Back to Journals » Nanotechnology, Science and Applications » Volume 14
Graphene and Graphene Oxide as a Support for Biomolecules in the Development of Biosensors
Authors Shahriari S , Sastry M, Panjikar S, Singh Raman RK
Received 26 August 2021
Accepted for publication 2 October 2021
Published 16 November 2021 Volume 2021:14 Pages 197—220
DOI https://doi.org/10.2147/NSA.S334487
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Video abstract of "Graphene and graphene oxide as biomolecules" [ID 334487].
Views: 2233
Shiva Shahriari,1 Murali Sastry,2 Santosh Panjikar,3,4 RK Singh Raman1
1Department of Mechanical & Aerospace Engineering, Monash University, Melbourne, Victoria, Australia; 2Department of Materials Science and Engineering, Monash University, Melbourne, Victoria, Australia; 3ANSTO, Australian Synchrotron, Melbourne, Victoria, Australia; 4Department of Molecular Biology and Biochemistry, Monash University, Melbourne, Victoria, Australia
Correspondence: RK Singh Raman
Department of Mechanical and Aerospace Engineering, Monash University, 17 College Walk (Building 31), Melbourne, Victoria, 3800, Australia
Tel +61 3 9905 3545
Fax +61 3 9905 1825
Email [email protected]
Santosh Panjikar
ANSTO, Australian Synchrotron, 800 Blackburn Road, Melbourne, Victoria, 3168, Australia
Tel +61 3 8540 4276
Email [email protected]
Abstract: Graphene and graphene oxide have become the base of many advanced biosensors due to their exceptional characteristics. However, lack of some properties, such as inertness of graphene in organic solutions and non-electrical conductivity of graphene oxide, are their drawbacks in sensing applications. To compensate for these shortcomings, various methods of modifications have been developed to provide the appropriate properties required for biosensing. Efficient modification of graphene and graphene oxide facilitates the interaction of biomolecules with their surface, and the ultimate bioconjugate can be employed as the main sensing part of the biosensors. Graphene nanomaterials as transducers increase the signal response in various sensing applications. Their large surface area and perfect biocompatibility with lots of biomolecules provide the prerequisite of a stable biosensor, which is the immobilization of bioreceptor on transducer. Biosensor development has paramount importance in the field of environmental monitoring, security, defense, food safety standards, clinical sector, marine sector, biomedicine, and drug discovery. Biosensor applications are also prevalent in the plant biology sector to find the missing links required in the metabolic process. In this review, the importance of oxygen functional groups in functionalizing the graphene and graphene oxide and different types of functionalization will be explained. Moreover, immobilization of biomolecules (such as protein, peptide, DNA, aptamer) on graphene and graphene oxide and at the end, the application of these biomaterials in biosensors with different transducing mechanisms will be discussed.
Keywords: functionalization, immobilization, oxygen functional groups, bioconjugate
Introduction
Biosensors have attracted significant attention due to their potential impact on human life, from diagnosis and treatment of diseases to the detection of environmental safety issues such as water contaminants.1 Graphene (G) (a two-dimensional allotrope of carbon) and its derivatives are promising candidates in designing rapid, accurate, and stable biosensors with high sensitivity and selectivity which provide low detection limits.2
Graphene is a hexagonal carbon lattice with a thickness of only one atom which has a variety of superior features such as large surface area, high thermal and electrical conductivity, and mechanical strength. The lack of defects in pure graphene causes low chemical reactivity and makes graphene inert in organic solvents. On the other hand, fabrication of graphene at high temperatures produces defects but damages its structure.3 However, these structural defects are the active sites for electron transfer and subsequently promote electrostatic interactions.1
Some of the special properties of graphene can only be accessible if it is functionalized with organic groups such as hydroxyl, carboxyl, or amino.4 For instance, graphene family materials will acquire dispersibility and colloidal stability in aqueous solutions when their surfaces are functionalized.5 However, some derivatives of graphene, such as graphene oxide (GO) and reduced graphene oxide (rGO), have abundant functional groups in their structures which help them interact with other molecules and disperse easily (Figure 1).
