Back to Journals » OncoTargets and Therapy » Volume 12
Identification of candidate biomarkers correlated with the diagnosis and prognosis of cervical cancer via integrated bioinformatics analysis
Authors Dai F, Chen G, Wang Y, Zhang L, Long Y, Yuan M, Yang D , Liu S , Cheng Y, Zhang L
Received 27 December 2018
Accepted for publication 15 April 2019
Published 12 June 2019 Volume 2019:12 Pages 4517—4532
DOI https://doi.org/10.2147/OTT.S199615
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Fangfang Dai,1,2,* Gantao Chen,3,* Yanqing Wang,1 Li Zhang,1 Youmei Long,1 Mengqin Yuan,1 Dongyong Yang,1 Shiyi Liu,1 Yanxiang Cheng,1 Liping Zhang2
1Department of Obstetrics and Gynecology, Renmin Hospital of Wuhan University, Wuhan, Hubei 430060, People’s Republic of China; 2Department of Obstetrics and Gynecology, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, People’s Republic of China; 3Department of Gastroenterology, Third People’s Hospital of Xiantao in Hubei Province, Wuhan 430060, People’s Republic of China
*These authors contributed equally to this work
Background: Cervical carcinoma is one of the most common malignant gynecological tumors and is associated with high rates of morbidity and mortality. Early diagnosis and early treatment can reduce the mortality rate of cervical cancer. However, there is still no specific biomarkers for the diagnosis and detection of cervical cancer prognosis. Therefore, it is greatly urgent in searching biomarkers correlated with the diagnosis and prognosis of cervical cancer.
Results: The mRNA and microRNA expression profile datasets (GSE7803, GSE9750, GSE63514, and GSE30656) were downloaded from the Gene Expression Omnibus database (GEO). The three microarray datasets were integrated to one via integrated bioinformatics. Differentially expressed genes (DEGs) and microRNAs (DEMs) were obtained by R software. The protein–protein interaction (PPI) networks of the DEGs were performed from the STRING database and further visualized by Cytoscape software. A total of 83 DEGs and 14 DEMs were screened from the microarray expression profile datasets. The miRNAs validated to be associated with cervical cancer were obtained using HMDD online website and the target genes of DEMs were identified using the miRWalk2.0 online database. ESR1, PPP1R3C, NSG1, and TMPRSS11D were the gene targets of hsa-miR-21; the targets of hsa-miR-16 were GYS2, ENDOU, and KLF4. These targets were all downregulated in cervical cancer. Finally, we verified the expression of those targets in cervical tissues from TCGA and GTEx databases and analyzed their relationship with survival of cervical cancer patients. In the end, the expression of key genes in cervical cancer tissues was verified via experiment method, we found KLF4 and ESR1 were downregulated in tumor tissues.
Conclusion: This study indicates that KLF4 and ESR1 are downregulated by the upregulated miR21 and miRNA16 in cervical cancer, respectively, using bioinformatics analysis, and the lower expression of KLF4 and ESR1 is closely related to the poor prognosis. They might be of clinical significance for the diagnosis and prognosis of cervical cancer, and provide effective targets for the treatment of cervical cancer.
Keywords: cervical cancer, integrated bioinformatics, differentially expressed genes, differentially expressed miRNAs, biomarkers
Introduction
Globally, cervical cancer is the fourth frequently occurring malignancy in women in the world, accounting for almost 12% of all female cancers. There are nearly 5,27,000 new cases and 2,65,000 deaths owing to cervical cancer annually. In fact, nearly 84% of cervical cancer cases occurred in developing countries, including China. Cervical cancer is a high burden for China, in virtue of its large population with geographical and socioeconomic inequities.1 At present, cervical cancer screening methods commonly used in clinical practice include Pap Smear, TCT, Hybrid Capture 2 (HC2) test, and colposcopy. The therapy of cervical cancer mainly includes surgical treatment, radiotherapy, and chemotherapy. These screening and treatment tests have greatly decreased the incidence and mortality rates for cervical cancer.2 However, the 5-year survival rate is still low, especially for advanced patients, 5-year disease-free survival rate (DFS) is only 50%.3 More importantly, there is still no specific biomarkers for the diagnosis and detection of cervical cancer prognosis. Therefore, it is extremely urgent to study the potential molecular mechanisms of malignant biological behavior of cervical cancer, and find more reliable and prospective biomarkers for diagnosing and monitoring recurrence and evaluating prognosis.
