Back to Journals » Clinical Interventions in Aging » Volume 15
Identification of Aberrantly Expressed Long Non-Coding RNAs and Nearby Targeted Genes in Male Osteoporosis
Authors Fei Q , Li X, Lin J, Yu L, Yang Y
Received 10 July 2020
Accepted for publication 16 August 2020
Published 24 September 2020 Volume 2020:15 Pages 1779—1792
DOI https://doi.org/10.2147/CIA.S271689
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Zhi-Ying Wu
Qi Fei,* Xiaoyu Li,* Jisheng Lin,* Lingjia Yu,* Yong Yang*
Department of Orthopedics, Beijing Friendship Hospital, Capital Medical University, Beijing 100050, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qi Fei Tel +86 10 6313 8353
Fax +86 10 8391 1029
Email [email protected]
Purpose: To investigate different expression profiles of long non-coding RNAs (lncRNAs) and mRNAs between male osteoporosis and normal control by high throughput RNA sequencing.
Methods: We obtained the different expression profiles of long non-coding RNAs (lncRNAs) and mRNAs between male osteoporosis and normal control by high throughput RNA sequencing. Compared to normal control, we identified the differentially expressed genes (DEGs), differentially expressed lncRNAs (DElncRNAs) and the nearby targeted DEGs of DElncRNAs in male osteoporosis. Functional annotation was used to further study the functions of DEGs in male osteoporosis. The DElncRNAs–DEGs interaction network was constructed. One DElncRNA-nearby targeted DEG interaction pair of LINC02009-CCR2 was validated in vitro.
Results: Totally, 3296 DEGs, 204 DElncRNAs and 168 DElncRNAs-nearby targeted DEGs pairs were obtained. The most significantly up-regulated and down-regulated DElncRNAs in male osteoporosis were Loc105372801 and KCNQ1OT1, respectively. Osteoclast differentiation and chemokine signaling pathway were significantly enriched pathways in male osteoporosis. Based on the DElncRNAs–DEGs interaction network in male osteoporosis, we obtained several interaction pairs including SNHG5-SYNCRIP-HBA1-HBB, HCG27-HLA-C, LINC02009-CCR2, and LOC101926887-IFIT1-IFIT2/IFIT3/IFIT5. The expression of LINC02009 and CCR2 was down-regulated in keeping with the RNA sequencing data.
Conclusion: Identified DElncRNAs–DEGs interaction pairs may be involved in the development of male osteoporosis, which make a contribution to underlying the mechanism of male osteoporosis. Among which, the validated DElncRNAs-nearby targeted DEGs interaction pair of LINC02009-CCR2 may be important regulators in the development of male osteoporosis.
Keywords: male osteoporosis, RNA sequencing, differentially expressed lncRNAs, differentially expressed genes, osteoclast differentiation, in vitro validation
Introduction
Primary osteoporosis is characterized by decreased bone mass, low bone mineral density (BMD) and loss of microarchitectural integrity with consequent increases the risk of fracture.1 Primary osteoporosis mainly occurs in postmenopausal women and senile men. Like postmenopausal osteoporosis, male osteoporosis is becoming a serious health problem worldwide with aging.2 The loss of BMD is approximately 1% per year in senile men and 20% of senile man aged over 50 years suffer an osteoporosis-related fracture.3 One third of men with osteoporosis suffer a hip fracture, a major risk factor for osteoporotic fracture-related death.4
In the last decades, there has been considerable effort to uncover the mechanisms underlying osteoporosis, especially the postmenopausal osteoporosis. Bone loss of osteoporosis was induced by remodeling imbalance between bone resorption and bone formation.4 Calcium,5 parathyroid hormone,6 estrogen7 and vitamin D8 are well-recognized regulators in osteoporosis. Multiple cytokines such as interleukin-1 (IL-1), IL-6, macrophage colony-stimulating factor (M-CSF), tumor necrosis factor-α (TNF-α) and receptor activator of nuclear factor-kB ligand (RANKL) were found to be involved in osteoporosis.9,10 Immune system plays a crucial role in osteoporosis as well.4,8 However, the molecular mechanisms of osteoporosis in men are not fully known.
