paraspeckles is still unclear, Choudhry et al23 demonstrated that NEAT1 was required for the hypoxic induction and nuclear retention of F11R/JAM1 mRNA. F11R mRNA encodes a regulator of tight junction assembly and was previously known to be subjected to increased RNA editing under hypoxic conditions.24 Consistent with our findings, HIF-2 rather than HIF-1 induced NEAT1 in hypoxic breast cancer cell lines and solid tumor models.23 Interestingly, in a recent report, it has been shown that MALAT1 is hypoxically induced in endothelial cells in vitro and is involved in balancing proliferative and migratory properties of these cells during vascular growth in vivo.25
Several additional recent studies identified specific lncRNAs that are regulated by oxygen and often play crucial roles in cancer progression. Among others, the lncRNAs AK058003, H19, HINCUT-1, UCA1, linc-RoR, lincRNA-p21, HOTAIR, and EFNA3-derived lncRNAs have all been reported to be induced by hypoxia, usually in a HIF-1 dependent manner.26–33 Finally, the HIFα loci themselves have been reported to generate lncRNAs. The HIF1A antisense lncRNA HIF1A-AS1 has been implicated in apoptosis regulation.34 HIF1A-AS2 lncRNA is overexpressed in clear cell renal cell carcinoma, can be hypoxically induced in lymphocytes, affects HIF signaling, and was later also found in several other cancer types as well as in normal tissues.35–38 The EPAS1 (HIF2A) promoter upstream transcript HIF2PUT lncRNA has been reported to be a regulatory factor for osteosarcoma stem cells.39
However, none of these reports addressed the question of whether these lncRNAs are induced by physiological hypoxia in normal tissues. Therefore, to the best of our knowledge, our study is the first to describe hypoxic induction of lncRNA in mouse tissues in vivo. Of note, kidney and testis were the two organs with highest hypoxic levels of MALAT1 lncRNA. Common to both organs are physiologically low tissue oxygen partial pressures.22,40 Typically, bioluminescence in normoxic mice constitutively expressing a luciferase protein fused to the oxygen-dependent degradation domain of HIF-1α can be readily observed in kidney and testis but not in other organs,41 consistent with the role of HIF in the oxygen-dependent regulation of MALAT1 lncRNA in vivo.
Conclusion
High expression levels and acute hypoxic induction of MALAT1 in several mouse organs suggest a hitherto unrecognized role of this lncRNA in systemic adaptation hypoxia.
Acknowledgments
The authors thank P Spielmann and the Zurich Integrative Rodent Physiology facility for technical assistance. This work was funded by the Swiss National Science Foundation grant 31003A_146203 (RHW), by the Klinischer Forschungsschwerpunkt (KFSP) Tumor Oxygenation project of the University of Zurich (RHW), by the National Center of Competence in Research (NCCR) Kidney Control of Homeostasis (Kidney.CH) (RHW, IJF, and DH), and by the European Union’s Seventh Framework Programme for research, technological development, and demonstration under grant agreement number 608847 (KAN).
Disclosure
The authors report no conflicts of interest in this work.
References
Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. | |
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. | |
Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. | |
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. | |
Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407–4416. | |
Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16:1151–1162. | |
Kaelin WG Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. | |
Aprelikova O, Wood M, Tackett S, Chandramouli GV, Barrett JC. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 2006;66:5641–5647. | |
Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1α, HIF-2α, and other pathways. J Biol Chem. 2006;281: 15215–15226. | |
Stiehl DP, Bordoli MR, Abreu-Rodríguez I, et al. Non-canonical HIF-2α function drives autonomous breast cancer cell growth via an AREG-EGFR/ErbB4 autocrine loop. Oncogene. 2012;31: 2283–2297. | |
Wollenick K, Hu J, Kristiansen G, et al. Synthetic transactivation screening reveals ETV4 as broad coactivator of hypoxia-inducible factor signaling. Nucleic Acids Res. 2012;40:1928–1943. | |
Choudhry H, Schödel J, Oikonomopoulos S, et al. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15:70–76. | |
Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. | |
Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. | |
Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175–3181. | |
Albers J, Rajski M, Schonenberger D, et al. Combined mutation of Vhl and Trp53 causes renal cysts and tumours in mice. EMBO Mol Med. 2013;5:949–964. | |
Stiehl DP, Wirthner R, Köditz J, Spielmann P, Camenisch G, Wenger RH. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J Biol Chem. 2006;281:23482–23491. | |
Kaufmann MR, Barth S, Konietzko U, et al. Dysregulation of hypoxia-inducible factor by presenilin/γ-secretase loss-of-function mutations. J Neurosci. 2013;33:1915–1926. | |
Barth S, Nesper J, Hasgall PA, et al. The peptidyl prolyl cis/trans isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol Cell Biol. 2007;27: 3758–3768. | |
Marti HH, Katschinski DM, Wagner KF, Schäffer L, Stier B, Wenger RH. Isoform-specific expression of hypoxia-inducible factor-1α during the late stages of mouse spermiogenesis. Mol Endocrinol. 2002;16:234–243. | |
Fuady JH, Bordoli MR, Abreu-Rodríguez I, et al. Hypoxia-inducible factor-mediated induction of WISP-2 contributes to attenuated progression of breast cancer. Hypoxia. 2014;2:23–33. | |
Wenger RH, Katschinski DM. The hypoxic testis and post-meiotic expression of PAS domain proteins. Semin Cell Dev Biol. 2005;16: 547–553. | |
Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2014;34(34):4482–4490. | |
Ben-Zvi M, Amariglio N, Paret G, Nevo-Caspi Y. F11R expression upon hypoxia is regulated by RNA editing. PLoS One. 2013;8:e77702. | |
Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. | |
Wang Y, Liu X, Zhang H, et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia. 2014;16:1094–1106. | |
Matouk IJ, Mezan S, Mizrahi A, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803: 443–451. | |
Ferdin J, Nishida N, Wu X, et al. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013;20: 1675–1687. | |
Xue M, Li X, Li Z, Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol. 2014;35:6901–6912. | |
Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127: 1585–1594. | |
Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53: 88–100. | |
Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. Epub January 19, 2015. | |
Gomez-Maldonado L, Tiana M, Roche O, et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene. 2015;34:2609–2620. | |
Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7:7643–7652. | |
Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. | |
Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1α are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. | |
Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, Capranico G. Characterization of novel antisense HIF-1α transcripts in human cancers. Cell Cycle. 2011;10:3189–3197. | |
Chen WM, Huang M, Kong R, et al. Antisense long noncoding RNA HIF1A-AS2 is upregulated in gastric cancer and associated with poor prognosis. Dig Dis Sci. 2015;60(6):1655–1662. | |
Wang Y, Yao J, Meng H, et al. A novel long non-coding RNA, hypoxia-inducible factor-2α promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. 2015;11: 2534–2540. | |
Wenger RH, Hoogewijs D. Regulated oxygen sensing by protein hydroxylation in renal erythropoietin-producing cells. Am J Physiol Renal Physiol. 2010;298:F1287–F1296. | |
Safran M, Kim WY, O’Connell F, et al. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci U S A. 2006;103:105–110. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.