Back to Journals » International Journal of Nanomedicine » Volume 13
Imaging of extracellular vesicles derived from human bone marrow mesenchymal stem cells using fluorescent and magnetic labels
Authors Dabrowska S, Del Fattore A, Karnas E, Frontczak-Baniewicz M, Kozlowska H , Muraca M, Janowski M, Lukomska B
Received 9 December 2017
Accepted for publication 20 January 2018
Published 19 March 2018 Volume 2018:13 Pages 1653—1664
DOI https://doi.org/10.2147/IJN.S159404
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Sylwia Dabrowska,1 Andrea Del Fattore,2 Elzbieta Karnas,3,4 Malgorzata Frontczak-Baniewicz,5 Hanna Kozlowska,6 Maurizio Muraca,7 Miroslaw Janowski,1,8 Barbara Lukomska1
1NeuroRepair Department, Mossakowski Medical Research Centre, Polish Academy of Sciences, Warsaw, Poland; 2Multifactorial Disease and Complex Phenotype Research Area, Bambino Gesù Children’s Hospital, IRCCS, Rome, Italy; 3Department of Cell Biology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Krakow, Poland; 4Malopolska Centre of Biotechnology, Krakow, Poland; 5Electron Microscopy Platform, Mossakowski Medical Research Centre, Polish Academy of Sciences, Warsaw, Poland; 6Laboratory of Advanced Microscopy Techniques, Mossakowski Medical Research Centre, Polish Academy of Sciences, Warsaw, Poland; 7Department of Women’s and Children’s Health, University of Padua, Padua, Italy; 8Russel H. Morgan Department of Radiology and Radiological Science, Division of MR Research, The Johns Hopkins University School of Medicine, Baltimore, MD, USA
Background: Mesenchymal stem cells have been shown therapeutic in various neurological disorders. Recent studies support the notion that the predominant mechanism by which MSCs act is through the release of extracellular vesicles (EVs). EVs seem to have similar therapeutic activity as their cellular counterparts and may represent an interesting alternative standalone therapy for various diseases. The aim of the study was to optimize the method of EV imaging to better understand therapeutic effects mediated by EVs.
Methods: The fluorescent lipophilic stain PKH26 and superparamagnetic iron oxide nanoparticles conjugated with rhodamine (Molday ION Rhodamine B™) were used for the labeling of vesicles in human bone marrow MSCs (hBM-MSCs). The entire cycle from intracellular vesicles to EVs followed by their uptake by hBM-MSCs has been studied. The identity of vesicles has been proven by antibodies against: anti-CD9, -CD63, and -CD81 (tetraspanins). NanoSight particle tracking analysis (NTA), high-resolution flow cytometric analysis, transmission electron microscopy (TEM), ELYRA PS.1 super-resolution microscopy, and magnetic resonance imaging (MRI) were used for the characterization of vesicles.
Results: The PKH26 and Molday ION were exclusively localized in intracellular vesicles positively stained for EV markers: CD9, CD63, and CD81. The isolated EVs represent heterogeneous population of various sizes as confirmed by NTA. The TEM and MRI were capable to show successful labeling of EVs using ION. Co-culture of EVs with hBM-MSCs revealed their uptake by cells in vitro, as visualized by the co-localization of PKH26 or Molday ION with tetraspanins inside hBM-MSCs.
Conclusion: PKH26 and Molday ION seem to be biocompatible with EVs, and the labeling did not interfere with the capability of EVs to re-enter hBM-MSCs during co-culture in vitro. Magnetic properties of IONs provide an additional advantage for the imaging of EV using TEM and MRI.