As an important component of bioinformatics, gene expression profiling is a promising and powerful tool in medical oncology with great potential clinical applications: from molecular diagnosis to molecular screening of cancers, from new targets discovery to tumor response prediction, from patients classification to prognosis prediction.4 Today, it has been widely used to study molecular changes involved in tumor progression to better understand the development processes in human carcinogenesis, and discover new prognostic markers and new therapeutic targets in various types of cancer.5 At present, a large number of studies on the expression profile of cervical cancer have been carried out to identify the differentially expressed genes (DEGs) related to the development process or prognosis of cancer. However, owing to different data processing methods and different sample sizes, the results of DEGs are incompatible and even contradictory.6,7 Therefore, the results obtained by different raw data processing methods still have some limitations and insufficiency. Comprehensive bioinformatics methods have been applied to cancer research, and a large amount of valuable biological information has been discovered, making it possible to find effective and reliable molecular markers.8
In this study, we downloaded three original mRNA microarray datasets from the NCBI-Gene Expression Omnibus (GEO) database (
Materials and methods
Microarray data
Using the keywords “cervical cancer” to search on the GEO database (
 | Table 1 Details for GEO cervical cancer data |
Screening for DEGs
The downloaded platform and series of matrix files were converted using the R language software and annotation package. The ID corresponding to the probe name was converted into an international standard name for genes (gene-symbol) and saved in a CSV file. z-SCORE is a commonly used standardization method that can normalize values to a matrix with a standard deviation of 1 and a mean of 0 for subsequent integration of multiple sets of data. After processing the original data through the RMA function and removing the background in the R/Bioconductor software, three gene expression profiles were normalized by the z-SCORE function and processed into one data set, which contained 58 normal cervical tissue samples and 82 tumor tissue samples. The differentially expressed genes (DEGs) and the differentially expressed miRNA (DEMs) were obtained and saved in CSV files by the limma package function. | log2FC| ≥1 and adjust P-value <0.05 were considered to indicate a significant difference.
PPI network
The STRING database (
Ascertain of miRNA target genes
The HMDD tool (
DEGs basic expression in normal and cancer tissues
The Cancer Genome Atlas (TCGA) is a collaboration between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) that has generated comprehensive, multi-dimensional maps of the key genomic changes in 33 types of cancer and corresponding normal tissues.14 GEPIA (
Survival analysis
OncoLnc database (
Immunohistochemistry
Six paraffin-embedded human cervix normal and tumor tissues were cut into 5 μm thick, 3 mm diameter sections to construct tissue microarrays, and then they were placed in 3% hydrogen peroxide solution to block endogenous peroxidase activity for 10 mins, immunohistochemical staining was carried out using a rabbit polyclonal primary antibody to KLF4 (1:200; Mitaka,11880–1-AP) and ESR1 (1:300; Mitaka, 21244–1-AP) for incubating at 4°C overnight. DAB solution was used for coloration, and hematoxylin was used for counterstaining. The final immunoreactive scores were evaluated independently by two pathologists blinded to the tissue parameters. The scores were determined by the sum of intensity and extent of the staining. The integrated option density (IOD) represents the score of intracellular ESR1 expression. The percentage of nuclear KLF4 staining of cervical tissue was grouped on a scale of <5%, 5–25%, 25–50%, and >50%.
Statistical analysis
GraphPad Prism5 was used for statistical analysis, independent samples t-test was used to analyze the correlation between KLF4, ESR1 expression, and tissue parameters. P≤0.05 was considered as statistical significant.
