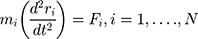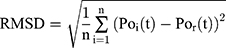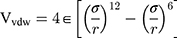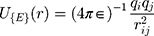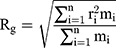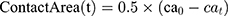Back to Journals » International Journal of Nanomedicine » Volume 15
Graphene-Based Nanoparticles as Potential Treatment Options for Parkinson’s Disease: A Molecular Dynamics Study
Authors Alimohammadi E , Khedri M, Miri Jahromi A, Maleki R , Rezaian M
Received 1 June 2020
Accepted for publication 8 September 2020
Published 18 September 2020 Volume 2020:15 Pages 6887—6903
DOI https://doi.org/10.2147/IJN.S265140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Ehsan Alimohammadi1 *,* Mohammad Khedri2 *,* Ahmad Miri Jahromi,3 Reza Maleki,4 Milad Rezaian5
1Neurosurgery Department, Kermanshah University of Medical Sciences, Kermanshah, Iran; 2Department of Chemical Engineering, Amirkabir University of Technology (Tehran Polytechnic), Tehran 1591634311, Iran; 3Department of Petroleum Engineering, Amirkabir University of Technology (Tehran Polytechnic), Tehran 1591634311, Iran; 4Department of Chemical Engineering, Sharif University of Technology, Tehran, Iran; 5Department of Pharmacology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran 19839-63113, Iran
*These authors contributed equally to this work
Correspondence: Reza Maleki
Department of Chemical Engineering, Sharif University of Technology, Tehran, Iran
Email [email protected]
Introduction: The study of abnormal aggregation of proteins in different tissues of the body has recently earned great attention from researchers in various fields of science. Concerning neurological diseases, for instance, the accumulation of amyloid fibrils can contribute to Parkinson’s disease, a progressively severe neurodegenerative disorder. The most prominent features of this disease are the degeneration of neurons in the substantia nigra and accumulation of α-synuclein aggregates, especially in the brainstem, spinal cord, and cortical areas. Dopamine replacement therapies and other medications have reduced motor impairment and had positive consequences on patients’ quality of life. However, if these medications are stopped, symptoms of the disease will recur even more severely. Therefore, the improvement of therapies targeting more basic mechanisms like prevention of amyloid formation seems to be critical. It has been shown that the interactions between monolayers like graphene and amyloids could prevent their fibrillation.
Methods: For the first time, the impact of four types of last-generation graphene-based nanostructures on the prevention of α-synuclein amyloid fibrillation was investigated in this study by using molecular dynamics simulation tools.
Results: Although all monolayers were shown to prevent amyloid fibrillation, nitrogen-doped graphene (N-Graphene) caused the most instability in the secondary structure of α-synuclein amyloids. Moreover, among the four monolayers, N-Graphene was shown to present the highest absolute value of interaction energy, the lowest contact level of amyloid particles, the highest number of hydrogen bonds between water and amyloid molecules, the highest instability caused in α-synuclein particles, and the most significant decrease in the compactness of α-synuclein protein.
Discussion: Ultimately, it was concluded that N-Graphene could be the most effective monolayer to disrupt amyloid fibrillation, and consequently, prevent the progression of Parkinson’s disease.
Keywords: α-synuclein, amyloid, graphene, Parkinson’s disease, molecular dynamics
Introduction
According to the World Health Organization, recent evidence has revealed the accumulation of amyloid molecules to be the fundamental mechanism underlying many health care disorders. (KT= 2.479 kJ/mol) A significant number of diseases, including many neurodegenerative disorders, are known to be associated with the formation of stable, fibrillar protein aggregates called amyloids.1,3 These aggregations are made in specific steps, during which a specific protein or part of a protein changes from its natural soluble state to insoluble form and eventually evolves into giant fibrillar proteins that accumulate in various tissues and organs of the body.4–6 Parkinson’s disease, one of the common motor abnormalities, is the second most well-known neurodegenerative disorder after Alzheimer’s.7 It is caused by the destruction of dopamine-producing cells in the substantia nigra of the brain’s basal ganglia.8 As the brain’s dopamine reserves decrease in proportionate to acetylcholine, symptoms of the disease appear. So far, there has been no definitive cure for Parkinson’s, even though many drugs such as levodopa, amantadine, biperiden, and selegiline have been used to treat it9–13 As far as the patients take these drugs, their symptoms are controlled. If, however, these medications are stopped, symptoms of the disease will recur even more severely. This fact highlights the importance of developing medications that are targeted on more basic underlying mechanisms of the disease. It is believed that one pathophysiology beyond the dopamine-producing cell death involves the abnormal accumulation of amyloid fibrils in damaged cells. Amyloid fibrils consist of beta-sheeted α-synuclein monomers, in which the beta strands are perpendicular to the principal axis of the fibril. Alpha-synuclein is a member of the synuclein proteins which have critical intracellular functions. Even though the precise actions of natural α-synuclein protein in the nervous system are remained unclear, studies have revealed this protein to play critical roles in the synaptic transmission, including the synthesis, axonal transport, release, and recycling of neurotransmitters. Functional proteins usually do not form amyloids unless they lose their natural folding. The process can be self-generated, meaning that a misfolded protein can induce the same dysfunction in other similar proteins. The misfolded peptides can form soluble aggregates known as oligomers, which evolve into protofibrils, amyloid fibrils, and Lewy bodies, all of which have cytotoxic effects and lead to the death of neurons by a variety of intracellular mechanisms.14–17 Figure 1 illustrates the process of amyloid formation and Parkinson’s disease development.
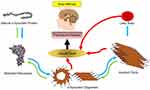 |
Figure 1 The mechanism underlying the development of Parkinson’s disease. |
Proteins may bind to monolayers in natural or denatured states, depending on the surface properties of the protein (such as load, hydrophobicity, stability) and the monolayer (hydrophobicity, size, coating). The properties of monolayers can affect the self-assembly and biological functions of proteins.18 According to studies,19–23 monolayers can affect the fibrillation process with different mechanisms which could be listed as follows:
- Monolayers can increase the number of hydrogen bonds between amyloid fibrils and water molecules, and therefore, block the active elongation site of the fibrils;18
- Monolayers can prevent the fibrillation of proteins by disrupting their secondary structure;
- The high local concentration of proteins on the surface of some monolayers can lead to the formation of critical nuclei, and in such cases, facilitate the formation of amyloid fibrils;18
- By their strong interactions with proteins, the monolayer can reduce the concentration of proteins in the solution, which will delay the appearance of critical nuclei, and hence, prevent fibril formation;18
- Specific monolayers can act as chaperons.24 Chaperons can bind to proteins through hydrophobic and electrostatic interactions, prevent their aggregation, and help them return to their original folding state.25
Several studies have been performed to investigate the use of monolayers as potential agents to prevent amyloid fibrillation. In these studies, quantum dots (QDs),20 gold nanoparticles,21–23 superparamagnetic iron oxide nanoparticles (SPION), graphene-based structures with positive and negative charges19 have been investigated.