Keywords: mesenchymal stem cells, extracellular vesicles, cell tracking, fluorescent dye, iron oxide, MRI
Introduction
Mesenchymal stem cells (MSCs) are considered as a promising therapeutic agent in various disorders. The therapeutic value of MSCs was evident in the number of experimental studies1–6 and clinical trials.7 Although it was initially thought that the therapeutic effects of MSCs depend on their capabilities for cell replacement, many studies have demonstrated that paracrine activity is the predominant mechanism by which MSCs participate in tissue repair.8–10 It was shown that by the secretion of different bioactive molecules, MSCs may support endogenous stem cell proliferation, reduce apoptosis, promote angiogenesis, and modulate immune reactions.11–15 Previous studies have focused on characterizing MSC-produced soluble factors, ie, cytokines, chemokines, and growth factors, but it has been recently documented that MSCs release also large numbers of extracellular vesicles (EVs).16–18 It seems that many therapeutic effects of MSCs might be mediated by the EVs.19–22 Increasing evidence indicates that EVs play also an important role in cell–cell communications. There are three main classes of EVs, such as exosomes, microvesicles, and apoptotic bodies with distinct size, density, and composition.23,24 However, the dimensions and density of different subtypes of vesicles can also overlap. Moreover, lack of specific markers for each type of vesicle makes it technically challenging to analyze their specific properties. EVs are released from cells under physiological and pathological conditions such as during cell activation, stress, and apoptosis.25 The subset of bioactive molecules harbored in EVs represents the content of parental releasing cell. The external stimulation from the surrounding microenvironment has been shown to regulate protein packaging into EVs and to affect their functional properties.26
The recent experimental studies reveal that EVs have the advantage as therapeutic agents over MSCs in many aspects. Cell free, EV-based therapy circumvents the risk associated with MSC-based therapies, such as replication competence, accumulation of genomic alterations, and genetic instability, and clinical-grade EVs have been recently manufactured.27–30 Moreover, systemically injected MSCs have been shown to be an object of immunomediated rejection. In addition, higher doses of MSCs delivered intra-arterially may produce adverse events such as micro-strokes.31,32 Despite the lack of MHC class II expression on the surface of MSCs, their iv infusion leads to immediate activation of innate immune reaction resulting in cell lysis and their clearance from the circulation within 60 minutes.33,34 In this regard, it will be interesting to determine whether EVs derived from MSCs infused systemically become detected by the host and rapidly cleared as their parental cells, as well as what is the final destinations of injected EVs. For that purpose, the noninvasive imaging of EVs in vivo would be beneficial. Advanced microscopy of fluorescently labeled EVs using several types of PKH-based tracers is a most frequently employed imaging method. The labeling with PKH-based tracers is relatively simple and fast and enables membranes of already isolated EVs to be stained. In recent times, superparamagnetic iron oxide (SPIO) nanoparticles have been extensively used preclinically35,36 and clinically37 for noninvasive stem cell tracking using magnetic resonance imaging (MRI). SPIO nanoparticles were also used to label exosomes from mouse B16-F10 melanoma cells labeled with SPIO nanoparticles were tracked in MRI after injection into the footpad of C57BL/6 mice.38 The ultrasmall SPIO (USPIO) was also recently incorporated into ASCs, and released EVs were shown to be loaded with this magnetic label.39 While the USPIO nanoparticles were easily detectable by MRI, the need for using electron microscopy (EM) for their postmortem identification makes the analysis of EV fate complex, and even more challenging when the co-localization of nanoparticles with certain proteins via immunostaining is needed. Therefore, the goal of our study was to investigate the feasibility and value of using fluorescent and magnetic double-labeled SPIO nanoparticles as a tool to image the EV trafficking in vitro as compared to PKH-based fluorescent labels as well as a potential to use the same label for the monitoring of EV delivery.
Methods
Cell culture
Commercially available human bone marrow MSCs (hBM-MSCs) (Lonza, Walkersville, MD, USA) were plated in 75 cm2 polystyrene tissue culture flasks (Thermo Fisher Scientific, Waltham, MA, USA) at a density of 5×103 cells/cm2 with 10 mL of Mesenchymal Stem Cell Growth Medium (MSCGM™, BulletKit™; Lonza). Cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2. Cells were subsequently maintained in MSCGM™ medium and passaged at 80% confluence in a ratio of 1:2 in trypsin/EDTA solution (Thermo Fisher Scientific) with the culture medium changed thrice a week.
Labeling of hBM-MSCs
The labeling of cells with Molday ION consisted of SPIO nanoparticles, and rhodamine purchased from BioPAL (Worcester, MA, USA) was performed as previously described by us.35 Briefly, 100 μL of Molday ION was added to the 5×105 hBM-MSCs cultured in 10 mL of MSCGM and incubated over 16 hours at 37°C in a humidified atmosphere containing 5% CO2. After that, medium with label was removed, cells were washed with phosphate-buffered saline (PBS), fresh medium was added, and cells were cultured for 48 or 72 hours. The labeling of cells with PKH26 (Red Fluorescent Cell Linker Kits MINI26; Sigma-Aldrich Co., St Louis, MO, USA) was performed at room temperature (RT) for 5 minutes in the dark and blocked with fetal bovine serum (FBS), according to manufacturer’s instructions. The unincorporated stains were removed by hBM-MSCs centrifugation at 400× g for 10 minutes at 20°C–25°C using Eppendorf Centrifuge 5804R. hBM-MSCs were washed with Dulbecco’s PBS (DPBS) without Ca++ and Mg++ (Lonza) and subjected to additional centrifugation. The pellet was re-suspended, and cells were plated in 75 cm2 polystyrene tissue culture flasks as described earlier.