 |
Figure 1 (A) The hexagonal honeycomb structure of graphene, (B) The amorphous structure of graphene oxide, and (C) Reduced graphene oxide. Reproduced from Thangamuthu M, Hsieh KY, Kumar PV, Chen GY. Graphene- and graphene oxide-based nanocomposite platforms for electrochemical biosensing applications. Int J Mol Sci. 2019;20(12):2975.8 |
Nonetheless, in some applications, for example in dimethylformamide (DMF)-exfoliated graphene-based nicotinamide adenine dinucleotide hydrogen (NADH) electroanalytical sensor, the attachment of oxygen functionalities to the electrode often causes fouling of NAD+. To solve this problem, unfunctionalized graphene which extensively covers the edge-plane sites and defects is applied and encourages the electro-oxidation of NADH and prevents its fouling.6
Oxygen‐containing groups and sp2 domains allow graphene and its derivatives to attach to the other molecules through covalent bonding and non‐covalent bonding.7 These properties make graphene nanomaterials the perfect supports for biomolecule immobilization and improve their stability in numerous environments. Immobilization of enzyme (for instance) on functionalized graphene or its derivatives will enhance the catalysis performance and reusability of the enzyme.8 Immobilized biomolecules act as sensing elements in biosensors due to their interaction with the target analyte.
A biosensor detects a target analyte by transforming the biological reaction to detectable signals. It is composed of three elements: bioreceptor, transducer, and output system. The signals attributed to the interaction between the biomolecule and the analyte are converted to visible signals which appear in the output system. The quality of a biosensor is initially determined by the biochemical specificity of the biomolecule as well as the quality of the transducer. Graphene-based nanomaterials play the role of transducers in biosensors (Figure 2).9–11
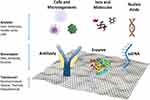 |
Figure 2 Examples of biosensors and components on a graphene platform. Reproduced from Peea-Bahamonde J, Nguyen HN, Fanourakis SK, Rodrigues DF. Recent advances in graphene-based biosensor technology with applications in life sciences. J Nanobiotechnology. 2018;16(1):1–7.9 |
Several advantages of graphene and its derivatives are attributed to their ordered pore structure, high surface area with numerous active sites, excellent electrical conductivity and electrochemical accessible surface area, light weight, and perfect mechanical strength,12–14 which make them suitable candidates for several applications; some of them are mentioned below:
- Core/shell-like cobalt phosphide decorated nitrogen and sulphur co-doped three-dimensional graphene (CoP@N, S-3D-GN) hybrid electrocatalyst demonstrated the best hydrogen evolution reaction (HER) catalytic activity amongst the as-synthesized electrocatalysts due to its synergistic effect of co-doping and the metal phosphide contribution. This specific morphology of the hybrid electrocatalyst stops the agglomeration and increases the pore sizes, which encourages the diffusion of electrons and maintains the structural stability during the HER process.13
- Heteroatom-doped or single/double-vacancy-modified graphenes have been widely studied in the electrochemical cells’ productions for energy storage because of their specific physicochemical properties. Karaman et al synthesized various graphene-like porous carbon networks (GPCs) with 3D hierarchically ordered “ion highways” through the carbonization/activation of orange-peel wastes which were used as electrode material in high-energy supercapacitors. The unique surface morphology and the well-tuned pore size distribution of the GPC eased diffusion of ions by reducing the ion transfer pathway through ion highways and accommodating a large number of charges, resulting in higher energy storage.15
- Karaman et al designed a sandwich-type electrochemical immunosensor for aflatoxin B1 (AFB1 stimulates cellular metabolism, producing large amounts of reactive oxygen species and free radicals) detection by porous graphene nanoribbon decorated with gold nanoparticles as a sensor platform and nanosheets of MoS2 incorporated with Ag nanocubes as a signal amplifier. The incorporation of metal nanoparticles promoted the sensing ability of porous graphene-based nanomaterials. This sensor offers a great method for the detection of AFB1 in wheat samples.16
- In a direct-methanol fuel cell (DMFC) application, a practical electrocatalyst containing graphene quantum dots and functionalized multi-walled carbon nanotubes (f-MWCNTs) was prepared by Gizem et al. Chronoamperometry (CA) analysis exhibited that the GQDs/f-MWCNTs/glassy carbon electrode (GCE) has higher current density compared to the other electrocatalysts as well as performing an important electrochemical activity against methanol.17
- Graphene quantum dot as a member of the 0-dimensional graphene family attracts remarkable attention to sensing applications due to its optoelectronic properties and high conductivity.18 A quartz crystal microbalance (QCM) immunosensor was developed based on Au-NPs decorated sulfur-doped graphene quantum dot and hollow ZnS-CdS nanocage for the detection of interleukin 6 (IL-6). IL-6 performs as a proinflammatory and anti-inflammatory cytokine and works as an important diagnostic biomarker for sepsis. The developed QCM immunosensor has the following features: i) it is very stable owing to the strong ester bonds; ii) no crosslinker was used; and iii) compared to conventional techniques like enzyme-linked immunosorbent assay (ELISA), the IL-6 analysis was done in short time. This immunosensor can be employed as a substitute method for clinical diagnosis.19