With the discovery of graphene, the use of two-dimensional structures in nanomaterial-based systems came into consideration.26–30 Generally, each carbon atom can make covalent bonds with a maximum of four other carbon atoms, forming a three-dimensional network, known as the diamond. If, however, each carbon atom only bonds with three other carbon atoms, a two-dimensional carbon allotrope is formed, called graphene. The surface area of graphene is much larger than that of carbon black and activated carbon. The hexagonal pattern of carbon atoms in graphene has turned this nanostructure into a giant aromatic molecule with unique electrical, optical, and thermal properties and excellent mechanical strength despite its small thickness and relatively low molecular weight. These features have facilitated the use of graphene in a variety of fields, including electronics, pharmaceutics, and the textile industry.31–33 In order to improve its physicochemical properties as to fit nanomaterial practice better, the basic structure of graphene has undergone novel modifications through which new generations of this carbon allotrope have been developed over time. Second-generation graphene molecules were established by chemical functionalization. This method has been vastly employed to improve the properties of this monolayer for many years. However, despite their advantageous features, chemically functionalized graphenes presented certain limitations as their potential utilities for innovative purposes began to emerge. Attempts to further adjust graphene molecules as to be suited for novel implementations gave rise to the emergence of last-generation carbon sheet allotropes or -doped graphenes. Such structures are designed as a proportion of carbon atoms in the mainstay composition are replaced with other elements such as bromine, nitrogen, phosphorus, etc.1,34,35
Graphene-based nanomaterials like all of the engineered biomaterials, have been shown to have cytotoxic effects. However, in order to reach the toxic level, they have to be administered more than their toxic dose, so future experimental animal studies and clinical trials should be conducted in a way that the administered doses of graphene -based nanomaterials do not reach the toxic dose.36–40 Functionalized graphene, such as carboxylated graphene, has shown much lower toxicity and better hydrophilicity than pristine graphene. However, doped graphene represents a newer generation of graphene-based nanomaterials which has been shown to have more desirable chemical properties and better biocompatibility36,41–44.
The implementation of graphene-based nanomaterials for the treatment of Parkinson’s disease have been the topic of a number of previously published computational studies, some of which have also been backed up by experimental evidence. Such researches contributed to the development of the main concept of this study which was performed.19,36,41,44–47 This study has investigated four types of last-generation graphene-based nanostructures as potential agents to disrupt the process of α-synuclein amyloid formation in Parkinson’s disease by using molecular dynamics (MD) simulation techniques. Different parameters involved in the interactions between α-synuclein proteins and the nanostructures have been analyzed. These monolayers include graphene, nitrogen-doped graphene (N-Graphene), phosphorus-doped graphene (P-Graphene), and graphene co-duped with bromine and nitrogen (BCN). Moreover, in order to validate the methods and algorithms used in this paper, an additional simulation of graphene polyglycerol interactions with α-synuclein protein was performed with conditions identical to a previously published similar study by Mohammad-Beigi et al, and the results were compared with each other.
Materials and Methods
Molecular Dynamics Simulation
Molecular dynamics (MD) is a method for analyzing the physical motion of atoms and molecules. Molecular dynamics simulations produce information at the microscopic level (position and velocity of atoms). In simulating the molecular dynamics, atoms and molecules interact with each other over a period of time.48 The trajectory of atoms and molecules is determined by the numerical solution of Newton’s motion equation. A molecular system typically contains a large number of components, and the properties of such complex systems cannot be determined analytically. For this reason, numerical methods are used.
These data are converted to macroscopic values (such as pressure and energy) using statistical mechanics. MD and statistical mechanics link microscopic concepts and observable macroscopic quantities. In molecular dynamics simulations, Newton’s second law is used to examine the temporal evolution of systems. By solving Newton’s motion equations, the position of the atom (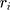 ) in a
) in a 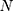 atomic system is obtained:
atomic system is obtained:
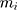 is the mass of the atom, and
is the mass of the atom, and 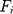 is the force that other atoms exert on atom
is the force that other atoms exert on atom  . In fact, in MD simulation methods with initial positions and velocities, the temporal evolution of the system can be examined. By integrating Newton’s motion equations, a path is obtained that shows how the velocity, position, and acceleration of particles change over time. In molecular dynamics, to measure a physical quantity, the average of that quantity is calculated at the time of simulation in the system path.49–51
. In fact, in MD simulation methods with initial positions and velocities, the temporal evolution of the system can be examined. By integrating Newton’s motion equations, a path is obtained that shows how the velocity, position, and acceleration of particles change over time. In molecular dynamics, to measure a physical quantity, the average of that quantity is calculated at the time of simulation in the system path.49–51
Simulation Method
Alpha-protein-synuclein contains 140 amino acids. This protein is made up of alpha, beta, gamma-synuclein and synutrin structures. The amount of alpha-synuclein protein in the central nervous system (CNS) is very high. Alpha-synuclein is naturally present in the cell cytoplasm. In this study we used from molecular structure of the alpha-synuclein protein that has been studied by Ulmer et al.52
The graphene structure is constructed by Nanotube_Modeler_1.7.9,53,54 with a size of 70 \AA \times 170 \AA in X-Y Cartesian coordinates. Then, with the help of Avogadro software, the molecular structures of BCN monolayers, a combination of nitrogen and bromine atoms with graphene carbon atoms, N-Graphene monolayers, a combination of nitrogen atoms with graphene carbon atoms, P-Graphene monolayers, a combination of phosphorus atoms with graphene carbon atoms, and pure graphene were made. The molecular structures of nitrogen, bromine, and phosphorus-graphene graphene have been optimized by Gaussian 09 software.55,56
To perform the simulation, the monolayer topology was designed using the x2top command and the amyloid topology by the pdb2gmx command for the optimized potentials for liquid simulations-All Atoms (OPLS-AA) force field.57,58 The simulation is performed in 4 steps, with energy level optimization and 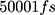 time steps. The simulation system is coupled with the Berendsen thermostat, considering the equilibrium temperature of
time steps. The simulation system is coupled with the Berendsen thermostat, considering the equilibrium temperature of 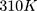 in the nvt step. The execution time of the nvt step was 100 picoseconds.59–61 Moreover, the npt step is balanced by the Parrinello_Rahman algorithm in 10 nanoseconds at 1 bar.62–64 At the end of the simulation, with considering the cut-off radius is 1.4 nanometers and h_bond idone by the lincs algorithm in 40 nanoseconds. Last but not least, simulation images were taken by vmd software.
in the nvt step. The execution time of the nvt step was 100 picoseconds.59–61 Moreover, the npt step is balanced by the Parrinello_Rahman algorithm in 10 nanoseconds at 1 bar.62–64 At the end of the simulation, with considering the cut-off radius is 1.4 nanometers and h_bond idone by the lincs algorithm in 40 nanoseconds. Last but not least, simulation images were taken by vmd software.