Immunocytochemical analysis
Immunocytochemistry was used to identify labeled intracellular vesicles prior to their isolation or after uptake of labeled EVs. For phenotypic analysis, indirect immunocytochemistry was performed on Molday ION-labeled hBM-MSCs and nonlabeled hBM-MSCs previously incubated with labeled hBM-MSC-EVs. The direct red fluorescence was used to capture the presence of labels and co-localize with immunocytochemical staining. The cells were fixed with 4% paraformaldehyde, blocked, and permeabilized with the mixture of 10% goat serum (Thermo Fisher Scientific), 0.1% bovine serum albumin (BSA) (Sigma-Aldrich Co.), and 0.25% Triton (Sigma-Aldrich Co.) for 1 hour at RT. Cells were incubated with the following primary mouse antihuman monoclonal antibodies: anti-CD73 (1:100; Santa Cruz Biotechnology Inc., Dallas, TX, USA), anti-CD90 (1:100; Santa Cruz Biotechnology Inc.), anti-CD44 (1:100; Santa Cruz Biotechnology Inc.), anti-STEM121 (1:100; Cellartis, Takara Bio Europe, France), anti-CD63 (1:100; BD Pharmingen), anti-CD9 (1:100; BD Pharmingen, New Jersey, USA), and anti-CD81 (1:100; Santa Cruz Biotechnology Inc.) overnight at 4°C. Then, the secondary goat antimouse antibodies conjugated with Alexa Fluor 488 nm/green (Thermo Fisher Scientific) were added and the slides were exposed for 60 minutes at RT in the dark. In addition, cell nuclei were counterstained with 5 μL (1.33 μg/1 mL) Hoechst 33258 (Sigma-Aldrich Co.). After washing with PBS, the slides were mounted with Fluorescent Mounting Medium (Dako Denmark A/S, Glostrup, Denmark). Negative controls were performed with the same procedure omitting the primary antibodies. Imaging was performed by super-resolution structured illumination microscopy (SR-SIM) on LSM 780/ELYRA PS.1 (Carl Zeiss Meditec AG, Jena, Germany) platform equipped with the ZEN 2012 software, lasers (488 or 561 nm), and 405 nm diode lamp with a 100×, NA 1.46 oil objective. Spherical aberration was minimized by choosing an immersion oil with a refractive index giving symmetrical point spread functions, and image stacks of several micrometer thicknesses were taken with 0.100 μm z-steps, five phases, five rotations per z-section. The slides analyzed with SR-SIM were registered, and the positive cells were counted.
EVs’ isolation from hBM-MSCs
The isolation of EVs was performed from conditioning media of Molday ION-labeled and nonlabeled hBM-MSCs. A total of 5×106 of hBM-MSCs (passages 4–6) were cultured in 75 cm2 polystyrene tissue flasks to reach 50%–60% confluence, then the culture medium was changed, and the cells were incubated for additional 48–72 hours to the confluence of 70%–80%. Cell culture supernatants were collected and centrifuged at 200× g for 10 minutes and then at 500× g for 10 minutes at 4°C, aliquoted, and frozen at −70°C for further use. In order to isolate EVs, hBM-MSCs’ culture supernatants were thawed, spun down at 2,000× g for 20 minutes to remove cellular debris, and then centrifuged at 100,000× g for 75 minutes at 4°C using a Thermo Scientific Type 865 Fixed Angle Rotor. The pellets were washed with DPBS and subjected to an additional centrifugation at 100,000× g for 75 minutes at 4°C using a Thermo Scientific Type 865 Fixed Angle Rotor. Then, the supernatant was discarded and the pellet was re-suspended in 100 μL of DPBS and stored at −70°C until needed.
Labeling of EVs using PKH26
EVs isolated from nonlabeled hBM-MSCs were tagged with PKH26 at RT for 5 minutes in the dark and blocked with FBS, according to manufacturer’s instructions. The unincorporated labels were removed by hBM-MSC-EVs centrifugation at 100,000× g for 75 minutes at 4°C using a Thermo Scientific T-865 Fixed Angle Rotor, Thermo Scientific Sorvall WX Ultracentrifuge Series. hBM-MSC-EVs were washed with DPBS and subjected to additional centrifugations. Then, the pellet was re-suspended in 1 mL of DPBS for further use.
NanoSight particle tracking analysis (NTA) of EVs isolated from hBM-MSCs
The size and concentration of hBM-MSC-EVs were analyzed using the NanoSight NS300 system (Malvern Instruments, Malvern, UK) and configured with scientific CMOS camera and blue 488 nm laser. For NanoSight analysis, EVs were diluted in 1 mL of DPBS and collected and analyzed by the NTA software Version 3.2. Each of six hBM-MSC-EV samples from the different isolations was recorded three times for 60 seconds at constant temperature 23°C creating three replicable histograms, which were averaged.