Recently, accumulated evidence has indicated that long non-coding RNAs (lncRNAs) are involved in various biological progress by regulating the gene expression at transcriptional, epigenetic, or post-transcriptional levels.11–13 It is reported that lncRNAs can alter the modification and location of transcription factors, act as precursor for siRNAs or miRNAs, regulate the alternative splicing of pre-mRNAs through SR complex, regulate histone modification by interacting with modification factors and bind to chromatin modification complexes to regulate chromatin remodeling and structure.14 Previous studies have reported that several lncRNAs such as DNACR and MEG3 are associated with postmenopausal osteoporosis.12,13,15
In this study, we investigated different expression profiles of lncRNAs and mRNAs between male osteoporosis and normal control by high throughput RNA sequencing. Differentially expressed lncRNAs (DElncRNAs) and differentially expressed genes (DEGs) in male osteoporosis were further identified. Since lncRNAs can regulate the expression of their nearby gene by cis-regulatory effects,11 the nearby targeted DEGs of DElncRNAs in male osteoporosis were identified as well. To further study the functions of DEGs in male osteoporosis, functional annotation was carried out.
Materials and Methods
Patients and Samples
We collected full blood samples from three men with osteoporosis and two healthy men. Detailed full blood collection was as follows: (1) the full blood was collected with BD tube (2.5 mL per tube) at room temperature (18–25°C); (2) the full blood was slowly reversed and mixed 8 to 10 times to mix well with the anticoagulant in the BD tube; (3) the full blood was placed vertically at room temperature for 12–24 h to fully contacted with the RNA stabilizer in the BD tube; (4) the full blood was transferred to the refrigerator at −20°C for 24 h, then transferred to the refrigerator at −80°C for long-term preservation.
The inclusion criteria of male osteoporosis were as follows: (1) according to world health organization (WHO) standards, patients whose T-score of the femoral neck or lumbar vertebrae or total hip is ≤-2.5 were diagnosed as osteoporosis; (2) patients had a new vertebral compression fracture with no trauma history; (3) patients had no prior history of drug use that might affect bone metabolisms such as glucocorticoids, thyroid hormone, parathyroid hormone, fluoride, calcitonin, thiazides, barbiturates, antiepileptic drugs, vitamin D and calcium. The exclusion criteria of male osteoporosis were as follows: (1) patients had diseases that affect bone metabolic such as kidney disease, liver disease, thyroid disease, diabetes, hyperprolactinemia, rheumatoid arthritis, ankylosing spondylitis, chronic abdominal bay absorption adverse symptoms, malignant tumor, blood disease and abnormal reproductive system and related medical history; (2) patients had traumatic fractures. In addition, the inclusion criteria of normal controls (men with degenerative osteoarthritis of the lumbar spine) were as follows: (1) individuals were matched by gender, ageand frequency of the case group; (2) T-scores of the femoral neck and lumbar vertebrae (L1-4) and total hip are all ≥-1. The exclusion criteria of normal controls were as follows: (1) People with a previous history of orthopedic system disease. The detailed information of participants is shown in Table 1. All the participants submitted the signed informed consent. The protocol for this study has been approved by the Ethics Committee of Beijing Friendship Hospital, Capital Medical University (2017-P2-084-01).
 |
Table 1 Patients Characteristics in RNA Sequencing |
RNA Sequencing
We isolated total RNA from these five blood samples through the TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). Nanodrop ND-2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) was used to test the concentration and purity of total RNA. 2% agarose gel was used to check the integrity of RNA. We also obtained the RNA integrity number (RIN) value by an Agilent 2100 Bioanalyzer. The threshold for cDNA library construction was as follows: (1) the total amount of RNA was more than 5μg; (2) the concentration of RNA was more than 200ng/mL; (3) the value of OD260/280 was 1.8–2.2.