Root-Mean-Square Deviation Analysis
Root-Mean-Square Deviation (RMSD) values are calculated by Equation (2). In Equation (2),  represents the number of particles,
represents the number of particles, 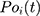 and
and 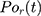 show the position of the particle
show the position of the particle  and the reference particle at time
and the reference particle at time  .
.
Energy Analysis
Van Der Walls (VDW) energy is calculated based on the Lenard-Jones equation (Equation 3). The energy of electrostatic interactions follows Columbus’ law. Colum’s law is shown in Equation 4.
where  is the distance between these two particles,
is the distance between these two particles, 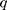 is the amount of charge mentioned in the topology for each particle,
is the amount of charge mentioned in the topology for each particle, 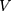 is the potential between the two atoms, and is the depth of the potential well. In this paper, energy analysis for different simulations is taken by mmpbsa software.65,66
is the potential between the two atoms, and is the depth of the potential well. In this paper, energy analysis for different simulations is taken by mmpbsa software.65,66
Gyration Radius
The gyration radius is calculated by Equation (5):
where  is the number of particles,
is the number of particles, 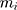 , the mass of the particle
, the mass of the particle  , and
, and 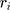 indicate the distance of the particle
indicate the distance of the particle  from the center of gravity. To measure the density of amyloid particles, an analysis of the radius of amyloid gyration was performed in all simulations.
from the center of gravity. To measure the density of amyloid particles, an analysis of the radius of amyloid gyration was performed in all simulations. 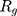 , helps to understand protein density during the simulation, where for higher
, helps to understand protein density during the simulation, where for higher 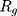 , higher density is expected.
, higher density is expected.
SASA
In this paper, the solvent accessible surface area (SASA) analysis for amyloid particles is taken in all simulations. The contact level of amyloid particles in this paper is calculated by Equation 6.
where 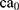 and
and 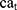 show the amount of SASA analysis at zero and at time
show the amount of SASA analysis at zero and at time  , respectively. Mohammad-Beigi et al19 have simulated graphene polyglycerol and amyloid in water using the OPLS-AA force field by Gromacs. In this study, the simulation algorithms, force field, molecular structures and simulation time were selected according to Mohammad-Beigi et al19 and the LINCS algorithm with the cut-off radius of 1.4 nm was implemented.
, respectively. Mohammad-Beigi et al19 have simulated graphene polyglycerol and amyloid in water using the OPLS-AA force field by Gromacs. In this study, the simulation algorithms, force field, molecular structures and simulation time were selected according to Mohammad-Beigi et al19 and the LINCS algorithm with the cut-off radius of 1.4 nm was implemented.
Results and Discussion
Formation of α-Synuclein Amyloids
Figure 2–3 illustrate how amyloid formation is affected by monolayers during 40 nanoseconds simulation. In order to form amyloids, α-synuclein proteins first become misfolded and then gradually rise to amyloids. Figure 2E shows the misfolded form of α-synuclein protein at the end of the simulation in the absence of monolayers. As can be seen in Figure 2A-D, the presence of monolayers disrupts the misfolding of α-synuclein proteins, giving rise to other forms which are not suitable for amyloid formation. Among these monolayers, N-Graphene provides better prevention of α-synuclein misfolding.
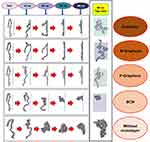 |
Figure 3 Changes in the α-synuclein structure during the simulation time in the presence of each monolayer. |
Evaluating the Interaction Energies
The amount and type of interaction energy are essential parameters as far as the interactions between monolayers are concerned. In general, the greater the absolute amount of interaction energy, the stronger are the interactions between the particles. This fact could play an important role in the prevention of amyloid formation. More absolute amounts of the interaction energy between the monolayers and α-synuclein proteins indicate better prevention of amyloid fibrillation. The type of interaction energy, whether electrostatic or VDW, is also essential.67–70
Figure 4 shows the energies of interactions between α-synuclein proteins and each of the monolayers versus the simulation time. These diagrams reveal the existence of considerable interactions between α-synuclein proteins and all monolayers. Negative energy values in most parts of the diagrams reveal the existence of attractive forces between the particles. It is also noted that VDW energy plays a vital role in the simulated interactions than electrostatic energy. Among these diagrams, Figure 4B and C present the highest and the lowest absolute energy values, respectively, which means that N-Graphene makes stronger interactions with α-synuclein proteins than other monolayers. The absolute total energy values in diagrams b and c increase monotonically, which reveals that N-Graphene and P-Graphene possess more stable interactions with increasing strength with α-synuclein proteins. On the other hand, the fluctuations in diagrams a and d reveal that the strength of interactions made between graphene and BCN and α-synuclein proteins suddenly changes during the simulation.
 |
Figure 4 Van der Waals, electrostatic, and total energy of interactions between α-synuclein protein and each monolayer versus time: (A) Graphene; (B) N- Graphene; (C) P- Graphene; (D) BCN. |
Table 1 shows the average values for electrostatic, VDW, and the total energies of the interactions between α-synuclein proteins and each of the monolayers. According to the values reported in the table, the highest absolute interaction energy belongs to N-Graphene, followed by graphene, BCN, and P-Graphene. Based on Figure 4 and Table 1, it can be concluded that N-Graphene makes stronger bonds with α-synuclein proteins, and therefore, can prevent the formation of α-synuclein amyloids more effectively than the other monolayers.
 |
Table 1 Average Values for VDW, Electrostatic, and the Total Energy of Interactions Between α-Synuclein Protein and Monolayers During the Simulation Process Monolayer |
Evaluating the Compactness of α-Synuclein
In order to form critical nuclei and make aggregations, particles need to become more compact. A critical indicator of the compactness of protein structures is the Gyration Radius (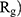 . The smaller the value of
. The smaller the value of 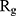 , the more compact is the macromolecule being investigated. Therefore, concerning α-synuclein proteins, the variation of
, the more compact is the macromolecule being investigated. Therefore, concerning α-synuclein proteins, the variation of 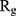 during the simulation indicates the possibility of prevention of amyloid formation.71–75
during the simulation indicates the possibility of prevention of amyloid formation.71–75
Figure 5 shows 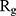 of α-synuclein proteins versus time with and without the presence of monolayers. According to the diagrams,
of α-synuclein proteins versus time with and without the presence of monolayers. According to the diagrams, 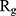 has been increased in the presence of monolayers, which means that they have decreased compactness of α-synuclein proteins, making the process of amyloid formation less likely. It is also noted that N-Graphene and graphene have caused higher values of
has been increased in the presence of monolayers, which means that they have decreased compactness of α-synuclein proteins, making the process of amyloid formation less likely. It is also noted that N-Graphene and graphene have caused higher values of 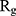 compared to P-Graphene and BCN.
compared to P-Graphene and BCN.