Structures and Properties of Graphene and Graphene Oxide
Graphene, a monolayer of sp2-bonded carbon atoms arranged in a honeycomb lattice, was produced in 2004, by separation of single layers of three-dimensional structure of graphite through mechanical exfoliation.4,20,21 Graphene possesses an extremely high surface area, high aspect ratio, and superior physical properties and biocompatibility which make it a great material for nanocomposites with high-performance and biological applications.2,21,22 Distribution of the benzene rings in the graphene structure contributes to the covalent or non-covalent bonds with the other atoms, which generally results in modifications of the electrical or chemical properties of graphene.3 Oxygen functional groups distort the graphene structure because planar sp2 converts to sp3 hybridization.23
The properties of GO, an oxidized form of graphene, include fluorescence-quenching capability, high energy transfer efficiency, biocompatibility, and facile chemical modification. The combination of these interesting properties along with simple and scalable synthesis preserves the potential applications of GO in many areas.24 GO is one of graphene’s derivatives which apart from superior properties of graphene contains oxygen functional groups on its surface. Hydroxyl and epoxy groups are spread in the shape of isolated islands on the top and bottom of GO’s basal plane causing a slight distortion in the lattice, while carboxyl groups are attached to the edges of the GO’s plane.23,25 One of the interesting properties of GO is amphiphilicity which means it has both hydrophobicity from pristine graphite and hydrophilicity from oxygen functionalities.24 Hydrophilicity and dispersion ability of GO in a variety of organic solvents is attributed to the hydrogen bonding between hydroxyl groups of GO and solvent interface.11,25 Due to the presence of functional groups, GO possesses two different regions: sp3 oxidized region and sp2 non-oxidized region of graphite, which has not been changed during GO synthesis.26
Synthesis of Graphene and Graphene Oxide
Synthesis of Graphene
Mechanical Exfoliation (Top-Down Approach)
Mechanical exfoliation involves weakening the van der Waals forces between graphite layers in order to separate them (top-down procedure).3,27 Technically, an adhesive such as a scotch-tape is employed to peel thin atomic layers of graphene from graphite.8
Chemical Vapor Deposition (CVD) (Bottom-Up Approach)
A complete CVD process is composed of three relatively independent stages: 1) the dissociation of carbon species and the formation of C atoms or small C clusters on a thin metal surface while a fluid (gas or liquid sprays) is decomposed at high temperature; 2) the nucleation of graphene while carbon atoms are dissolved into the metal crystalline surface (Ni or Cu); and 3) the lateral expansion of graphene islands.20,28 The carbon precursor in CVD is usually methane, but other sources such as ethanol, isopropanol, ethylene, and acetylene are applied as well.20
Synthesis of Graphene Oxide
Chemical Exfoliation
In this process, graphite converts to GO electrochemically in an acidic solution.29 At first, graphite oxide is produced by oxidation of graphite; after that, graphite oxide is exfoliated easily in a solvent because of weak van der Waals forces between the layers of graphite oxide. Following the separation of individual sheets, single layers of graphite oxide called graphene oxide are obtained, and reduction of GO with reducing agents such as hydrazine hydrate (N2H4) at high temperature (100°C) will produce reduced graphene oxide (rGO) (Figure 3).8 Exfoliation will be accomplished successfully when several parameters such as sonication time, energy, and solvents are employed properly.30
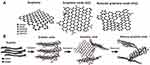 |
Figure 3 (A) Schematic chemical structures of graphene, graphene oxide, and reduced graphene oxide. (B) Route of graphite to reduced graphene oxide. Reproduced from Jimenez-Cervantes E, Lopez Barroso J, Martínez–Hernández A, Velasco–Santos C. Graphene–based materials functionalization with natural polymeric biomolecules. Recent Adv Graphene Res. 2016;1:257–298.7 |
Brodie and Staudenmaier Methods
Brodie was the first person who used a combination of fuming HNO3 and KClO3 as an oxidation media for producing GO. After him, Staudenmaier added H2SO4 to the acid mixture in order to improve the process efficiency by performing the reaction in one single vessel.29,31 The lower carbon-to-oxygen ratio of Brodie’s method compared to Staudenmaier’s method demonstrates that the GO produced by Brodie contains higher oxygen functional groups.31
Hummers Method