EVs isolated from hBM-MSCs’ characterization by high-resolution flow cytometric analysis
Flow cytometry analysis of stained hBM-MSC-EVs was performed with Apogee A50-Micro cytometer, unique high-resolution system dedicated for the reliable characterization of small particles. To ensure the specificity of obtained data, appropriate isotype controls were also included in gating strategy. hBM-MSC-EVs were stained with the following mouse monoclonal antibodies: FITC-conjugated anti-CD9 clone M-L13, anti-CD63 clone H5C6, and anti-CD81 clone JS-81 (all from BD Biosciences, San Jose, CA, USA) and PE-conjugated anti-CD44 clone BJ18, anti-CD73 clone AD2, and anti-CD90 clone 5E10 (all from BioLegend, San Diego, CA, USA) or appropriate isotype-match controls. Briefly, hBM-MSC-EVs suspended in 0.2 μm filtered PBS were incubated with antibodies for 30 minutes at 4°C. Prior to addition, all antibodies were centrifuged at 21,000× g for 20 minutes at 4°C to remove potential protein aggregates. Additionally, hBM-MSC-EVs were stained with PKH26 membrane dye at RT for 5 minutes in the dark and blocked with FBS, according to manufacturer’s instructions. The sample of PBS only with PKH26 and FBS was also prepared as a control. Stained EV samples were analyzed by Apogee A50-Micro flow cytometer (Apogee Flow Systems, Hemel Hempstead, UK), and the percentage of gated positive events was calculated by the Histogram software (Apogee Flow Systems).
Transmission electron microscopy (TEM)
For ultrastructural studies, hBM-MSC-EV pellets were fixed with 2% paraformaldehyde (Sigma-Aldrich Co.) and 2.5% glutaraldehyde (Merck KGaA, Darmstadt, Germany) in 0.1 M cacodylate buffer, pH 7.4, overnight at 4°C, postfixed with 1% osmium tetroxide (Sigma-Aldrich Co.), dehydrated in a graded series of ethanol and propylene oxide (abcr GmbH, Karlsruhe, Germany), embedded in Agar 100 resin kit R1031 (Agar Scientific Ltd, Stansted, UK), and sectioned (50 nm) using ultramicrotome (RMC Boeckeler, Tucson, AZ, USA). Ultrathin sections were stained with uranyl acetate and lead citrate. Grids were examined with a JEM 1200EX electron microscope (Oxford, UK).
MRI analysis
MRI acquisition was performed using the 7T scanner (BioSpec 70/30USR; Bruker AXS Inc., Madison, WI, USA). For the detection of hBM-MSC-EVs stained with iron nanoparticles, T2-weighted sequence (TR =2,500 ms, TE =36 ms, factional anisotropy (FA) =180°, TA =2 minutes 40 seconds) was used. For MR imaging of Molday ION-labeled hBM-MSC-EVs, the pellet of ~1.3×109 EVs diluted in 200 μL of DPBS (Lonza) was deposited in PCR tubes and placed in the phantom consisting of 2% agar to stabilize the tubes and avoid artifacts from surrounding air. hBM-MSC-EVs labeled with Molday ION and non-labeled EVs were examined. The T2 maps were calculated from the acquired data.
Internalization of EVs into hBM-MSCs
To analyze the transfer of hBM-MSC-EVs into cells, hBM-MSCs at passage 5 were plated to poly-L-lysine-coated coverslips at initial density 103 cells/cm2 maintained at 37°C overnight and then incubated with EVs labeled with PKH26 or Molday ION for 24 hours. After removing media and thorough washout of cells, they were fixed and subjected to immunocytochemical analysis as described earlier.
Results
Characterization of hBM-MSCs
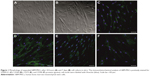
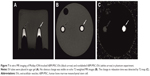
hBM-MSCs have grown in vitro as spindle-shaped cells adherent on tissue culture plastic (Figure 1A and B). High expansion capacity of hBM-MSCs gives a steady increase in total counts of cells reaching 90% confluence after 7 days in culture. The human origin of obtained cells was confirmed through positive staining by STEM121™ antibody (Figure 1C), and their biological identity was proved by the abundant expression of combination of surface markers specific for MSCs: CD44, CD73, and CD90 (Figure 1D–F). This profile remained stable at passages P4, P5, and P6.
Imaging of intracellular vesicles
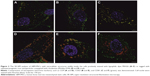
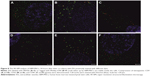
We have demonstrated that both labels PKH26 and Molday ION were successfully uptaken by hBM-MSCs and deposited in the intracellular vesicles, which co-localized with EV markers including CD9, CD63, and CD81 (Figure 2). Interestingly, the different image of labeled vesicles is seen using these two tracers. The PKH26-labeled vesicles were seen yellow indicating the direct co-localization of membranous PKH26 label (red) with tetraspanins (green) present on the membrane of vesicles. In contrast, the intracellular vesicles labeled with Molday ION express red dots visible inside EVs surrounded by green areola of positively stained tetraspanins present on the membrane of vesicles.