 |
Figure 5 Rg of α-synuclein proteins versus the simulation time in the presence of (A) Graphene; (B) N- Graphene; (C) P- Graphene; (D) BCN; (E) Without monolayers. |
Table 2 indicates the variation of 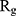 during 40 nanoseconds. This table shows that in the presence of monolayers, slighter decrease in
during 40 nanoseconds. This table shows that in the presence of monolayers, slighter decrease in 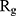 occurs, where among the monolayers, N-Graphene has the least variation, followed by graphene, P-Graphene, and BCN. Based on Figure 5 and Table 2, although all the monolayers can reduce the compactness of α-synuclein proteins and decrease the likelihood of amyloid formation, N-Graphene has the most desired effect followed by graphene, P-Graphene, and BCN Table 2.
occurs, where among the monolayers, N-Graphene has the least variation, followed by graphene, P-Graphene, and BCN. Based on Figure 5 and Table 2, although all the monolayers can reduce the compactness of α-synuclein proteins and decrease the likelihood of amyloid formation, N-Graphene has the most desired effect followed by graphene, P-Graphene, and BCN Table 2.
 |
Table 2 Values of the Gyration Radius Variation for α-Synuclein Proteins in the Presence of Each Monolayer |
Evaluating the Hydrogen Bonds
Hydrogen bonding is the strongest intermolecular bond.76–78 Concerning the formation of α-synuclein amyloids, the existence of abundant hydrogen bonds between α-synuclein proteins and water molecules can interrupt their movement in the aqueous media and disrupt the process of amyloid formation.
Figure 6 shows the number of hydrogen bonds versus time during the simulation. By comparing the diagrams, it is noticed that in the presence of N-Graphene, the number of hydrogen bonds between α-synuclein proteins and water molecules is substantially increased compared to other monolayers.In order to make a quantified comparison of the effectiveness of monolayers to increase the number of hydrogen bonds between α-synuclein proteins and water molecules, the average number of hydrogen bonds for each simulation is shown in Table 3 Although all monolayers increased the number of hydrogen bonds between α-synuclein proteins and water molecules, the most significant effect is for N-Graphene. Therefore N-Graphene is the best in preventing the amyloid formation and ceasing the progress of Parkinson’s disease Table 3.
 |
Table 3 The Average Number of Hydrogen Bonds Between α-Synuclein Protein and Water Molecules During the Simulation Time in the Presence of Each Monolayer |
Evaluating the Secondary Structure of α-Synuclein
Intramolecular hydrogen bonds form the secondary structure of proteins. Depending on the position of the bonds and the types of involved amino acids, the secondary structure of proteins can be classified into several categories. The most important types of the secondary structure consist of the α-helix and β-sheet. Other less common structures include 3-helices, π-helices, β-bridges, turns, bends, and coils. One single protein could have a combination of these secondary structures throughout its residues. Nevertheless, the secondary structure of a protein and its functionality can change in different cellular conditions.
The α-synuclein protein is naturally a membrane-bound protein consisting of 140 amino acids. The analysis of the structure of α-synuclein reveals that the entire length of the protein can be divided into three domains with different structures and functions. The N-terminal, consisting of amino acids 1–60, is an amphipathic domain that interacts with phospholipid membranes. It consists of two α-helices in the membrane-bound state, while parts of which may turn into random coils under certain circumstances. Amino acids 61–95 make the hydrophobic middle domain, also refers to non-amyloid component (NAC), which has a tendency to form β-sheets and is mostly involved in α-synuclein protein aggregation and amyloid formation. Finally, the C-terminal, consisting of amino acids 96–140, is the region that makes interactions with other proteins and causes the water solubility of the natural α-synuclein protein. This domain contains five prolines, which are responsible for the presence of coils, or the lack of secondary structure, in the C-terminal. During the process of aggregation and formation of oligomers, amyloid fibrils, and Lewy bodies, the secondary structures of α-synuclein proteins undergo significant transformations, resulting in an increased proportion of β-sheets and α-helices, and decreased proportion of coils, bends, and turns.79–84
In order to further investigate the effectiveness of monolayers in preventing amyloid formation, the secondary structure of α-synuclein proteins has been analyzed with and without monolayers. In Figure 7, different colours represent different structures. The numerical results are in Table 4 Decreased helices and β-sheets, and increased turns, bends, and coils favour the prevention of α-synuclein aggregation and amyloid formation. Based on Figure 7 and Table 4, it can be concluded that all monolayers have increased the proportion of turns, bends and coils, and reduced the proportion of helices and β-sheets, among which the most favourable result belongs to N-Graphene, followed by graphene, P-Graphene, and BCN.
 |
Table 4 The Secondary Structure of the α-Synuclein Protein in the Presence of Each Monolayer |
Evaluating the Stability of α-Synuclein
The stability of proteins is an important indicator to understand the aggregation of particles and amyloid formation. α-synuclein protein stability reduces the motility of the particles and creates the conditions for deformation in its chain. Any agent which causes an increase in the molecular motions of α-synuclein proteins interferes with the process of amyloid fibrillation. In order to assess the stability of α-synuclein proteins, the following two parameters have been analyzed.
Analysis of RMSD
RMSD is the most commonly used parameter to evaluate the stability of a system in MD simulations. The RMSD curve reflects fluctuations of the particle calculated at different points of the simulation time relative to a constant reference. The greater the oscillations of the particles, the more unstable the system and the greater the slope of the RMSD curve will be.85–90 Therefore, higher fluctuation rates reflected by the RMSD curve reveal better prevention of α-synuclein amyloid formation by the monolayer.
Figure 8 shows the RMSD curves of α-synuclein proteins in the presence and absence of monolayers. Apparent instability of the system is noted in the first 15 nanoseconds of all simulations, as reflected by a sudden surge at the first five nanoseconds, followed by moderate oscillations in the next ten nanoseconds of simulation. Figure 8C (P-Graphene), D (BCN), and -E (without monolayers) maintain the instability of α-synuclein proteins after nanosecond 15; however, in Figure 8A (Graphene) and B (N-Graphene), fluctuations exist until the end of the simulation process. Therefore, it can be concluded that graphene and N-Graphene cause more sustained instability in the system.Besides, the geometric means of the derivatives of RMSD are shown in Table 5, which shows that N-Graphene has the highest value with the most significant instability of α-synuclein particles. Hence, N-Graphene is the best to prevent the formation of amyloids, followed by graphene, P-Graphene, and BCN.
 |
Table 5 Values of the Geometric Means of α-Synuclein RMSD Derivatives in the Presence of Each Monolayer |
 |
Figure 8 RMSD values of α-synuclein protein versus the simulation time in the presence of (A) Graphene; (B) N- Graphene; (C) P- Graphene; (D) BCN; (E) Without monolayers. |
Analysis of RMSF
In the RMSF diagram, the average fluctuation during the whole simulation process is shown for each atom. Higher RMSF values indicate more instability of the system and hence, better prevention of α-synuclein amyloid formation by monolayers.81,91–94 The RMSF curves of α-synuclein proteins in the presence and absence of each monolayer are shown in Figure 9. A comparison of the curves reveals that all monolayers increase RMSF values, while N-Graphene and graphene cause the most instability.Furthermore, to compare the effectiveness of each monolayer, the average RMSF values are shown in Table 6, which shows the highest value for N-Graphene, followed by graphene, P-Graphene, and BCN. It can be concluded that N-Graphene has the highest potential can prevent amyloid fibrillation more effectively by disrupting the stability of the system.