A mixture of concentrated H2SO4, NaNO3, and KMnO4 is used in the Hummers–Offeman method to obtain GO from graphite oxides’ stacked layers.25,32 There are three main stages in Hummers method: 1) intercalation of layers at low temperature (below 5°C); 2) oxidation of the intercalated graphite layers at medium temperature (~35°C); and 3) hydrolysis of products at high temperature (98°C).29 The consumption of large amounts of oxidants and intercalating agents as well as the long processing time are some of the reasons why scientists considered modifying Hummers method.32
Tour’s Method
Tour’s group developed a new method in which using sodium nitride has been avoided while more potassium permanganate was used. In addition, phosphoric acid as a new acid was introduced to the reaction vessel, and GO with a higher degree of oxidation was produced. Substitution of nitric acid with phosphoric acid hindered the release of toxic gases such as NO2, N2O4, or ClO2. Moreover, more graphitic surfaces were offered by phosphoric acid, and in comparison the efficiency was much higher than Hummers method.33
In a similar procedure, Yu et al modified Hummers method by pre-oxidizing graphite to increase the interlayer spaces. Instead of NaNO3, the amount of KMnO4 was enhanced, and in the end KMnO4 was replaced by K2FeO4 when no NaNO3 was in the system. Removing NaNO3 from the system prevented release of toxic gases like NO2 and N2O4.32
Oxygen Functional Groups (OFGs)
Quite a lot of graphene nanomaterials properties are attributed to the presence of oxygen functional groups and their distribution in the structure. In addition, OFGs play a crucial role in the functionalization of graphene-based materials and their interaction with other molecules (immobilization of molecules on graphene), which makes the final compound an appropriate candidate for several applications (Figure 3).
Distribution of OFGs
Among different OFGs, hydroxyl and epoxy groups are spread on the surface of GO and other oxidized graphene materials. Therefore, the geometry of hydroxyl and epoxide distribution as isolated areas or evenly spread, or other possible forms, impacts the properties of graphene nanomaterials. To investigate the distribution of OFGs, Shin et al deposited titanium oxide on oxygen groups of GO by atomic deposition layer method (ADL) at various temperatures. They claimed that epoxy and hydroxyl groups could sit in three different areas: the center of hexagons (hollow-site), C-C bonds (bridge-site), and directly above C atoms (top-site). Epoxide groups prefer to sit in bridge-site, while hydroxyl groups prefer top-site as their stable adsorption site.23
Attachment of functional groups on both sides of the GO layer is more favorable than one side because structural vertical distortion is removed. Uniform (evenly distributed with similar distances from each other) and island structures (the highly oxidized domain-like distribution) are two selected patterns to define two-sided hydroxyl and epoxide groups. Both epoxy and hydroxyl groups tend to accommodate themselves on graphene in domain-like distribution rather than uniform because of their binding energy (Eb) (change in the energy when GO is formed from its components [ie graphene and functional groups]). Hydroxyl groups show more attraction towards graphene; therefore, instead of distinct epoxide units, epoxide-hydroxyl compounds appear in the structure (Figures 4 and 5).8,23
 |
Figure 4 Dominant functional groups present in GO and rGO along with close-ups of various other functional groups and defects in rGO. Reproduced from Thangamuthu M, Hsieh KY, Kumar PV, Chen GY. Graphene- and graphene oxide-based nanocomposite platforms for electrochemical biosensing applications. Int J Mol Sci. 2019;20(12):2975.8 |
 |
Figure 5 Top and side views of the optimized atomic structures for uniform- and island-type distribution of (A) epoxy groups and (B) hydroxyl groups. Red and blue spheres denote the oxygen and hydrogen atoms, respectively. Eb versus the number of (C) epoxide and (D) hydroxyl species for two configurations. Reproduced from Shin DS, Kim HG, Ahn HS, et al. Distribution of oxygen functional groups of graphene oxide obtained from low-temperature atomic layer deposition of titanium oxide. RSC Adv. 2017;7(23):13979–13984.23 |
Liu et al proposed a new structural model analyzed by IR mapping and discovered that disorders in GO structure are preferable sites for OFGs attachment. Chemical composition inspection revealed that the C-O bonds were mostly from hydroxyl, epoxy, and some ether groups, while the C=O bonds came from carboxyl and ketone compounds. There were a few lactones and anhydrides in the structure, which could be because of the combination of ketone and ether. C-O and C=O bonds are spread on the planes; however, C=O mostly prefers to sit on the edges and folds (that have vacancy defects). Lactone and anhydride can accommodate at the edges of the GO sheet.34 Ketone and quinone are mainly found at defects; however, other functional groups might be created, relating to the defect size.35
Liu et al continued their discoveries on spatial distribution of OFGs by applying a novel AFM-IR (atomic force microscopy–infrared radiation) technique to distinguish ketone and carboxyl groups in the overlapping regions of GO multilayers. They found that ketone and carboxyl are more stable at the edges and folded areas, whereas ketone groups show inhomogeneous distribution on the plane. Carbonyl groups prefer to accommodate themselves mostly on the edges and less on the folded areas and distinct regions of the basal planes. C-O functional groups spread homogeneously on the surface and exhibit an almost uniform distribution.36