Results
Identification of DEGs and DEMs in cervical cancer
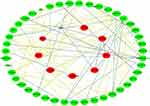
The cervical cancer raw expression microarray datasets GSE7803, GSE9750, and GSE63514 were processed and normalized, and the results are shown in Figure 1. Three datasets were integrated into one, including 58 normal cervical samples and 82 cervical cancer specimens, which were screened by the limma package, 83 differentially expressed genes (DEGs) were obtained (Data are shown in Table 2). Total 12 upregulated genes and 71 downregulated genes were identified. Among them, CDKN2A, SPP1, SYCP2, MMP12, and CXCL8 were upregulated genes, and ESR1, PPP1R3C, NSG1, TMPRSS11D, GYS2, CDKN2A, ENDOU, and KLF4 were downregulated. The log FC of top 10 up and down-DEGs is shown in Table S1. Volcano plots of gene expression profile data in 140 cervical samples are shown in Figure 1SA. The cluster heatmaps of 83 DEGs in all samples are shown in Figure S2A. The miRNA dataset GSE30656 comprised 10 normal cervix tissue samples, and 19 squamous cell carcinoma or adenocarcinoma tissue samples, 14 differentially expressed miRNAs (DEMs) were obtained using the limma package, including 3 upregulated miRNAs and 11 downregulated miRNAs (Table 3). Volcano plot of miRNA expression profile data and heatmap of DEMs are shown in Figures S1B and S2B. The PPI network complex contains 52 nodes and 100 edges (Figure 2).
 | Table 2 Screening DEGs in cervical cancer by integrated microarray |
 | Table 3 Screening DEMs in cervical cancer |
Validated miRNAs in cervical cancer
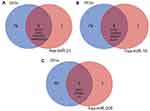
The HMDD tool was fully utilized for searching the validated cancer-related miRNAs, there are total 47 miRNAs which have been reported and validated in cervical cancer (Table 4). The ‘Calculate and draw custom Venn diagrams’ tool was used to identify the DEMs within the list of validated miRNAs, as shown in Figure 3, there are three identified DEMs within the validated miRNAs, including hsa-miR-21, hsa-miR-16, and hsa-miR-205. In the three DEMs, hsa-miR-21 and hsa-miR-16 are upregulated and hsa-miR-205 is downregulated.
 | Table 4 miRNAs which have been reported and validated in cervical cancer |
 | Figure 3 The identified DEMs within the list of validated miRNAs. |
miRNA‑DEG pairs
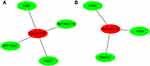
MicroRNAs (miRNAs) are non-coding RNAs of approximately 20–25 nucleotides in length that recognize target mRNA by base pairing and up- or downregulate translation of target mRNA. The target genes of the validated DEMs in cervical cancer were performed by online databases miRWALK2.0. It was identified that three validated DEM targets were DEGs. ESR1, PPP1R3C, NSG1,and TMPRSS11D were the gene targets of hsa-miR-21 (Figure 4A); GYS2, CDKN2A, ENDOU, and KLF4 were the targets of hsa-miR-16 (Figure 4B), while hsa-miR-205 targets were DSG1, SPINK5, and IVL (Figure 4C). Among them, CDKN2A was upregulated, while other target genes were downregulated in the DEGs; hsa-miR-21 and hsa-miR-16 were upregulated, while hsa-miR-205 was downregulated in the DEMs. In view of miRNAs negatively regulating the expression of their targets, we analyzed the correlation of upregulated genes with downregulated miRNAs, downregulated genes and upregulated miRNAs. Then, we made the visual networks of hsa-miR-21 (Figure 5A), hsa-miR-16 (Figure 5B), and their targets by cytoscape software.
DEGs expression state in normal and cervical cancer samples
GEPIA, an open-access website of the genome-wide expression in normal and cancer tissues from TCGA and GTEx, was used to identify the basic expression level of differentially expressed genes in 319 cervix uteri tissues. In the present study, we would like to validate whether the expression of miRNA target genes from the GEO database is consistent with their expression in these cervical tissues from TCGA and GTEx. As expected, KLF4, the target genes of hsa-miR-16 was significantly downregulated in cervical cancer samples, as were ESR1 and PPP1R3C. However, the targets ENDOU, GYS2 and TMPRSS11D had no statistical significance, and the gene NSG1 cannot be detected, as shown in Figure 6. Therefore, we chose KLF4, ESR1, and PPP1R3C for subsequent research.