 |
Table 6 Average RMSF Values of α-Synuclein Protein Atoms in the Presence of Each Monolayer |
 |
Figure 9 RMSF values of α-synuclein protein versus the simulation time in the presence of (A) Graphene; (B) N- Graphene; (C) P- Graphene; (D) BCN; (E) Without monolayers. |
Evaluating the Contact Area of α-Synuclein Molecules
Amyloid formation increases if α-synuclein proteins are in close contact. So, the contact surface of α-synuclein proteins indicates the probability of amyloid formation. SASA is an essential parameter for evaluating the contact surface of biomolecules.95–97 In order to analyze the effects of monolayers on preventing amyloid fibrillation, SASA values have been calculated, and the contact area of α-synuclein proteins plotted as a function of time (Figure 10 and Table 7), which shows that N-Graphene has the least values of the contact area.
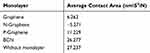 |
Table 7 Average Values of Contact Area for α-Synuclein Proteins During the Simulation in the Presence of Each Monolayer |
 |
Figure 10 The contact area of α-synuclein proteins versus time in the presence of (A) Graphene; (B) N- Graphene; (C) P- Graphene; (D) BCN; (E) Without monolayers. |
Table 7 shows the average contact area of α-synuclein proteins during the simulations. The values of the table reveal that although all monolayers reduce the contact surface of α-synuclein proteins and disrupt amyloid formation, the most desirable effect is seen in the simulation of N-Graphene, followed by graphene, P-Graphene, and BCN.
Validation Analysis
Mohammad-Beigi et al19 simulated the molecular dynamics of graphene polyglycerol and amyloid in water. Ultimately, as a validation of the algorithms used in this paper, an additional simulation of the van der Waals energy of interactions between graphene polyglycerol and α-synuclein proteins have been performed with conditions identical to a previously published study by Mohammad-Beigi et al.19 The results are shown in Figure 11 and indicate that the values of the curve produced by this paper and Mohammad-Beigi et al19 posses similar results.
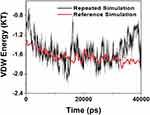 |
Figure 11 Comparison of the values of VDW between the α-synuclein protein and Graphene polyglycerol versus time in two different studies; black curve: the current study; red curve: the previously published study of Mohammad-Beigi et al.19 |
Conclusion
Alpha-synuclein proteins can aggregate and rise to oligomers, amyloids, and Lewy bodies, which is a vital sign of Parkinson’s disease pathophysiology. Preventing this pathological cascade has been the target of many conducted studies so far. In this study, the effects of four graphene-based nanostructures on preventing α-synuclein amyloid formation were investigated. For this purpose, important parameters involved in the interactions of α-synuclein proteins with themselves and with the monolayers were analyzed. Ultimately, it was concluded that, although all monolayers changed the evaluated parameters in the way which helps in the prevention of α-synuclein amyloid formation, the most significant results were reported for N-Graphene and graphene. N-Graphene and graphene are the most effective particle for the prevention of amyloid fibrillation and hence, ceasing the progression of Parkinson’s disease.
Such results could have important implications in the future of Parkinson’s disease treatment. For such a purpose, small-scale computational studies must be followed by gross-scale simulations in which more variables and factors are being considered as part of the simulation process. The clinical implementation of such a potential treatment option for Parkinson’s disease undoubtedly requires experimental in-vitro analyses followed by animal studies which would be strongly suggested by the authors of this paper. Ultimately, the results of such experimental studies must be put into a series of single or double-blind clinical trials in which already-proven effective nanomaterials could be administered as therapeutic agents or used as carries for conventional medications of Parkinson’s disease.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Yavarpour-Bali H, Pirzadeh M, Ghasemi-Kasman M. Curcumin-loaded nanoparticles: a novel therapeutic strategy in treatment of central nervous system disorders. Int J Nanomed. 2019;14:4449–4460. doi:10.2147/IJN.S208332
2. Xiong B, Zhou Y, Zhao Y, et al. The use of nitrogen-doped graphene supporting Pt nanoparticles as a catalyst for methanol electrocatalytic oxidation. Carbon N Y. 2013;52:181–192. doi:10.1016/j.carbon.2012.09.019
3. Ross C, Taylor M, Fullwood N, Allsop D. Liposome delivery systems for the treatment of Alzheimer’s disease. Int J Nanomed. 2018;13:8507–8522. doi:10.2147/IJN.S183117
4. Lee CC, Nayak A, Sethuraman A, Belfort G, McRae GJ. A three-stage kinetic model of amyloid fibrillation. Biophys J. 2007;92:3448–3458. doi:10.1529/biophysj.106.098608
5. Javdani N, Rahpeyma SS, Ghasemi Y, Raheb J. Effect of superparamagnetic nanoparticles coated with various electric charges on α-synuclein and β-amyloid proteins fibrillation process. Int J Nanomed. 2019;14:799–808. doi:10.2147/IJN.S190354
6. Zand Z, Afarinesh Khaki P, Salihi A, et al. Cerium oxide NPs mitigate the amyloid formation of α-synuclein and associated cytotoxicity. Int J Nanomed. 2019;14:6989–7000. doi:10.2147/IJN.S220380
7. Bi CC, Wang A, Chu Y, et al. Intranasal delivery of rotigotine to the brain with lactoferrin-modified PEG-PLGA nanoparticles for Parkinson’s disease treatment. Int J Nanomed. 2016;11:6547–6559. doi:10.2147/IJN.S120939
8. Yan X, et al. Lactoferrin-modified rotigotine nanoparticles for enhanced nose-to-brain delivery: LESA-MS/MS-based drug biodistribution, pharmacodynamics, and neuroprotective effects. Int J Nanomed. 2018;13:273–281. doi:10.2147/IJN.S151475
9. Pedrosa Carrasco AJ, Timmermann L, Pedrosa DJ. Management of constipation in patients with Parkinson’s disease. Npj Park Dis. 2018;4:1–10.