Effect of pH, Temperature, and Reaction Time on OFGs
pH
Hui et al discovered that the carboxyl groups located at the edges of GO sheets will protonate in acidic environments and if pH value drops under 2, high protonation of the -COO- groups occurs. On the contrary, higher pH in basic media causes detachment of several functional groups.37 X-ray photoelectron spectroscopy (XPS) analysis shows, in acidic environment (pH between 3 and 7), the C/O ratio enhances a little, which could be related to the size of GO nanosheets, because the edges of small sheets accommodate more functional groups fraction. Conversely, the ratio of C/O increases significantly in an alkaline environment (pH between 9 and 12) because of considerable detachment of the OFGs. At pH values between 5 and 9, in contrast to the C-O groups, the amount of -C=O and -O-C=O groups reduces. All attached functional groups will leave the surface when pH value is adjusted to 12.37
Aliyev et al exhibited that OFGs can be categorized to acidic and basic where acidic ones prefer to attach to the edges of graphene layers, while basal planes are more favorable for basic groups. Carboxylic, lactonic, and phenolic -OH groups are among the possible acidic oxides.35
Temperature
Luo et al investigated the preparation of graphite oxide containing different OFGs and sensitivity of ammonia gas. At 0°C while the oxidation has not been completed, the total number of hydroxyl and carboxyl groups was low. The oxidation of graphite progressed slowly as temperature elevated, and the number of hydroxyl groups at the edge and surface of graphite increased while the number of carboxyl groups decreased.38 At 100°C, due to the further oxidation of graphite, hydroxyl groups detached from the surface, and at 150°C they decomposed to separate water molecules from oxygen atoms which then combined with graphene to form epoxide.23,38 Table 1 demonstrates the content of OFGs at different temperatures.39 With increasing reaction temperature, at first, the ratio of C/O and content of C=C decreased and then increased. The reason lies in the fact that, by decreasing the content of C=C in graphite, the content of OFGs gradually increased, therefore the amount of oxygen increased, which caused a decrease in the C/O ratio.38 At the same time, by increment in the reaction temperature, at first, the content of C-OH increased and then decreased. This is because, at low reaction temperatures, C-OH prefers to accommodate itself on the graphite surface, and by increment in the reaction temperature, the content of C-OH gradually increased. At 100°C, graphite oxide initially lost a part of its C-OH.38,40 The amount of C-O-C and O-C=O increased slowly with increase in reaction temperature, pointing out that the higher reaction temperature during the high temperature stage is beneficial for the formation of C-O-C and O-C=O. However, the amount of O-C=O increased slowly with the increment in the reaction temperature; the rate of increase was way less than the rate of decrease of C-OH, which indicates that graphite oxide at 100°C has a lower amount of water and a bigger contact angle compared to at 50°C.38 Raising the temperature up to 300°C causes the length of the C-O bond in epoxide to increase, which thereby assists in the removal of functional groups from graphene.39
 |
Table 1 Content of Each Functional Group of Graphite Oxide Prepared at Different Reaction Temperature |
Reaction Time
Nazri et al examined the effect of reaction time on OFGs of GO produced by modified Hummers method. In their study, the structure of GO has been analyzed by various characterization methods at different process times of 24, 48, 72, and 96 hours. They discovered that a minimum of 72 hours is the necessary time for oxidizing graphite to GO with Hummers method. X-ray diffraction (XRD) results demonstrated that the intercalation between GO layers and attachment of OFGs to the edges and surface of GO during chemical reaction causes an increase in the layer distance. Scanning electron microscopy (SEM) results showed the plate-like surface, wavy-wrinkled, and layered-flakes (crumpled structure) on GO after 24, 72, and 96 hours, respectively.41 In longer reaction times, a higher amount of GO was produced because of longer exposure to the oxidants.41
Functionalization of Graphene and Graphene Oxide
Despite several unique properties of pure graphene, the lack of band gap, chemical reactivity and poor water dispersibility and agglomeration during the reduction process are some drawbacks in its application.7 This problem can be solved by modifying graphene-based nanomaterials through different methods, some of which are exerted during the synthesis and some of which are implemented by adding polymers, metal nanoparticles, or biomolecules.11,42 Modifying graphene family nanomaterials by functional groups preserves their perfect properties, as well as adding the characterization of functional groups to them.43
Functionalization of Graphene
Interactions contributed to the functionalization of graphene are divided into covalent and non-covalent bindings. Each of these bonds are applied when a special type of graphene properties is needed.