We removed ribosomal RNA by Ribo-Zero Magnetic kit (EpiCentre, Madison, WI, USA). RNA was fragmented into about 200 base pairs randomly. By using Truseq RNA sample Prep Kit (Illumina, Inc., San Diego, CA, USA), cDNA library was constructed. Firstly, the first cDNA strand was synthesized with RNA fragments primed with random hexamer primers. Secondly, we synthesized the second cDNA strand with dUTP instead of dTTP. Thirdly, end repair, 3ʹend adenylation and adapter ligation of double-stranded cDNAs were performed with End Repair Mix. Lastly, the second cDNA was digested by UNG enzyme (Illumina, Inc., San Diego, CA, USA). After the construction of the cDNA library, we amplified the library by performing polymerase chain reaction (PCR) for 15 cycles. Then, the amplified cDNA library was purified and quantified by Certified Low Range Ultra Agarose (Bio-Rad) and Picogreen (Molecular probes) on TBS380 (Turner Biosystems), respectively. The bridge PCR on cBot was performed by Truseq PE Cluster Kit v3-cBot-HS. In the end, sequencing of each library was performed by Illumina HiSeq 2000.
Data Processing
The FASTQ sequence data derived from RNA-seq results were obtained through Base Calling. After removed the low-quality reads by cutadapt (http://cufflinks.cbcb.umd.edu/), we obtained the high-quality clean data. Fasta and gff files of human reference genome (Ensemble GRCh38.p7) were downloaded from NCBI. TopHat (http://tophat.cbcb.umd.edu/) and Ensemble gene annotation were used to align clean reads to Ensemble GRCh38.p7. Cuffquant (http://cufflinks.cbcb.umd.edu/) was used to determine the expression of both mRNAs and lncRNAs and outputted the normalized data. The DEGs and DElncRNAs in male osteoporosis compared to normal control were obtained by using an R package, DEGseq. P-value <0.05 was the threshold of DEGs and DElncRNAs in male osteoporosis. The heat-maps of top 50 up- and down-regulated DEGs and DElncRNAs expression profile were performed with cluster in R language.
Protein-Protein Interaction (PPI) Network of DEGs
Based on Biological General Repository for Interaction Datasets (BioGRID) (http://thebiogrid.org/), we explored the proteins interacted with proteins encoded by the top 10 up-regulated and down-regulated DEGs in male osteoporosis. After removing these proteins that were not belonging to DEGs, we constructed the PPI network of top 10 up- and down-regulated DEGs in male osteoporosis.
Nearby Targeted DEGs of DElncRNAs
In order to identify the target DEGs of DElncRNAs, the DEGs transcribed within a 100 kbp window upstream or down-stream of DElncRNAs were researched and considered as nearby cis targets of DElncRNAs. The network of DElncRNA-near targeted DEGs was constructed by using Cytoscape.
Functional Annotation of All DEGs and Nearby Targeted DEGs of DElncRNAs
To further study the biological functions of identified all of DEGs and nearby targeted DEGs of DElncRNAs, we performed the Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs by online GeneCoDis3 software. False discovery rate (FDR) <0.05 was regarded as the cut-off for significant GO terms and KEGG pathways.
DElncRNAs-Nearby Targeted DEGs Interaction Network
Firstly, we constructed the DElncRNAs-nearby targeted DEGs interaction network by using all of the DElncRNAs and DEGs in male osteoporosis compared to normal control. Then, this DElncRNAs-nearby targeted DEGs interaction network and the male osteoporosis-specific PPI network were merged with the top 10 up- and down-regulated DEGs.
In vitro Validation
One DElncRNA-nearby targeted DEGs interaction pair of LINC02009-CCR2 was selected randomly for validation. A total of 36 full blood samples (16 cases) from male osteoporosis patients and healthy individuals (20 normal controls) were used for validation of CCR2 and LINC02009. The clinical information of these individuals was shown in the supplementary Table 1. We performed QRT-PCR in an ABI 7500 real-time PCR system. Relative gene expression was analyzed using 2−ΔΔCt method. The human GAPDH gene was used as an endogenous control for mRNA expression in the analysis. All the participants submitted the signed informed consent. The protocol for this study has been approved by the Ethics Committee of Beijing Friendship Hospital, Capital Medical University (2017-P2-084-01).