Survival analysis
In this study, the relationship between the expression of KLF4, ESR1, PPP1R3C, and the overall survival of patients with cervical cancer was analyzed by OncoLnc tool. We found that the low mRNA expression of KLF4 was associated with poor overall survival rate (log-rank P=0.00966; Figure 7A). However, there was no significant in ESR1 and PPP1R3C (log-rank P<0.05; Figure 7C, Figure 7E). At the same time, the 5-year survival of patients with cervical cancer was analyzed by the HPA dataset; it was obvious that the low mRNA expression of KLF4 and ESR1 was associated with poor 5-year survival rate (P<0.05; Figure 7B and D). However, the high mRNA expression of PPP1R3C had a poor 5-year survival rate (Figure 7F). Overall, KLF4 is closely related to overall survival, and ESR1 is closely related to 5-year survival. KLF4 and ESR1 might be critical biomarkers for the prognosis of cervical cancer.
The expression of KLF4 and ESR1 in cervical tissues by immunohistochemistry
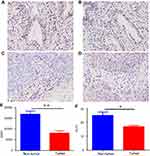
To further determine whether the two key genes obtained above were downregulated in cervical cancer, the protein expression in cervical tissues was detected by immunohistochemistry method. The results were consistent with those obtained by bioinformatics, the expression of KLF4 and ESR1 in cervical cancer tissues was significantly lower than that in normal cervical tissues (P<0.05; Figure 8).
Discussion
As one of the most common malignant gynecological tumors, cervical cancer is one of the leading causes of cancer death in women. According to the latest statistics released in 2015, China ranks second in new cases of cervical cancer every year in the world.16 According to the statistics, 80% of patients have developed aggressive cancers at the time of diagnosis, and the age of diagnosis is slowly decreasing. In addition, cervical cancer still carries a high risk of morbidity and mortality on account of the metastasis and recurrence.17 Meanwhile, reliable and specific biomarkers for diagnosis and prognosis of cervical cancer are deficient and unexplored. Therefore, it is essential and urgent to search for biomarkers of diagnosis and novel treatment targets to predict the survival of cervical cancer. As an indispensable part of bioinformatics, gene expression microarrays have been extensively used to study cancer-related genes and provide broad prospects for drug-based molecular targeting, molecular prediction, and molecular therapy.18 A series of biomarkers have been presented potentially as the diagnosis and prognosis in cervical cancer and other cancers.19,20
In the present study, three raw mRNA and one miRNA expression profile datasets were used to identify potential biomarkers in cervical cancer. The expression patterns obtained from the GEO database included 3 human cervical cancer raw mRNA profiles and 1 human cervical cancer miRNA expression dataset, all of which contained a comparison between cervical cancer and normal cervix tissue samples. After using R software and bioinformatics to integrate and deeply analyze these datasets, there are total of 83 DEGs and 14 DEMs that were identified.
MiRNA, a non-coding RNA of approximately 22 nucleotides in length, regulates expression of various oncogenes and tumor suppressor genes by targeting the 3’UTR of the target mRNA, resulting in degradation or inhibition of translation.21 In the study, we obtained the validated miRNAs by reference to existing studies using HMDD and identified target genes of the DEMs by miRWALK2.0. As miRNAs negatively regulate the expression of their target genes, we identified that ESR1, PPP1R3C, NSG1, and TMPRSS11D were potentially the targets of miRNA-21, KLF4, ENDOU, and GYS2 were the gene targets of miRNA −16. Among them, miRNA-21 and miRNA-16 were upregulated, while the targets ESR1, PPP1R3C, NSG1, KLF4, ENDOU, and GYS2 were downregulated.