Non-Covalent Functionalization of Graphene
Non-covalent attachments are not disruptive and save graphene’s initial properties like their electrical conductivity and large surface area.30,44 Physical mixing, in situ polymerization, and solvent and melt processing are the primary procedures that use van der Waals interactions for production of graphene–polymer composites.44 The planar groups of the material and the carbon atoms of graphene are attached to each other via van der Waals forces; however, the effects of these physicochemical interactions are weak.42 Electrostatic interaction is an easy mechanism for self-assembly of graphene to other materials.44 Gue et al developed paraffin-graphene microcapsules for thermal energy storage due to the electrostatic interaction between the paraffin particles and graphene sheets. This interaction promotes paraffin entrapment in the shell and prevents its leakage during phase transformation.45 A simple hydrogen bond is not strong enough for supporting the attachment, whereas several hydrogen bonds provide enough strength as seen in DNA hybridization with graphene.44 π–π stacking occurs when π orbitals of two rather big non-polar aromatic rings overlap. These bonds are as strong as covalent bonds but compared to them, no conjugations of graphene sheets are disrupted, thus the electronic properties of graphene are saved.44
Covalent Functionalization of Graphene
Mechanical properties and stability of graphene are improved through covalent interactions; however, they disrupt the graphene structure.44 Direct doping can modify the band structure of graphene.44 Annealing heat treatment, ion bombardment, and arc discharge are some techniques that embody different elements (nitrogen, phosphorus, boron, …) into graphene defects (substitutional, vacancy) without disturbing the 2D structure of graphene.43,46,47 If graphene is synthesized through reduction of GO, some functional groups remain in the structure which are accessible for covalent interactions.44 Molecules of aryl diazonium salt can modify carbon materials through C-C covalent attachments. Simple agitation and electrochemical reduction adsorption are two strategies involved in aryl diazonium salts attachment to graphene, while N2 is released as the procedure moves forward.44 The aryl radical is extracted from diazonium salt through the single electron transfer (SET) process and addition of graphene sheet. Reaction between the radical species due to addition of aryl radicals is a route to proper functionalization of graphene.48
Functionalization of Graphene Oxide
Compared to graphene, a broad range of chemical mechanisms are available for absorption of functional groups to the GO surface.25 Surface polarity, hydrophilicity, and easy exfoliation in aqueous media are some of the properties obtained due to the functionalization of GO.35
Covalent Functionalization of Graphene Oxide
Co‐precipitation or hydrothermal methods are used for covalent functionalization of GO which increase the activity, selectivity, and stability of it.30 Covalent functionalization is carried out through three steps: 1) ring opening reaction of epoxy groups to create amine groups; 2) diazonium reactions or cycloaddition of rGO; and 3) nucleophilic reaction of carboxylic groups by hydroxyl or amine groups.26,30,44
Click Chemistry
In this technique, small organic units are joined rapidly to the surface of GO and facilitate the attachment of nanomaterials. For instance, through a cycloaddition reaction between an azide and alkyne group which is catalyzed by Cu, GO can be functionalized effectively (the azide-alkyne click chemistry).7,49
Linker Reaction
Denaturation of some biomolecules (like proteins) in direct contact with GO and lack of affinity of some materials towards GO could be solved by application of small functional molecules that work as linkage or bridge for covalent attachment.7 For example, in cryo-electron microscopy for analyzing the protein structure, protein needs to attach to a GO-coated gold grid. Protein bonds covalently to its cognate catcher which is connected to the grid by a polyethylene glycol (PEG) spacer. PEG is used to protect the protein from contacting with the surface of GO and the air and water interface. In addition, it prevents the protein from denaturation and aggregation.50
Direct Chemical Attachment