Results
LncRNAs and mRNAs Expression Profile
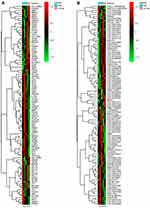
A total of 3296 DEGs (1428 up-regulated DEGs and 1868 down-regulated DEGs) were obtained. A total of 204 DElncRNAs including 57 up-regulated DElncRNAs and 147 down-regulated DElncRNAs were obtained. The top 10 up- and down-regulated DEGs and top 10 up- and down-regulated DElncRNAs were shown in Tables 2 and 3, respectively. The heat-maps of the top 50 up- and down-regulated DElncRNAs and DEGs expression profile are illustrated in Figure 1.
 |
Table 2 Top 10 Up- and Down-Regulated DEGs in Male Osteoporosis |
 |
Table 3 Top 10 Up- and Down-Regulated DElncRNAs in Male Osteoporosis |
PPI Network of DEGs in Male Osteoporosis
The PPI network consisted of 101 nodes and 105 edges (Figure 2). Based on this network, HLA-C (degree=18), KRT1 (degree=17) and IFIT1 (degree=15) were three hub proteins in male osteoporosis.
Nearby Targeted DEGs of DElncRNAs
After searching the DEGs transcribed within a 100 kbp window upstream or down-stream of DElncRNAs, we obtained 168 DElncRNAs-nearby targeted DEGs pairs which consisted of 116 DElncRNAs and 162 DEGs. The DElncRNAs-nearby targeted DEGs network is illustrated in Figure 3. LOC101926887 (degree=5), LOC100131626 (degree=5) and TMX2-CTNND1 (degree=5) were the top three DElncRNAs covered most nearby targeted DEGs.
Functional Annotation of All DEGs and Nearby Targeted DEGs of DElncRNAs
Firstly, we performed the functional enrichment analysis of all DEGs. In the GO enrichment analysis (Figure 4A–C), apoptotic process (FDR=4.78E-14), regulation of apoptotic process (FDR=8.32E-10), protein binding (FDR=3.42E-86), nucleotide binding (FDR=1.69E-22), nucleus (FDR=2.15E-73), cytoplasm (FDR=3.48E-72) were significantly enriched GO terms in male osteoporosis. Oxidative phosphorylation (FDR=3.52E-11), pyrimidine metabolism (FDR=8.20E-09), osteoclast differentiation (FDR=8.54E-09) and chemokine signaling pathway (FDR=5.61E-15) were significantly enriched pathways in male osteoporosis (Figure 4D). The detailed signaling pathway of osteoclast differentiation is shown in Figure 5. Then, we performed the functional annotation of nearby targeted DEGs of DElncRNAs. Based on the GO enrichment analysis (Figure 6), type I interferon-mediated signaling pathway (FDR=2.66E-05), cytokine-mediated signaling pathway (FDR=3.78E-05), protein binding (FDR=4.34E-09), zinc ion binding (FDR=9.54E-06), cytoplasm (FDR=1.26E-09) and cytosol (1.30E-06) were significantly enriched GO terms of nearby targeted DEGs of DElncRNAs in male osteoporosis. However, we did not obtain an enriched pathway for nearby targeted DEGs of DElncRNAs in male osteoporosis based on the KEGG enrichment analysis.
DElncRNAs-Nearby Targeted DEGs Interaction Network
After merged the DElncRNAs-nearby targeted DEGs interaction network and the PPI network (involved top 10 up- and down-regulated DEGs), we obtained key DElncRNA-nearby targeted DEGs (top 10 up- and down-regulated DEGs) interaction network (Figure 7).
QRT-PCR Confirmation of DEGs
To manifest the expression of high throughput RNA sequencing for male osteoporosis, we compared the results of qRT-PCR and RNA sequencing of DElncRNA-nearby targeted DEGs interaction pair LINC02009-CCR2. In Figure 8, expressions of LINC02009 and CCR2 were all down-regulated and consistent with our RNA sequencing results.
 |
Figure 8 QRT-PCR verification results. The X-axis was indicated different samples and the Y-axis was indicated the relative level of CCR2 mRNA (A) and LINC02009 lncRNA (B) for qRT-PCR results. |
Discussion
Accumulated evidence has emphasized the importance of lncRNAs in various cancers and other diseases, such as osteoporosis.11–13 Herein, we obtained the lncRNA and mRNA expression profiles between male osteoporosis and normal control by high throughput RNA sequencing. Based on functional annotation, we identified several DElncRNA-DEGs interaction pairs, which may be associated with the process of male osteoporosis.