10. Qu M, Lin Q, He S, et al. A brain targeting functionalized liposomes of the dopamine derivative N-3,4-bis(pivaloyloxy)-dopamine for treatment of Parkinson’s disease. J Control Release. 2018;277:173–182. doi:10.1016/j.jconrel.2018.03.019
11. Kim K, Bohnen NI, Müller MLTM, Lustig C. Compensatory dopaminergic-cholinergic interactions in conflict processing: evidence from patients with Parkinson’s disease. Neuroimage. 2019;190:94–106. doi:10.1016/j.neuroimage.2018.01.021
12. McKinley JW, Shi Z, Kawikova I, et al. Dopamine deficiency reduces striatal cholinergic interneuron function in models of Parkinson’s disease. Neuron. 2019;103(1056–1072.e6):1056–1072.e6. doi:10.1016/j.neuron.2019.06.013
13. Moayeri A, Darvishi M, Amraei M. Homing of super paramagnetic iron oxide nanoparticles (SPIONs) labeled adipose-derived stem cells by magnetic attraction in a rat model of parkinson’s disease. Int J Nanomed. 2020;15:1297–1308. doi:10.2147/IJN.S238266
14. Li J, Uversky VN, Fink AL. Effect of familial Parkinson’s disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human α-synuclein. Biochemistry. 2001;40:11604–11613. doi:10.1021/bi010616g
15. Wu WH, Sun X, Yu Y-P, et al. TiO2 nanoparticles promote β-amyloid fibrillation in vitro. Biochem Biophys Res Commun. 2008;373:315–318. doi:10.1016/j.bbrc.2008.06.035
16. Härd T, Lendel C. Inhibition of amyloid formation. J Mol Biol. 2012;421:441–465. doi:10.1016/j.jmb.2011.12.062
17. Gazit E. A possible role for π‐stacking in the self‐assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi:10.1096/fj.01-0442hyp
18. Colvin VL, Kulinowski KM. Nanoparticles as catalysts for protein fibrillation. Proc Natl Acad Sci U S A. 2007;104:8679–8680. doi:10.1073/pnas.0703194104
19. Mohammad-Beigi H, et al. Mechanistic understanding of the interactions between nano-objects with different surface properties and α-synuclein. ACS Nano. 2019;13:3243–3256. doi:10.1021/acsnano.8b08983
20. Waxman EA, Giasson BI. Molecular mechanisms of α-synuclein neurodegeneration. Biochim Biophys Acta - Mol Basis Dis. 1792;616–624:2009.
21. Gaumet M, Vargas A, Gurny R, Delie F. Nanoparticles for drug delivery: the need for precision in reporting particle size parameters. Eur J Pharm Biopharm. 2008;69:1–9. doi:10.1016/j.ejpb.2007.08.001
22. Yang JA, Lin W, Woods WS, George JM, Murphy CJ. α-Synuclein’s adsorption, conformation, and orientation on cationic gold nanoparticle surfaces seeds global conformation change. J Phys Chem B. 2014;118:3559–3571. doi:10.1021/jp501114h
23. Álvarez YD, Fauerbach JA, Pellegrotti JV, et al. Influence of gold nanoparticles on the kinetics of α-synuclein aggregation. Nano Lett. 2013;13:6156–6163. doi:10.1021/nl403490e
24. Sanfins E, Dairou J, Rodrigues-Lima F, Dupret JM. Nanoparticle-protein interactions: from crucial plasma proteins to key enzymes. J. Phys. Conf. Ser. 2011;304:012039. doi:10.1088/1742-6596/304/1/012039
25. Vertegel AA, Siegel RW, Dordick JS. Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir. 2004;20:6800–6807. doi:10.1021/la0497200
26. Bond M, Pitt M, Akoh J, et al. The effectiveness and cost-effectiveness of methods of storing donated kidneys from deceased donors: a systematic review and economic model. Health Technol Assess (Rockv). 2009;13.
27. Wearable T, Kidney A. for End-Stage Renal Disease. 2005;149:325–333.
28. Kuitunen A, Vento A, Suojaranta-Ylinen R, Pettilä V. Acute renal failure after cardiac surgery: evaluation of the RIFLE classification. Ann Thorac Surg. 2006;81:542–546. doi:10.1016/j.athoracsur.2005.07.047
29. Lassen M, Gallus A, Raskob G. New England Journal NFL.pdf. N Engl J Med. 2010;363:2487–2498. doi:10.1056/NEJMoa1006885
30. Davenport A. Dialysis: a wearable dialysis device: the first step to continuous therapy. Nat Rev Nephrol. 2016;12:512–514. doi:10.1038/nrneph.2016.100
31. Huang L, Cao Y, Diao D. N-doped graphene sheets induced high electrochemical activity in carbon film. Appl Surf Sci. 2019;470:205–211. doi:10.1016/j.apsusc.2018.11.075
32. Science S Ac ce us pt. 2019.
33. Khan FA, Almohazey D, Alomari M, Almofty SA. Impact of nanoparticles on neuron biology: current research trends. Int J Nanomed. 2018;13:2767–2776. doi:10.2147/IJN.S165675
34. Yang L, Jiang S, Zhao Y, et al. Boron-doped carbon nanotubes as metal-free electrocatalysts for the oxygen reduction reaction. Angew. Chemie - Int. Ed. 2011;50:7132–7135. doi:10.1002/anie.201101287
35. Yang DS, Bhattacharjya D, Inamdar S, Park J, Yu JS. Phosphorus-doped ordered mesoporous carbons with different lengths as efficient metal-free electrocatalysts for oxygen reduction reaction in alkaline media. J Am Chem Soc. 2012;134:16127–16130. doi:10.1021/ja306376s
36. Carrero-Sánchez JC, Elías AL, Mancilla R, et al. Biocompatibility and toxicological studies of carbon nanotubes doped with nitrogen. Nano Lett. 2006;6:1609–1616. doi:10.1021/nl060548p
37. Baldea I, Olteanu D, Filip GA, et al. Cytotoxicity mechanisms of nitrogen-doped graphene obtained by electrochemical exfoliation of graphite rods, on human endothelial and colon cancer cells. Carbon. 2020;158:267–281. doi:10.1016/j.carbon.2019.12.011
38. Liao C, Li Y, Tjong SC. Graphene nanomaterials: synthesis, biocompatibility, and cytotoxicity. Int. J. Mol. Sci. 2018;19(11):3564. doi:10.3390/ijms19113564
39. Pelin M, Fusco L, León V, et al. Differential cytotoxic effects of graphene and graphene oxide on skin keratinocytes. Sci Rep. 2017;7:1–12.