The functional groups of GO are chemically adsorbed to the other molecules by assistance of a catalyst or without it. Stability and reproducibility are the advantages of direct chemical attachment.7 Niyogi et al investigated the amide bond of octadecyl amine (ODA) with carboxylic groups of GO. Initially, by the aid of thionyl chloride, carboxylic groups of GO are converted into acyl chlorides. After that, the acyl chloride and the alkylamine of GO react directly, and an amide bond is created.51,52 Zhang et al applied a diazonium reaction on GO to increase its carboxyl groups. The large surface area and enhanced electrocatalytic activity of the functionalized GO provided a low anodic oxidation potential.53 Li et al modified GO by further oxidation in the mixture of H2SO4/HNO3 to obtain functionalized graphene oxide (GO-COOH). They immobilized lipase on GO-COOH through the interaction between carboxyl groups of functionalized GO and amino groups of lipase.54 In another study, Li et al modified the carboxyl groups of GO by epoxy chloropropane (GO-EC) which facilitated the immobilization of (+) γ-lactamase on GO. The enzyme was immobilized on the carrier directly via chemical covalent bond between the amino groups of the enzyme and the epoxy groups of GO.55
In covalent bonding between GO and other chemicals, usually more than one functional group is involved, which improves the activity and loading ability of GO. The properties and applications of functionalized G and GO are illustrated in Table 2.43
 |
Table 2 Properties and Applications of Functionalized Graphene and Graphene Oxide |
Non‐Covalent Functionalization of GO
Non-covalent functionalization of GO can be carried out through hydrogen bonding, π–π stacking, van der Waals interactions, electrostatic interactions, and cation-π interactions.43 Aromatic rings of GO are the place where π orbitals stack and cause the attachment. Moreover, these bonds stabilize the aromatic systems.7 Most polymers, surfactant “tails”, quantum dots, and mainly hydrophobic polypeptides do not have aromatic rings or charged groups in their structure. They bond to GO by van der Waals forces and hydrophobic interactions.7 Hydroxyl and epoxy groups on GO’s surface absorb polar molecules via hydrogen bonding, whereas carbonyl and carboxyl groups attract charged molecules through electrostatic interactions.1 The materials with amine or hydroxyl groups in their structure interact with GO through hydrogen bonding.7
The simultaneous functionalization and reduction of GO and metal nanoparticles through the natural agent tannic acid (TA) has been employed in glucose biosensors. TA reduces both GO and metal ions to rGO and metal NPs. Electron transfer is enhanced owing to the oxidation of TA to quinone, which is easily reducible at negative potentials. The deposition of metal NPs improved the electrical conductivity and biocompatibility of rGO. The π–π interactions between benzene rings of rGO and the phenolic groups of TA caused the deposition of TA on rGO. Covalent assisted hydrogen bonds attract glucose oxidase (GOx) on TA-coated rGO, which practically occurs through submerging the rGO-(Pt/Au) NPs-GOx/GCE in the enzyme solution.56,57
Modification of Graphene-Based Nanomaterials with Different Materials
Polymers
Modifying GO with polymers such as chitosan and dextran provides more functional groups for binding with other molecules and prevents the aggregation of GO sheets because of electrostatic charges.42 Mao et al fabricated a label-free amperometric electrochemical immunosensor to detect prostate-specific antigen (PSA) by employing graphene sheet–methylene blue–chitosan nanocomposite (GS-MB-CS) as electrode. This film demonstrated high binding affinity towards the electrode and was used for immobilization of PSA antibody. The principle of this biosensor relies on the response of peak current to capturing PSA due to the reactions between antigen and antibody.8,58
Metal and Metal Oxide Nanoparticles
The bonding energies of the adatom determine three sites with high symmetry for adsorbing metal adatoms: the center of hexagons (the hollow site), C-C bonds (the bridge site), and directly above C atoms (the top site). The adsorption, which is dependent on the adatom’s bonding energies, may occur via chemisorption or physisorption.59 Metal and metal oxide nanoparticles (Cu, Co, Ti, Au, Ag, Pd, Pt, Fe3O4, ZrO2, Co3O4) bond to graphene nanoparticles to supply a hydrophilic microenvironment for supporting the activities of immobilized biomolecules.42,60 Metal nanoparticles can improve the electrical communications of interlayers and fast conductivity of graphene and present lowest detection limits in biosensors.9
Decoration of the GO structure with gold nanoparticles showed synergistic effect and good biocompatibility, which were applied by Song et al to produce a disposable biosensor with a fast amperometric response and good storage stability for sensing catechol. They designed a screen-printed electrode (SPE) by covalent bonding of 1-pyrenebutanoic acid, succinimidyl ester (PASE) adsorbed on GO sheets and amines of tyrosinase-protected gold nanoparticles (Tyr-Au) (immobilization of Tyr-Au).61