Compared to normal control, HBB (hemoglobin subunit beta) was the most significantly up-regulated gene, while HBA1 was a significantly down-regulated gene in male osteoporosis. HBB and HBA1 encode the β and α 1 subunit of hemoglobin, respectively. As a major component of hemoglobin, iron is an important factor of bone metabolism. Iron can disorder the process of bone formation, make toxic effects on osteoblasts and reduce the recruitment of osteoblastic-lineage cells.16 It is pointed out that hemoglobin level is related to BMD and bone mass negatively and independently.17 Moreover, higher bone loss was found in men with lower hemoglobin level than women with lower hemoglobin level.17 Moreover, we found an interaction of “SNHG5-SYNCRIP-HBA1-HBB” based on the DElncRNA-nearby targeted DEGs interaction network. SYNCRIP (synaptotagmin binding cytoplasmic RNA interacting protein) was a down-regulated gene in male osteoporosis compared to normal control. SYNCRIP has been reported to be involved in bone morphogenesis protein signaling pathway.18 These three DEGs (HBA1, HBB and SYNCRIP) may be involved in the pathogenesis of male osteoporosis by their interactions. Small nucleolar RNA host gene 5 (SNHG5) is a member of the non-protein-coding multiple snoRNA host gene family which was significantly down-regulated in male osteoporosis in our study. SNHG5 has been reported to be associated with various cancers.19,20 SNHG5 is involved in gastric cancer cell proliferation and migration.19 SNHG5 can be served as a tumor marker of malignant melanoma20 and promotes tumor cell survival in colorectal cancer.19 However, no association between SNHG5 and male osteoporosis has been reported. Our study suggested that down-regulated SNHG5 may play an important role in male osteoporosis through regulating the expression of SYNCRIP, HBB and HBA1.
Based on the male osteoporosis-specific PPI network, major histocompatibility complex, class I, C (HLA-C) was a hub gene, which was up-regulated in male osteoporosis compared to normal control. HLA-C is a member of the HLA class I heavy chain paralogues. HLA class I molecule consists of a heavy chain and a light chain which plays a key role in the immune system. Accumulated evidence has demonstrated the relationship between the immune system and bone.21 T cells play a crucial role in the formation and activity of osteoclast and osteoblast in multiple skeletal diseases, such as osteoporosis.22 The production of T-lymphocyte, T-helper and T-cytotoxic cells is decreased with aging.23 Additionally, other osteoporosis-related factors including estrogen, calcium, parathyroid hormone, and vitamin D play a regulatory in immune system.23 The role of immune system in the pathogenesis of osteoporosis is mainly induced by decreased estrogen and secondary hyperparathyroidism.23 In this study, HLA-C was a targeted nearby cis-DEG of HCG27 (HLA complex group 27). In this study, HCG27 was a significantly up-regulated lncRNA in male osteoporosis. Our finding emphasized the importance of HCG27 in the process of male osteoporosis.
Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) was another hub gene based on PPI network which was up-regulated in male osteoporosis. Depending on the DElncRNAs-nearby targeted DEGs interaction network, IFIT1 could be interacted with three other up-regulated DEGs including IFIT2, IFIT3 and IFIT5. These four genes were interferon-induced proteins that contain tetratricopeptide repeats.24 As an innate immune bottleneck, IFIT1 is expressed during the response of osteoblasts to immune cytokine interferon-β.25 IFIT2 is a regulator of innate immunity as well. Both IFIT1 and IFIT2 play a role in the fracture healing process in osteoporosis,26 which provides new clues for osteoporotic fracture repair therapies. LOC101926887 can interact with the IFIT1-IFIT2, IFIT1-IFIT3 and IFIT1-IFIT5 interaction pairs. In addition, LOC101926887 was one of the up-regulated lncRNA covered most nearby targeted DEGs. Through regulating the expression of targeted nearby DEGs (IFIT1, IFIT2, IFIT3, IFIT15 and LIPA), LOC101926887 may be an important regulator of male osteoporosis. LOC100131626 and TMX2-CTNND1 were the other two DElncRNAs that covered most nearby targeted DEGs in male osteoporosis. LOC100131626 was a down-regulated lncRNA in male osteoporosis containing five targeted nearby DEGs (KMT2A, ATP5L, UBE4A, CD3D and CD3E). TMX2-CTNND1 was an up-regulated lncRNA in male osteoporosis containing five targeted nearby DEGs (CTNND1, TMX2, ZDHHC5, CLP1 and YPEL4). These two DElncRNAs may play crucial roles in male osteoporosis through regulating their nearby targeted DEGs as well.