To further study whether there is a difference in the expression of above genes in normal and tumor tissues, we analyzed their expression in 319 cervical tissue samples from the TCGA and GTEx databases, including 306 tumor samples and 13 normal tissue samples. As expected, the target gene KLF4 was significantly downregulated in cervical cancer samples, as were ESR1 and PPP1R3C. However, the expression of ENDOU, GYS2, TMPRSS11D, and NSG1 is upregulated or missing in cancer tissues. Subsequently, further analysis was performed using the survival data from TCGA via OncoLnc and HPA website to identify the relationship between the postoperative survival of patients and the expression of above three genes. It suggested that the low expression of KLF4 was closely associated with poor overall survival and 5-year survival time of cervical cancer patients (P<0.05), the low expression of ESR1 and the high expression of PPP1R3C were closely associated with poor 5-year survival rate (P<0.05). However, ESR1 and PPP1R3C expressions were not significantly associated with OS (P>0.05). Overall, KLF4 and ESR1 are closely related to overall survival and 5-year survival. They might be of great clinical significance for the prognosis of cervical cancer and provide effective targets for the treatment of cervical cancer.
Kruppel like factor 4 (KLF4) located at 9q31 is a critical DNA-binding transcription regulator.22 It is highly expressed in the gastrointestinal tract and other epithelial tissues, and it has been reported to regulate cell proliferation, apoptosis, differentiation and the maintenance of telomerase activity.23,24 A number of studies have suggested that KLF4 functions as a tumor suppressor in various cancers, such as human pancreatic cancer and colorectal cancer.25,26 Yang et al indicated that KLF4 was downregulated in the development and progression of cervical carcinoma, and the overexpression of exogenous KLF4 protein inhibited the cell growth and tumor formation by activating the cell cycle suppressor p27, which supports the hypothesis that KLF4 is a tumor suppressor in cervical carcinoma.27 However, there is still no report on the relationship between KLF4 and the prognosis of cervical cancer.
Estrogen receptor 1 (ESR1) is a receptor protein, which is usually enriched expression in breast, endometrium, and normal cervical tissue, which is necessary for sexual development and reproductive function, and performs an important role in cellular development and differentiation. ESR1 protein is closely related to a variety of gynecological oncology diseases. It is well known that loss of ESR in breast cancer patients indicates invasiveness and poor prognosis.28 In ovarian carcinoma, ESR1 methylation influences ovarian cancer development, and its promoter methylation was negatively correlated with survival in the subgroup of low-grade ovarian carcinoma patients.29 ESR1 also plays a critical and diverse role in cervical cancer. Many previous studies indicated that ESR1 was downregulated in cervical malignant cells and human papillomavirus (HPV) DNA positive cervical intraepithelial neoplasia tissues.30,31 On the other hand, ESR1 plays a major role in mediating cervical cancer invasion and progression, and loss of it enhances cervical cancer invasion.32 However, the expression of ESR1 in advanced cervical cancer and its impact on survival needs further study.
A variety of miRNAs have also been reported to downregulate KLF4 and ESR1 in various types of disease. In gastric cancer, KLF4 was a direct target of miR-103; And in non-small cell lung cancer cells, miR-25 could enhance cell migration and invasion via ERK signaling pathway by inhibiting KLF4.33,34Similarly, it has been reported that miR-301a-3p suppresses estrogen signaling by directly inhibiting ESR1 in ERα positive breast cancer.35 However, the relationship between miRNAs and KLF4 and ESR1 in cervical cancer is rarely reported.
miRNA-21 and miRNA-16 have been validated to be closely related to cervical cancer. The expression of miR-21 was upregulated by the HPV16 E7 oncoprotein in vivo, and the high expression of miR-21 in cervical cancer may play a possible role of oncogenes.36,37 miR-16-1 belongs to the miR-16 cluster and has been implicated in various aspects of carcinogenesis including cell proliferation and regulation of apoptosis. In human cervical cancer cells, miR-16-1 post-transcriptionally downregulates Cyclin E1 (CCNE1) gene expression to deregulate the cell cycle .38 The above indicates that miRNA-21 and miRNA-16 might act as oncogenes to promote carcinogenesis in a variety of pathways. In this study, using comprehensive bioinformatics analysis methods, we found that miRNA16 and miRNA-21 might lead to the pathogenesis of cervical cancer by inhibiting the expression of KLF4 and ESR1 in cervical tissues, respectively, and cervical cancer patients with low expression of KLF4 and ESR1 may be associated with poor overall survival and 5-year survival prognosis. At the end, we found the expression of KLF4 and ESR1 protein was significantly lower in cervical cancer tissues via experiment method.