TEM analysis of EVs isolated from hBM-MSCs
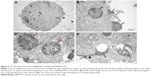
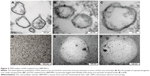
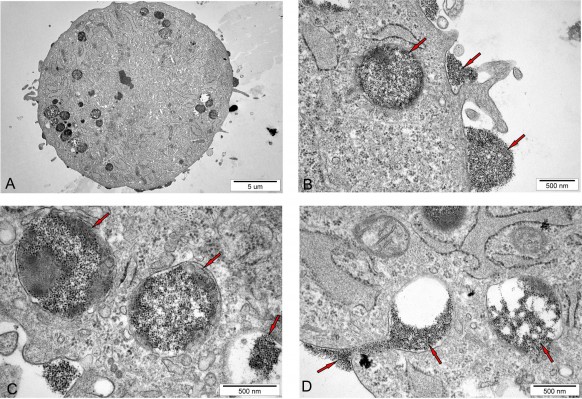
The ultrastructural analysis of hBM-MSCs confirmed the localization of Molday ION in intracellular vesicles (Figure 3A–D). Additionally, we have shown Molday ION attachment and accumulation at the extracellular membrane. Interestingly, some of nanoparticles were captured during their uptake by hBM-MSCs and deposition within intracellular vesicles (Figure 3D). The TEM also revealed that EVs isolated from hBM-MSCs consist of heterogeneous population of vesicles enriched for smaller exosomes and larger microvesicles with different sizes and shapes (Figure 4A–C). EVs obtained from Molday ION-tagged hBM-MSCs demonstrated that iron nanoparticles remained inside the vesicles after the isolation procedure (Figure 4D–F).
NTA of hBM-MSC-EVs
The NanoSight confirmed the heterogeneity of EV population (Figure 5A). The results obtained from six different samples and three recordings showed that hBM-MSC-EVs consisted of two subpopulations: exosomes/smaller microvesicles with peak at 111.67±11.55 nm and larger microvesicles with peak at 398.33±49.33 nm (Figure 5B). Furthermore, most of hBM-MSC-EVs (>80%) had an average diameter between 86.73±1.61 and 218.60±2.86 nm (Table 1). We have also demonstrated that 5×106 hBM-MSCs generated 1.26×109±1.84×107 exosome-enriched particles. The concentration corresponded to 110.17±24.00 particles/frame and 134.87±37.64 centers/frame.
EVs isolated from hBM-MSCs’ characterization by high-resolution flow cytometric analysis
Comparison of tested samples with the size-defined calibrating beads revealed that majority of visualized objects possessed size between ~100 and 250 nm. However, we observed that only a small subset of analyzed events was positive for tetraspanins, with similar amount for CD81 (2.85%±0.87%) and CD63 (2.58%±1.36%) but the lowest expression for CD9 (1.43%±0.46%). In contrast, the analysis of surface antigens’ characteristic for MSCs revealed that their hBM-MSC-EVs can harbor all three tested markers but with differential expression. In particular, the highest expression was observed for CD44, reaching 21.32%±3.27% of all analyzed particles. Moreover, 16.70±7.44 and 8.75%±2.03% of acquired EVs contained CD90 and CD73, respectively (Figure 6A).
We found that 9.7% of particles present in FBS were positive for PKH26, whereas 58.9% of events in EVs sample possessed cell-membrane content. Thus, taking into account nonspecific particle ballast derived from serum, 49.2% of analyzed hBM-MSC-EVs were specifically positive for PKH26 membrane staining (Figure 6B).
MR imaging of EVs isolated from hBM-MSCs
The threshold of hBM-MSC-EV visualization determined in phantom experiment provided satisfactory signal by MRI. While T2-weighted images showed no obvious difference between hBM-MSC-EVs labeled with Molday ION and non-labeled hBM-MSC-EVs, the T2-map revealed the change of relaxation time from 974 ms for nonlabeled hBM-MSC-EVs to 253 ms for hBM-MSC-EVs labeled with Molday ION (Figure 7).
Uptake of labeled EVs by hBM-MSCs
The SR-SIM analysis showed that labeled EVs were taken up by hBM-MSCs after 24 hours of co-culture. It seems that PKH26 or Molday ION labeling do not prevent the uptake of EVs by MSCs in vitro. The immunofluorescence revealed the co-localization of PKH26 or Molday ION with tetraspanins: CD9, CD63, and CD81 inside hBM-MSCs (Figure 8).
Discussion
In the present study, EVs were isolated from MSCs and their purity, quantity, and visualization in vitro after staining with different dyes were determined with currently available techniques. Comprehensive characterization of EVs is therefore a critical step in the further studies after their transplantation.
We have shown that EV labels are vigorously uptaken by the hBM-MSCs and trafficked to the intracellular vesicles, which co-localize with markers of EVs. It indicates that the process of intracellular localization of labels is actively regulated, and the deposited labels are ready to be released from cells inside EVs. Our observation also indicates that this trafficking phenomenon could be also used for screening for the new EV labels.