Compared to normal control, Loc105372801 and KCNQ1OT1 was the most significantly up-regulated and down-regulated DElncRNA in male osteoporosis, respectively. No previous studies have reported functions of Loc105372801 and KCNQ1OT1 in male osteoporosis. KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) is only expressed on the paternal allele.27 KCNQ1OT1 is closely related to the pathogenesis of familial Beckwith-Wiedemann syndrome.28 Loss of imprinting of KCNQ1OT1 locus by epigenetic disruption is related to colorectal carcinogenesis.29–31 A novel tetranucleotide repeat polymorphism (rs35622507) within KCNQ1OT1 confers risk for hepatocellular carcinoma.30 KCNQ1OT1 involves in breast cancer through the epigenetic suppression of the cyclin-dependent kinase inhibitor 1C.31 The precise function of KCNQ1OT1 in male osteoporosis needs further research.
In our study, chemokine signaling pathway was a significantly enriched pathway in male osteoporosis. As one of the DEGs enriched in chemokine signaling pathway, C-C motif chemokine receptor 2 (CCR2) is a receptor for ligands like monocyte chemoattractant protein-1 (MCP-1). CCR2 is served as a regulator of bone loss, which is expressed on preosteoclasts and osteoclasts.32 The MCP-1/CCR2 axis has been demonstrated to be involved in fusion and maturation of osteoclast and bone resorption.33,34 In addition, gene variants of MCP-1 and CCR2 are reported to be risk factors for osteoporosis.35 In young adult mice, knockout of the Ccr2 results in delay in fracture healing.36,37 In the present study, CCR2 and LINC02009 were all significantly down-regulated in male osteoporosis, which was validated by qRT-PCR. CCR2 was a nearby targeted DEG of a down-regulated DElncRNA, LINC02009. Our result suggested that LINC02009 may be associated with the process of osteoporosis by regulating the expression of CCR2 and other nearby targeted DEGs including lactotransferrin (LTF) and leucine-rich repeat containing 2 (LRRC2).
Osteoclast differentiation was another significantly enriched pathway for DEGs in male osteoporosis which was demonstrated to be crucial pathway of osteoporosis. These DEGs enriched in osteoclast differentiation may be important regulators in male osteoporosis as well.
Taken together, identified several DElncRNA-nearby targeted DEGs interaction pairs such as SNHG5-SYNCRIP-HBA1-HBB, HCG27-HLA-C, LINC02009-CCR2, LOC101926887-IFIT1-IFIT2, LOC101926887-IFIT1-IFIT3 and LOC101926887-IFIT1-IFIT5 may play an important role in the pathogenesis of male osteoporosis. The validated DElncRNA-nearby targeted DEGs interaction pair of LINC02009-CCR2 may be important regulators in the process of male osteoporosis as well. Our findings may contribute to underlying the function of lncRNAs and mechanism of male osteoporosis and provide new clues for novel diagnostic and therapeutic strategies for male osteoporosis. However, there are limitations to our study. Firstly, the sample size in the RNA sequencing is small, larger numbers of blood samples or bone samples are further needed for further validation of identified lncRNAs and genes. Secondly, we did not validate the expression of identified other DElncRNAs and DEGs in male osteoporosis or any cell lines. Thirdly, we did not perform the deeper mechanism study of male osteoporosis, some in vivo or in vitro experiments are further needed.
Abbreviations
BMD, bone mineral density; CCR2, C-C motif chemokine receptor 2; DEGs, differentially expressed genes; DElncRNAs, differentially expressed lncRNAs; FDR, false discovery rate; GO, Gene Ontology; HCG27, HLA complex group 27; HBB, hemoglobin subunit beta; HLA-C, histocompatibility complex, class I, C; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; IL-1, interleukin-1; IL-6, interleukin-6; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; LTF, lactotransferrin; LRRC2, leucine-rich repeat containing 2; lncRNAs, long non-coding RNAs; M-CSF, macrophage colony-stimulating factor; MCP-1, monocyte chemoattractant protein-1; PCR, polymerase chain reaction; RANKL, receptor activator of nuclear factor-kB ligand; RIN, RNA integrity number; SNHG5, small nucleolar RNA host gene 5; SYNCRIP, synaptotagmin binding cytoplasmic RNA interacting protein; TNF-α, tumor necrosis factor-α; WHO, world health organization.