Conclusion
In summary, in this study, we find that KLF4 and ESR1 are downregulated by the upregulated miR21 and miRNA16 in cervical cancer, respectively, and the lower expression of KLF4 and ESR1 is closely related to the poor prognosis. They might be critical biomarker for diagnosis and predicting cervical cancer progression. Our study has important clinical significance for better understanding the development and prognosis of cervical cancer. Of course, whether these genes can be used as the biomarkers for diagnosis and prognosis of cervical cancer needs further clinical study.
Acknowledgments
This work was supported by the Chinese Medical Association Clinical Research Special Fund Project (grant no 17020310700) and the Wuhan University Independent Research Project (grant no 413000117).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Di J, Rutherford S, Chu C. Review of the cervical cancer burden and population-based cervical cancer screening in China. Asian Pac J Cancer Prev. 2015;16(17):7401–7407. doi:10.7314/APJCP.2015.16.17.7401
2. Kessler TA. Cervical cancer: prevention and early detection. Semin Oncol Nurs. 2017;33(2):172–183. doi:10.1016/j.soncn.2017.02.005
3. Chopra S, Gupta M, Mathew A, et al. Locally advanced cervical cancer: a study of 5-year outcomes. Indian J Cancer. 2018;55(1):45–49. doi:10.4103/ijc.IJC_428_17
4. Wadlow R, Ramaswamy S. DNA microarrays in clinical cancer research. Curr Mol Med. 2005;5(1):111–120.
5. Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35(3):201–209. doi:10.1016/j.ctrv.2008.10.006
6. Wang S, Chen X. Identification of potential biomarkers in cervical cancer with combined public mRNA and miRNA expression microarray data analysis. Oncol Lett. 2018;16(4):5200–5208. doi:10.3892/ol.2018.9323
7. Ge Y, Zhang C, Xiao S, et al. Identification of differentially expressed genes in cervical cancer by bioinformatics analysis. Oncol Lett. 2018;16(2):2549–2558. doi:10.3892/ol.2018.8953
8. Yang X, Zhu S, Li L, et al. Identification of differentially expressed genes and signaling pathways in ovarian cancer by integrated bioinformatics analysis. Onco Targets Ther. 2018;11:1457–1474. doi:10.2147/OTT.S152238
9. Zhai Y, Kuick R, Nan B, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67(21):10163–10172. doi:10.1158/0008-5472.CAN-07-2056
10. Scotto L, Narayan G, Nandula SV, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47(9):755–765. doi:10.1002/gcc.20577
11. Den Boon JA, Pyeon D, Wang SS, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112(25):E3255–E3264. doi:10.1073/pnas.1509322112
12. Wilting SM, Snijders PJ, Verlaat W, et al. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene. 2013;32(1):106–116. doi:10.1038/onc.2012.20
13. von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261.
14. Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi:10.1016/j.cell.2015.12.028
15. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
16. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
17. Wang IT, Chou SC, Lin YC. Zoledronic acid induces apoptosis and autophagy in cervical cancer cells. Tumour Biol. 2014;35(12):11913–11920. doi:10.1007/s13277-014-2460-5
18. Petryszak R, Burdett T, Fiorelli B, et al. Expression atlas update–a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42(Databaseissue):D926–D932. doi:10.1093/nar/gkt1270
19. Ni M, Liu X, Wu J, et al. Identification of candidate biomarkers correlated with the pathogenesis and prognosis of non-small cell lung cancer via integrated bioinformatics analysis. Front Genet. 2018;9:469. doi:10.3389/fgene.2018.00173
20. Zhang GM, Goyal H, Song LL. Bioinformatics analysis of differentially expressed miRNA-related mRNAs and their prognostic value in breast carcinoma. Oncol Rep. 2018;39(6):2865–2872. doi:10.3892/or.2018.6393
21. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
22. Marrero-Rodriguez D, la Cruz HA, Taniguchi-Ponciano K, et al. Kruppel like factors family expression in cervical cancer cells. Arch Med Res. 2017;48(4):314–322. doi:10.1016/j.arcmed.2017.06.011
23. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi:10.1016/j.cell.2006.07.024
24. Foster KW, Liu Z, Nail CD, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24(9):1491–1500. doi:10.1038/sj.onc.1208307
25. Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68(12):4631–4639. doi:10.1158/0008-5472.CAN-07-5953
26. Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23(2):395–402. doi:10.1038/sj.onc.1207067
27. Yang WT, Zheng PS. Kruppel-like factor 4 functions as a tumor suppressor in cervical carcinoma. Cancer. 2012;118(15):3691–3702. doi:10.1002/cncr.26698
28. Li X, Yang J, Peng L, et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161(2):279–287. doi:10.1007/s10549-016-4059-6
29. Kirn V, Shi R, Heublein S, et al. Estrogen receptor promoter methylation predicts survival in low-grade ovarian carcinoma patients. J Cancer Res Clin Oncol. 2014;140(10):1681–1687. doi:10.1007/s00432-014-1729-9
30. Coelho FR, Prado JC, Pereira Sobrinho JS, et al. Estrogen and progesterone receptors in human papilloma virus-related cervical neoplasia. Braz J Med Biol Res. 2004;37(1):83–88.
31. Lopez-Romero R, Garrido-Guerrero E, Rangel-Lopez A, et al. The cervical malignant cells display a down regulation of ER-alpha but retain the ER-beta expression. Int J Clin Exp Pathol. 2013;6(8):1594–1602.
32. Zhai Y, Bommer GT, Feng Y, Wiese AB, Fearon ER, Cho KR. Loss of estrogen receptor 1 enhances cervical cancer invasion. Am J Pathol. 2010;177(2):884–895. doi:10.2353/ajpath.2010.091166
33. Zheng J, Liu Y, Qiao Y, Zhang L, Lu S. miR-103 promotes proliferation and metastasis by targeting KLF4 in gastric cancer. Int J Mol Sci. 2017;18(5):910. doi:10.3390/ijms18050910
34. Ding X, Zhong T, Jiang L, Huang J, Xia Y, Hu R. miR-25 enhances cell migration and invasion in non-small-cell lung cancer cells via ERK signaling pathway by inhibiting KLF4. Mol Med Rep. 2018;17(5):7005–7016. doi:10.3892/mmr.2018.8772
35. Lettlova S, Brynychova V, Blecha J, et al. MiR-301a-3p suppresses estrogen signaling by directly inhibiting ESR1 in ERalpha positive breast cancer. Cell Physiol Biochem. 2018;46(6):2601–2615. doi:10.1159/000489687
36. Liu L, Wang YL, Wang JF. [Differential expression of miR-21, miR-126, miR-143, miR-373 in normal cervical tissue, cervical cancer tissue and Hela cell]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2012;43(4):536–539.
37. Gomez-Gomez Y, Organista-Nava J, Ocadiz-Delgado R, et al. The expression of miR-21 and miR-143 is deregulated by the HPV16 E7 oncoprotein and 17beta-estradiol. Int J Oncol. 2016;49(2):549–558. doi:10.3892/ijo.2016.3575
38. Zubillaga-Guerrero MI, Alarcon-Romero Ldel C, Illades-Aguiar B, et al. MicroRNA miR-16-1 regulates CCNE1 (cyclin E1) gene expression in human cervical cancer cells. Int J Clin Exp Med. 2015;8(9):15999–16006.
Supplementary materials
 | Table S1 The log FC and P.Value of top 10 up and down-DEGs |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.