40. Liao KH, Lin YS, MacOsko CW, Haynes CL. Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts. ACS Appl Mater Interfaces. 2011;3:2607–2615. doi:10.1021/am200428v
41. Kim D, Yoo JM, Hwang H, et al. Graphene quantum dots prevent α-synucleinopathy in Parkinson’s disease. Nat. Nanotechnol. 2018;13:812–818. doi:10.1038/s41565-018-0179-y
42. Banerjee AN. Graphene and its derivatives as biomedical materials: future prospects and challenges. Interface Focus. 2018;8. doi:10.1098/rsfs.2017.0056
43. Lee H, Paeng K, Kim IS. A review of doping modulation in graphene. Synth Met. 2018;244:36–47. doi:10.1016/j.synthmet.2018.07.001
44. Wang H, Maiyalagan T, Wang X. Review on recent progress in nitrogen-doped graphene: synthesis, characterization, and its potential applications. ACS Catal. 2012;2:781–794. doi:10.1021/cs200652y
45. Ysselstein D, Krainc D. Untangling alpha synuclein fibrils by graphene quantum dots. Mov Disord. 2018;33:1673. doi:10.1002/mds.27511
46. Schroer ZS, Wu Y, Xing Y, et al. Nitrogen-sulfur-doped graphene quantum dots with metal ion-resistance for bioimaging. ACS Appl Nano Mater. 2019;2:6858–6865. doi:10.1021/acsanm.9b01309
47. Yang Y, Asiri AM, Tang Z, Du D, Lin Y. Graphene based materials for biomedical applications. Mater Today. 2013;16:365–373. doi:10.1016/j.mattod.2013.09.004
48. Frenkel D, Smit B, Tobochnik J, McKay SR, Christian W. Understanding molecular simulation. Comput Phys. 1997;11:351. doi:10.1063/1.4822570
49. Huang J, Lemkul JA, Eastman PK, Mackerell AD. Molecular dynamics simulations using the drude polarizable force field on GPUs with OpenMM: implementation, validation, and benchmarks. J Comput Chem. 2018;1–8. doi:10.1002/jcc.25339
50. Jones JE, et al. On the determination of molecular fields. —II. From the equation of state of a gas. Proc R Soc London Ser A, Contain Pap a Math Phys Character. 1924;106:463–477.
51. Rezvantalab S, Keshavarz Moraveji M, Khedri M, et al. An insight into the role of riboflavin ligand in the self-assembly of poly(lactic-co-glycolic acid)-based nanoparticles – a molecular simulation and experimental approach. Soft Matter. 2020;16(22):5250–5260. doi:10.1002/9783527682300
52. Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human α-synuclein. J Biol Chem. 2005;280:9595–9603. doi:10.1074/jbc.M411805200
53. Mortazavifar A, Raissi H, Akbari A. DFT and MD investigations on the functionalized boron nitride nanotube as an effective drug delivery carrier for Carmustine anticancer drug. J Mol Liq. 2019;276:577–587. doi:10.1016/j.molliq.2018.12.028
54. Shafiei F, Hashemianzadeh SM, Bagheri Y. Insight into the encapsulation of gemcitabine into boron- nitride nanotubes and gold cluster triggered release: a molecular dynamics simulation. J Mol Liq. 2019;278:201–212. doi:10.1016/j.molliq.2019.01.020
55. Wang L, Xu J, Wang X, Cheng Z, Xu J. Facile preparation of N-rich functional polymer with porous framework as QCM sensing material for rapid humidity detection. Sensors Actuators B Chem. 2019;288:289–297. doi:10.1016/j.snb.2019.02.058
56. Chen W, He J, Jiang Y, Zhang H, Zhou X. Experimental and theoretical studies on the atmospheric degradation of 1, 1, 2, 2, 3, 3, 4-heptafluorocyclopentane. Atmos Environ. 2019;196:38–43. doi:10.1016/j.atmosenv.2018.09.057
57. Robertson MJ, Qian Y, Robinson MC, Tirado-Rives J, Jorgensen WL. Development and Testing of the OPLS-AA/M force field for RNA. J Chem Theory Comput. 2019;15:2734–2742. doi:10.1021/acs.jctc.9b00054
58. Dalgicdir C, Van Der Vegt NFA. Improved temperature behavior of PNIPAM in water with a modified OPLS model. J Phys Chem B. 2019;123:3875–3883. doi:10.1021/acs.jpcb.9b01644
59. Itliong JN, Villagracia ARC, Moreno JLV, et al. Investigation of reverse ionic diffusion in forward-osmosis-aided dewatering of microalgae: a molecular dynamics study. Bioresour. Technol. 2019;279:181–188. doi:10.1016/j.biortech.2019.01.109
60. Kyrychenko A, Blazhynska MM, Slavgorodska MV, Kalugin ON. Stimuli-responsive adsorption of poly(acrylic acid) onto silver nanoparticles: role of polymer chain length and degree of ionization. J Mol Liq. 2019;276:243–254. doi:10.1016/j.molliq.2018.11.130
61. Herrera-Rodríguez AM, Miletić V, Aponte-Santamaría C, Gräter F. Molecular dynamics simulations of molecules in uniform flow. Biophys J. 2019;116:1579–1585. doi:10.1016/j.bpj.2018.12.025
62. Zhong W, Cai Y, Tománek D. Computer simulation of hydrogen embrittlement in metals. Nature. 1993;362:435–437. doi:10.1038/362435a0
63. Martoňák R, Donadio D, Oganov AR, Parrinello M. Crystal structure transformations in SiO2 from classical and ab initio metadynamics. Nat Mater. 2006;5:623–626. doi:10.1038/nmat1696
64. Carretero-González R, Kevrekidis PG, Kevrekidis IG, Maroudas D, Frantzeskakis DJ. A Parrinello-Rahman approach to vortex lattices. Phys Lett Sect a Gen at Solid State Phys. 2005;341:128–134.
65. Wang E, Sun H, Wang J, et al. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119:9478–9508. doi:10.1021/acs.chemrev.9b00055
66. Greene D, Qi R, Nguyen R, Qiu T, Luo R. Heterogeneous dielectric implicit membrane model for the calculation of MMPBSA binding free energies. J Chem Inf Model. 2019;59:3041–3056. doi:10.1021/acs.jcim.9b00363
67. Astrand P, Wallqvist A, Karlström G. Intermolecular interactions of urea and water. J Chim Phys. 1991;88:2457–2464. doi:10.1051/jcp/1991882457
68. Damghani T, Sedghamiz T, Sharifi S, Pirhadi S. Critical c-Met-inhibitor interactions resolved from molecular dynamics simulations of different c-Met complexes. J Mol Struct. 2020;1203:127456. doi:10.1016/j.molstruc.2019.127456
69. Dehury B, Behera SK, Mahapatra N. Structural dynamics of Casein Kinase I (CKI) from malarial parasite Plasmodium falciparum (Isolate 3D7): insights from theoretical modelling and molecular simulations. J Mol Graph Model. 2017;71:154–166. doi:10.1016/j.jmgm.2016.11.012
70. Maleki R, Khoshoei A, Ghasemy E, Rashidi A. Molecular insight into the smart functionalized TMC-Fullerene nanocarrier in the pH-responsive adsorption and release of anti-cancer drugs. J Mol Graph Model. 2020;100:107660. doi:10.1016/j.jmgm.2020.107660
71. Kharazian B, Lohse SE, Ghasemi F, et al. Bare surface of gold nanoparticle induces inflammation through unfolding of plasma fibrinogen. Sci. Rep. 2018;8:1–9. doi:10.1038/s41598-018-30915-7
72. Ajori S, Ansari R, Haghighi S. A molecular dynamics study on the buckling behavior of cross-linked functionalized carbon nanotubes under physical adsorption of polymer chains. Appl Surf Sci. 2018;427:704–714. doi:10.1016/j.apsusc.2017.08.049
73. Shariatinia Z, Mazloom-Jalali A. Chitosan nanocomposite drug delivery systems designed for the ifosfamide anticancer drug using molecular dynamics simulations. J Mol Liq. 2019;273:346–367. doi:10.1016/j.molliq.2018.10.047
74. Xu WS, Carrillo JMY, Lam CN, Sumpter BG, Wang Y. Molecular dynamics investigation of the relaxation mechanism of entangled polymers after a large step deformation. ACS Macro Lett. 2018;7:190–195. doi:10.1021/acsmacrolett.7b00900
75. Khoshoei A, Ghasemy E, Poustchi F, Shahbazi MA, Maleki R. Engineering the pH-sensitivity of the graphene and carbon nanotube based nanomedicines in smart cancer therapy by grafting trimetyl Chitosan. Pharm Res. 2020;37:1–13.