Acknowledgment
This work was supported by Beijing health system high-level health technical personnel training project (NO: 2015-3-009).
Disclosure
The authors report no conflicts of interest for this work.
References
1. National Institutes of Health. Nih consensus development panel on osteoporosis prevention, diagnosis, and therapy, march 7–29, 2000. South Med J. 2001;94:569–573.
2. Khosla S, Amin S, Orwoll E. Osteoporosis in men. Endocr Rev. 2008;29:441–464.
3. Hannan MT, Felson DT, Dawson-Hughes B, et al. Risk factors for longitudinal bone loss in elderly men and women: the framingham osteoporosis study. J Bone Miner Res. 2000;15:710–720. doi:10.1359/jbmr.2000.15.4.710
4. Johnell O, Kanis JA. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos Int. 2006;17:1726–1733. doi:10.1007/s00198-006-0172-4
5. Cano A, Chedraui P, Goulis DG, et al. Calcium in the prevention of postmenopausal osteoporosis: emas clinical guide. Maturitas. 2018;107:7–12. doi:10.1016/j.maturitas.2017.10.004
6. Wein MN, Kronenberg HM. Regulation of bone remodeling by parathyroid hormone. Cold Spring Harb Perspect Med. 2018;8:a031237. doi:10.1101/cshperspect.a031237
7. Liyuan L, Zhao W. Ovarian aging and osteoporosis. Adv Exp Med Biol. 2018.
8. Bellavia D, Costa V, De Luca A, et al. Vitamin d level between calcium-phosphorus homeostasis and immune system: new perspective in osteoporosis. Curr Osteoporos Rep. 2016. doi:10.1007/s11914-016-0331-2
9. Bai P, Sun Y, Jin J, et al. Disturbance of the OPG/RANK/RANKL pathway and systemic inflammation in COPD patients with emphysema and osteoporosis. Respir Res. 2011;12:157. doi:10.1186/1465-9921-12-157
10. Matuszewska A, Szechiński J. [Mechanisms of osteoporosis development in patients with rheumatoid arthritis]. Postepy Hig Med Dosw (Online). 2014;68:145–152. doi:10.5604/17322693.1088339
11. Lan X, Zhang H, Wang Z, et al. Genome-wide analysis of long noncoding RNA expression profile in papillary thyroid carcinoma. Gene. 2015;569:109–117. doi:10.1016/j.gene.2015.05.046
12. Wang Q, Li Y, Zhang Y, et al. LncRNA meg3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting mir-133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi:10.1016/j.biopha.2017.02.090
13. Tong X, Gu PC, Xu SZ, Lin XJ. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem. 2015;79:732–737. doi:10.1080/09168451.2014.998617
14. Wei MM, Zhou GB. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics. 2016;14:280–288. doi:10.1016/j.gpb.2016.03.007
15. Huynh NP, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58:116–141. doi:10.1080/03008207.2016.1194406
16. Shen GS, Yang Q, Jian JL, et al. Hepcidin1 knockout mice display defects in bone microarchitecture and changes of bone formation markers. Calcif Tissue Int. 2014;94:632–639. doi:10.1007/s00223-014-9845-8
17. Cesari M, Pahor M, Lauretani F, et al. Bone density and hemoglobin levels in older persons: results from the InCHIANTI study. Osteoporos Int. 2005;16:691–699. doi:10.1007/s00198-004-1739-6
18. Halstead JM, Lin YQ, Durraine L, et al. Syncrip/hnRNP Q influences synaptic transmission and regulates BMP signaling at the drosophila neuromuscular synapse. Biol Open. 2014;3:839–849. doi:10.1242/bio.20149027
19. Damas ND, Marcatti M, Côme C, et al. Snhg5 promotes colorectal cancer cell survival by counteracting stau1-mediated mRNA destabilization. Nat Commun. 2016;7:13875.