We have used an ultracentrifugation method to isolate EVs, which is still a gold standard, and allows getting ultrapure population of EVs but is very tedious, time-consuming, and cost-ineffective from the perspective of large-scale, clinic-driven EV production. Therefore, the specificity of EV labeling might be warranted in future for different methods of isolation, especially in case of low EV purity. For example, it is possible that labels are attached to the impurities and by this way falsely report on the EV destination.
TEM was capable to unequivocally prove the presence of Molday ION inside the EVs. Interestingly, Molday ION was uniformly distributed inside the EVs in our study, while the nonfluorescent USPIO nanoparticles reported by others looked rather clumped.40 Further studies to investigate if such difference in intravesicular distribution affects their therapeutic potential are warranted.
We have shown that labeling dramatically changes T2 relaxation time of EV suspension in vitro, which is very promising for future detailed studies on in vivo EV detection. While it has been previously shown that EVs can be detected in vivo by MRI, but no quantification of sensitivity or other MR parameters was performed.38,39
The distribution of EV size revealed two peaks at 111 and 398 nm, indicating the existence of two populations of EVs. The majority of EV population consists of small EVs (>80%), while the large EVs are in minority.
While high-resolution flow cytometry allowed counting the PKH26 labeled EVs, it was limited to show the presence of tetraspanins’ expression. Importantly, Apogee A50-Micro flow cytometer used in this study possesses ~110 nm detection limits for polystyrene beads and 180 nm for silica beads with refractive index more comparable to biological particles. Thus, we suppose that the sensitivity of this method is insufficient to detect such low intensity of fluorescence from the antibodies bound to the low-expressed antigens, especially present on exosomal fraction of EVs (<100 nm in diameter). In contrast, based on the Apogee analysis, we were able to detect the MSC surface markers on 10%–20% of EVs. Altogether, it may indicate that large EVs including ectosomes, which represent 20% of EVs, are shed from the cellular membrane and contain cell membrane surface markers. Since they are larger, they are detectable by high-resolution flow cytometry. The remaining small EVs, which derive from intracellular multivesicular bodies, could not capture enough antibodies to generate the signal detectable by the Apogee system. Nevertheless, despite some limitations considering the detection level, Apogee instrument is still one of the very few systems dedicated for the direct cytometric analysis of nanoparticles, and its detection is more sensitive compared to classical flow cytometers. Additionally, the EV array might be an interesting alternative for future studies.40
Although we have shown many important advances in EVs’ labeling and imaging, many questions are still needed to be answered in future studies. We have shown that labeling of hBM-MSCs with PKH26 or Molday ION is an effective way to isolate labeled EVs from them. Bright fluorescence staining of exosomes, i.e., PKH26 lipophilic dyes allows quantitative and qualitative analyses.41,42 However, this method is not ideal for the visualization of in vivo administration due to high absorption of visible light by the tissues. SPIO nanoparticles appear to be ideal cargo candidates for EVs due to their small size, biocompatibility, and unique magnetic properties. We demonstrated that EVs tagged previously by SPIO nanoparticles were visible within cells possibly because of their uptake. Our study suggests that SPIO nanoparticles do not interfere with the endocytosis of labeled hBM-MSC-EVs into cells and their diffusing in local microenvironments of cytoplasm. However, the release of free iron ions may affect distinct cellular processes and influences cell physiology. Therefore, further studies need to be performed to understand if they are equally therapeutically active as nonlabeled EVs.
Conclusion
This is one of the first articles focused on EV labeling to provide a convenient tool to study EV trafficking in vitro and in vivo. We have shown trafficking of PKH26 and Molday ION inside the hBM-MSCs to intracellular vesicles, which co-localized with markers of EVs. Then, we were able to confirm effective fluorescent labeling of EVs by high-resolution flow cytometry and magnetic labeling by TEM and MRI. Super-resolution microscopy was instrumental to detect EV uptake by hBM-MSCs. The fluorescent and magnetic double labeling of EVs by Molday ION is particularly appealing, as it allows to visualize EVs both in vitro and in vivo along with convenient detection during postmortem analysis and co-localization with specific intracellular structures detected by immunohistochemistry. The therapeutic activity as well as efficacy of uptake of labeled EVs versus nonlabeled EVs still needs to be examined in future studies.
Acknowledgments
We thank Jaroslaw Orzel for his help in performing MRI experiments. This work was supported by the Polish Ministry of Scientific Research and Higher Education grant, KNOW 06 project, STRATEGMED1/235773/19/NCBR/2016 “EXPLORE ME” project (SD, MJ, and BL) and R01 NS091110 project (MJ). MM gratefully acknowledges the financial support of “Fondazione Città dellà Speranza” (Padova, Italy). Ultracentrifuge Series Thermo Fisher Scientific used in these studies was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by The Innovative Economy, The National Cohesion Strategy of Poland. MRI experiments carried out with the use of the CePT infrastructure were financed by the European Union – the European Regional Development Fund in the Operational Programme “Innovative Economy” for 2007–2013.