76. Fang H, Tang Z, Liu X, Huang Y, Sun CQ. Discriminative ionic capabilities on hydrogen-bond transition from the mode of ordinary water to (Mg, Ca, Sr)(Cl, Br) 2 hydration. J Mol Liq. 2019;279:485–491. doi:10.1016/j.molliq.2019.01.147
77. Zhong Y, Chen Y, Feng X, et al. Hydrogen-bond facilitated intramolecular proton transfer in excited state and fluorescence quenching mechanism of flavonoid compounds in aqueous solution. J. Mol. Liq. 2020;302:112562. doi:10.1016/j.molliq.2020.112562
78. Bu R, Xiong Y, Wei X, Li H, Zhang C. Hydrogen bonding in CHON-containing energetic crystals: a review. Cryst Growth Des. 2019;19:5981–5997. doi:10.1021/acs.cgd.9b00853
79. Miceli M, Muscat S, Morbiducci U, Cavaglià M, Deriu MA. Ultrasonic waves effect on S-shaped β-amyloids conformational dynamics by non-equilibrium molecular dynamics. J Mol Graph Model. 2020;96:107518. doi:10.1016/j.jmgm.2019.107518
80. Turner M, Mutter ST, Kennedy-Britten OD, Platts JA. Molecular dynamics simulation of aluminium binding to amyloid-β and its effect on peptide structure. PLoS One. 2019;14:1–14. doi:10.1371/journal.pone.0217992
81. Nemaysh V, Luthra PM, Advances RSC. Computational analysis revealing that K634 and T681 mutations modulate the 3D-structure of PDGFR- b and lead to sunitinib resistance †. 2017:37612–37626. doi:10.1039/c7ra01305a
82. Turner M, Mutter ST, Kennedy-Britten OD, Platts JA. Replica exchange molecular dynamics simulation of the coordination of Pt(ii)-Phenanthroline to amyloid-β. RSC Adv. 2019;9:35089–35097. doi:10.1039/C9RA04637B
83. Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human α-synuclein: a possible molecular link between parkinson’s disease and heavy metal exposure. J Biol Chem. 2001;276:44284–44296. doi:10.1074/jbc.M105343200
84. Murray IVJ, Giasson BI, Quinn SM, et al. Role of α-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry. 2003;42:8530–8540. doi:10.1021/bi027363r
85. Khezri A, Karimi A, Yazdian F, et al. Molecular dynamic of curcumin/chitosan interaction using a computational molecular approach: emphasis on biofilm reduction. Int. J. Biol. Macromol. 2018;114:972–978. doi:10.1016/j.ijbiomac.2018.03.100
86. Saini RK, Thakur H, Goyal B. Effect of Piedmont mutation (L34V) on the structure, dynamics, and aggregation of Alzheimer’s Aβ40 peptide. J Mol Graph Model. 2020;97:107571. doi:10.1016/j.jmgm.2020.107571
87. Bertaccini EJ, Tradelia JR, Lindane E. Normal-mode analysis of the glycine alpha1 receptor by three separate methods. J Chem Inf Model. 2007;47:1572–1579. doi:10.1021/ci600566j
88. Hasanzade Z, Raissi H. Investigation of graphene-based nanomaterial as nanocarrier for adsorption of paclitaxel anticancer drug: a molecular dynamics simulation study. J Mol Model. 2017;23:1–8. doi:10.1007/s00894-017-3207-1
89. Rahmatabadi SS, Sadeghian I, Ghasemi Y, Sakhteman A, Hemmati S. Identification and characterization of a sterically robust phenylalanine ammonia-lyase among 481 natural isoforms through association of in silico and in vitro studies. Enzyme Microb Technol. 2019;122:36–54. doi:10.1016/j.enzmictec.2018.12.006
90. Farhadian S, Shareghi B, Asgharzadeh S, Rajabi M, Asadi H. Structural characterization of α‑chymotrypsin after binding to curcumin: spectroscopic and computational analysis of their binding mechanism. J Mol Liq. 2019;289:111111. doi:10.1016/j.molliq.2019.111111
91. Chairatana P, Niramitranon J, Pongprayoon P. Dynamics of human defensin 5 (HD5) self-assembly in solution: molecular simulations/insights. Comput Biol Chem. 2019;83:107091. doi:10.1016/j.compbiolchem.2019.107091
92. Liu J, Wei B, Che C, et al. Enhanced stability of manganese superoxide dismutase by amino acid replacement designed via molecular dynamics simulation. Int. J. Biol. Macromol. 2019;128:297–303. doi:10.1016/j.ijbiomac.2019.01.126
93. Zaboli M, Raissi H, Zaboli M, Farzad F, Torkzadeh-Mahani M. Stabilization of D-lactate dehydrogenase diagnostic enzyme via immobilization on pristine and carboxyl-functionalized carbon nanotubes, a combined experimental and molecular dynamics simulation study. Arch Biochem Biophys. 2019;661:178–186. doi:10.1016/j.abb.2018.11.019
94. Rezapour N, Rasekh B, Mofradnia SR, et al. Molecular dynamics studies of polysaccharide carrier based on starch in dental cavities. Int. J. Biol. Macromol. 2019;121:616–624. doi:10.1016/j.ijbiomac.2018.10.027
95. Pereira GRC, Da Silva ANR, Do Nascimento SS, De Mesquita JF. In silico analysis and molecular dynamics simulation of human superoxide dismutase 3 (SOD3) genetic variants. J Cell Biochem. 2019;120:3583–3598. doi:10.1002/jcb.27636
96. Arifuzzaman M, Mitra S, Das R, et al. In silico analysis of nonsynonymous single-nucleotide polymorphisms (nsSNPs) of the SMPX gene. Ann. Hum. Genet. 2020;84:54–71. doi:10.1111/ahg.12350
97. Da Silva ANR, Pereira GRC, Moreira LGA, Rocha CF, De Mesquita JF. SOD1 in amyotrophic lateral sclerosis development – in silico analysis and molecular dynamics of A4F and A4V variants. J Cell Biochem. 2019;120:17822–17830. doi:10.1002/jcb.29048
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.