20. Ichigozaki Y, Fukushima S, Jinnin M, et al. Serum long non-coding RNA, snoRNA host gene 5 level as a new tumor marker of malignant melanoma. Exp Dermatol. 2016;25:67–69. doi:10.1111/exd.12868
21. Faienza MF, Ventura A, Marzano F, Cavallo L. Postmenopausal osteoporosis: the role of immune system cells. Clin Dev Immunol. 2013;2013:575936. doi:10.1155/2013/575936
22. D’Amelio P, Grimaldi A, Di Bella S, et al. Estrogen deficiency increases osteoclastogenesis up-regulating t cells activity: a key mechanism in osteoporosis. Bone. 2008;43:92–100. doi:10.1016/j.bone.2008.02.017
23. Clowes JA, Riggs BL, Khosla S. The role of the immune system in the pathophysiology of osteoporosis. Immunol Rev. 2005;208:207–227. doi:10.1111/j.0105-2896.2005.00334.x
24. Wathelet MG, Clauss IM, Content J, Huez GA. The IFI-56K and IFI-54K interferon-inducible human genes belong to the same gene family. FEBS Lett. 1988;231:164–171. doi:10.1016/0014-5793(88)80724-5
25. McDermott JE, Vartanian KB, Mitchell H, et al. Identification and validation of ifit1 as an important innate immune bottleneck. PLoS One. 2012;7:e36465. doi:10.1371/journal.pone.0036465
26. Gao F, Xu F, Wu D, Cheng J, Xia P. Identification of novel genes associated with fracture healing in osteoporosis induced by krm2 overexpression or lrp5 deficiency. Mol Med Rep. 2017;15:3969–3976. doi:10.3892/mmr.2017.6544
27. Sunamura N, Ohira T, Kataoka M, et al. Regulation of functional kcnq1ot1 lncRNA by β-catenin. Sci Rep. 2016;6:20690. doi:10.1038/srep20690
28. H’Mida Ben-Brahim D, Hammami S, Haddaji Mastouri M, et al. Partial kcnq1ot1 hypomethylation: a disguised familial beckwith-wiedemann syndrome as a sporadic adrenocortical tumor. Appl Transl Genom. 2015;4:1–3. doi:10.1016/j.atg.2014.10.001
29. Nakano S, Murakami K, Meguro M, et al. Expression profile of lit1/kcnq1ot1 and epigenetic status at the kvdmr1 in colorectal cancers. Cancer Sci. 2006;97:1147–1154. doi:10.1111/j.1349-7006.2006.00305.x
30. Wan J, Huang M, Zhao H, et al. A novel tetranucleotide repeat polymorphism within kcnq1ot1 confers risk for hepatocellular carcinoma. DNA Cell Biol. 2013;32:628–634. doi:10.1089/dna.2013.2118
31. Rodriguez BAT, Weng YI, Liu TM, et al. Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1c in breast cancer cells. Carcinogenesis. 2011;32:812–821. doi:10.1093/carcin/bgr017
32. Binder NB, Niederreiter B, Hoffmann O, et al. Estrogen-dependent and c-c chemokine receptor-2-dependent pathways determine osteoclast behavior in osteoporosis. Nat Med. 2009;15:417–424. doi:10.1038/nm.1945
33. Sul OJ, Ke K, Kim WK, et al. Absence of MCP-1 leads to elevated bone mass via impaired actin ring formation. J Cell Physiol. 2012;227:1619–1627. doi:10.1002/jcp.22879
34. Mader TL, Novotny SA, Lin AS, et al. Ccr2 elimination in mice results in larger and stronger tibial bones but bone loss is not attenuated following ovariectomy or muscle denervation. Calcif Tissue Int. 2014;95:457–466. doi:10.1007/s00223-014-9914-z
35. Eraltan H, Cacina C, Kahraman OT, et al. Mcp-1 and ccr2 gene variants and the risk for osteoporosis and osteopenia. Genet Test Mol Biomarkers. 2012;16:229–233. doi:10.1089/gtmb.2011.0216
36. Xing Z, Lu C, Hu D, Iii TM, Marcucio RS. Rejuvenation of the inflammatory system stimulates fracture repair in aged mice. J Orthop Res. 2010;28(8):1000–1006. doi:10.1002/jor.21087
37. Xing Z, Lu C, Hu D, et al. Multiple roles for ccr2 during fracture healing. Dis Model Mech. 2010;3:451–458. doi:10.1242/dmm.003186
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.