The authors confirm that all data underlying the findings are fully available.
Author contributions
All authors declare the contribution in this article. SD performed hBM-MSC culture, cell characterization, cell labeling and immunocytochemistry, and EVs’ isolation and their NanoSight and MRI analyses and drafted the article. ADF performed EVs’ isolation and helped to revise the article. EK carried out flow cytometry of EV samples, data acquisition, analysis, and interpretation and helped to revise the article. MF-B performed the ultrastructural observations of EVs at the electron microscope, interpreted the data, and helped to revise the article. HK performed images of EVs using superresolution – structured illumination microscopy and helped to revise the article. MM was involved in the study design and data interpretation and helped to revise the article. MJ participated in data interpretation and manuscript editing. BL conceived the study, participated in its design, and helped with coordination, analysis, and interpretation of data and drafting the article. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Newman RE, Yoo D, LeRoux MA, Danilkovitch-Miagkova A. Treatment of inflammatory diseases with mesenchymal stem cells. Inflamm Allergy Drug Targets. 2009;8(2):110–123. | ||
Adas G, Arikan S, Karatepe O, et al. Mesenchymal stem cells improve the healing of ischemic colonic anastomoses (experimental study). Langenbecks Arch Surg. 2011;396(1):115–126. | ||
Xie MW, Gorodetsky R, Micewicz ED, et al. Marrow-derived stromal cell delivery on fibrin microbeads can correct radiation-induced wound-healing deficits. J Invest Dermatol. 2013;133(2):553–561. | ||
Mazzanti B, Lorenzi B, Lorenzoni P, et al. Treatment of experimental esophagogastric myotomy with bone marrow mesenchymal stem cells in a rat model. Neurogastroenterol Motil. 2013;2(10):e669–e679. | ||
Li DL, He XH, Zhang SA, Fang J, Chen FS, Fan JJ. Bone marrow-derived mesenchymal stem cells promote hepatic regeneration after partial hepatectomy in rats. Pathobiology. 2013;80(5):228–234. | ||
Casañas J, de la Torre J, Soler F, et al. Peripheral nerve regeneration after experimental section in ovine radial and tibial nerves using synthetic nerve grafts, including expanded bone marrow mesenchymal cells: morphological and neurophysiological results. Injury. 2014;45(suppl 4):S2–S6. | ||
clinicaltrials.gov. Available from: https://clinicaltrials.gov/ct2/results?cond=mesenchymal+stem+cells&term=&cntry=&state=&city=&dist=. Accessed February 3, 2018. | ||
Ranganath SH, Levy O, Inamdar MS, Karp JM. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell. 2012;10(3):244–258. | ||
Janowski M, Walczak P, Date I. Intravenous route of cell delivery for treatment of neurological disorders: a meta-analysis of preclinical results. Stem Cells Dev. 2010;19:5–16. | ||
Muhammad G, Xu J, Bulte JWM, Jablonska A, Walczak P, Janowski M. Transplanted adipose-derived stem cells can be short-lived yet accelerate healing of acid-burn skin wounds: a multimodal imaging study. Sci Rep. 2017;7(1):4644. | ||
Bao X, Wei J, Feng M, et al. Transplantation of human bone marrow-derived mesenchymal stem cells promotes behavioral recovery and endogenous neurogenesis after cerebral ischemia in rats. Brain Res. 2011;1367:103–113. | ||
Huang W, Mo X, Qin C, Zheng J, Liang Z, Zhang C. Transplantation of differentiated bone marrow stromal cells promotes motor functional recovery in rats with stroke. Neurol Res. 2013;35(3):320–328. | ||
Guo F, Lv S, Lou Y, et al. Bone marrow stromal cells enhance the angiogenesis in ischemic cortex after stroke: involvement of notch signaling. Cell Biol Int. 2012;36(11):997–1004. | ||
Le Blanc K, Mougiakakos D. Multipotent mesenchymal stromal cells and the innate immune system. Nat Rev Immunol. 2012;12(5):383–396. | ||
Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10(6):709–716. | ||
Collino F, Deregibus MC, Bruno S, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One. 2010;5(7):e11803. | ||
Mokarizadeh A, Delirezh N, Morshedi A, Mosayebi G, Farshid AA, Mardani K. Microvesicles derived from mesenchymal stem cells: potent organelles for induction of tolerogenic signaling. Immunol Lett. 2012;147(1–2):47–54. | ||
Baglio SR, Pegtel DM, Baldini N. Mesenchymal stem cell secreted vesicles provide novel opportunities in (stem) cell-free therapy. Front Physiol. 2012;3:359. | ||
Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4(3):214–222. | ||
Budoni M, Fierabracci A, Luciano R, Petrini S, Di Ciommo V, Muraca M. The immunosuppressive effect of mesenchymal stromal cells on B lymphocytes is mediated by membrane vesicles. Cell Transplant. 2013;22(2):369–379. | ||
Kordelas L, Rebmann V, Ludwig AK, et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28(4):970–973. | ||
Doeppner TR, Herz J, Görgens A, et al. Extracellular vesicles improve post-stroke neuroregeneration and prevent postischemic immunosuppression. Stem Cells Transl Med. 2015;4(10):1131–1143. | ||
Gyrgy B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. | ||
Koniusz S, Andrzejewska A, Muraca M, Srivastava AK, Janowski M, Lukomska B. Extracellular vesicles in physiology, pathology, and therapy of the immune and central nervous system, with focus on extracellular vesicles derived from mesenchymal stem cells as therapeutic tools. Front Cell Neurosci. 2016;10:109. | ||
Yuana Y, Sturk A, Nieuwland R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013;27(1):31–39. | ||
Kilpinen L, Impola U, Sankkila L, et al. Extracellular membrane vesicles from umbilical cord blood-derived MSC protect against ischemic acute kidney injury, a feature that is lost after inflammatory conditioning. J Extracell Vesicles. Epub 2013 Dec 10:2. | ||
Wang Y, Zhang Z, Chi Y, et al. Long-term cultured mesenchymal stem cells frequently develop genomic mutations but do not undergo malignant transformation. Cell Death Dis. 2013;4:e950. | ||
Binato R, de Souza Fernandez T, Lazzarotto-Silva C, et al. Stability of human mesenchymal stem cells during in vitro culture: considerations for cell therapy. Cell Prolif. 2013;46(1):10–22. | ||
Pan Q, Fouraschen SM, de Ruiter PE, et al. Detection of spontaneous tumorigenic transformation during culture expansion of human mesenchymal stromal cells. Exp Biol Med. 2014;239(1):105–115. | ||
Gimona M, Pachler K, Laner-Plamberger S, Schallmoser K, Rohde E. Manufacturing of Human Extracellular Vesicle-Based Therapeutics for Clinical Use. Int J Mol Sci. 2017;18(6):E1190. | ||
Janowski M, Lyczek A, Engels C, et al. Cell size and velocity of injection are major determinants of the safety of intracarotid stem cell transplantation. J Cereb Blood Flow Metab. 2013;33(6):921–927. | ||
Cui L, Kerkelä E, Bakreen A, et al. The cerebral embolism evoked by intra-arterial delivery of allogeneic bone-marrow mesenchymal stem cells in rats is related to cell dose and infusion velocity. Stem Cell Res Ther. 2015;6:11. | ||
von Bahr L, Batsis I, Moll G, et al. Analysis of tissues following mesenchymal stromal cell therapy in humans indicates limited long-term engraftment and no ectopic tissue formation. Stem Cells. 2012;30(7):1575–1578. | ||
Moll G, Rasmusson-Duprez I, von Bahr L, et al. Are therapeutic human mesenchymal stromal cells compatible with human blood? Stem Cells. 2012;30(7):1565–1574. | ||
Muhammad G, Jablonska A, Rose L, Walczak P, Janowski M. Effect of MRI tags: SPIO nanoparticles and 19F nanoemulsion on various populations of mouse mesenchymal stem cells. Acta Neurobiol Exp (Wars). 2015;75(2):144–159. | ||
Modo M, Hoehn M, Bulte JW. Cellular MR imaging. Mol Imaging. 2005;4(3):143–164. | ||
Janowski M, Walczak P, Kropiwnicki T, et al. Long-term MRI cell tracking after intraventricular delivery in a patient with global cerebral ischemia and prospects for magnetic navigation of stem cells within the CSF. PLoS One. 2014;9(2):e97631. | ||
Hu L, Wickline SA, Hood JL. Magnetic resonance imaging of melanoma exosomes in lymph nodes. Magn Reson Med. Epub 2014 Jul 22. | ||
Busato A, Bonafede R, Bontempi P, et al. Magnetic resonance imaging of ultrasmall superparamagnetic iron oxide-labeled exosomes from stem cells: a new method to obtain labeled exosomes. Int J Nanomedicine. 2016;11:2481–2490. | ||
Jørgensen MM, Bæk R, Varming K. Potentials and capabilities of the Extracellular Vesicle (EV) Array. J Extracell Vesicles. 2015;4:26048. | ||
Franquesa M, Hoogduijn MJ, Ripoll E, et al. Update on controls for isolation and quantification methodology of extracellular vesicles derived from adipose tissue mesenchymal stem cells. Front Immunol. 2014;5:525. | ||
van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, et al. Fluorescent labelling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–1326. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.