Graphene derivatives play a major role in photoelectrochemical systems as they are excellent photogenerated electron carrier materials. The OFGs in their structure enhance the electron transfer.62 Cakiroglu et al designed a photoelectrochemical (PEC) glucose sensor by utilizing Cu2O/rGO-coated TiO2 nanotubes (NTs)-arrayed titanium foil. Cu2O NPs accommodated themselves evenly on rGO for the photoelectrochemical glucose determination without using an enzyme. The perfect increased photocurrent can be related to the p-n heterojunction created between Cu2O NPs and TiO2 NTs, as well as perfect conductivity of rGO, which can increase the charge separation efficiency and at the same time promote electron transfer. rGO has been significantly utilized to intensify the separation of the photogenerated electron–hole pairs and decrease the recombination, due to its electron-capturing ability.62
Biomolecules
Modifying the layers of G and GO with biomolecules makes them a perfect substrate for immobilizing macromolecules.42 The modification principle of GO by biomolecules lies in the attaching of nitrogen atom of an amine functional group to the carboxylic acid of GO via a condensation reaction which produces a stable amide bond. In this process, epoxide groups are substituted by nucleophiles, and an amine and hydroxyl are produced accompanied by the ring opening.7
Laaksonen et al demonstrated a method for both exfoliation and functionalization of graphene by hydrophobins (HFBI). The surface energies of graphene and the hydrophobic side of HFBI are similar and different from the hydrophilic side of HFBI and solvent. These differences are smaller than an uncoated system; therefore, attachment of HFBI to graphene results in a stable dispersion of graphene.42,63 Zhang et al functionalized graphene oxide by adding abundant carboxyl groups through a diazonium reaction. After that, Nα, Nα-bis(carboxymethyl)-L-lysine hydrate (NTA-NH2), and Ni+2 were fastened to GO. Histidine (His)-tagged acetylcholinesterase (AChE) was captured by functionalized GO due to the specific interaction between His-tag and Ni-NTA. This bioelectrode was used for fabrication of paraoxon biosensors.53
Immobilization of Biomolecules on Graphene and Graphene Oxide
Immobilization Methods
During the immobilization procedure, the aim is to keep active sites accessible for targets as well as preserving conformational stability and bioactivity. The size, shape, and polarity of the biomolecule, working area, storage conditions, and presence of functional groups are all important parameters in immobilization that must be taken into account.11 Immobilization methods can be classified into reversible and irreversible due to the detachment ability of biomolecules from their substrates.
Irreversible Immobilization
Irreversible immobilization is a covalent bonding of biomolecules to the support which is a strong chemisorption that prevents the leakage of biomolecule. Crosslinking (free ends of crosslinkers such as glutaraldehyde, glyoxal, or hexamethylenediamine can hold biomolecules on the transducer surface by forming covalent bonds)64,65 and entrapment in a polymeric gel network (this gel traps the enzyme but substrate and product can exit the system) are examples of this method.11 In irreversible immobilization, OFGs present in the GO structure provide several reaction sites for interaction with small molecules, polymers, biomacromolecules, and inorganic nanoparticles with no need of adding crosslinkers or even functionalization of GO.66 Srivastava et al employed functionalized graphene sheets and glutaraldehyde (crosslinker) to covalently immobilize fenugreek β-amylase. They discovered that non-toxicity of the functionalized graphene and high thermal stability of immobilized enzymes can be utilized for production of maltose in food and pharmaceutical industries.67 Loo et al investigated the covalent attachment of the thrombin aptamer II (NH2‐THR‐APT) to disposable electrical printed (DEP) electrodes which were modified with GO. To activate carboxylic acid groups, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were deposited onto GO‐modified electrodes in phosphate buffer solution (PBS). To remove non-bonded EDS and NHS, the electrodes were washed with PBS. After that, NH2‐THR‐APT with a suitable concentration in PBS was developed on the electrodes.68
Reversible Immobilization
Physical attachment (hydrogen binding, van der Waals forces, electron transition complexes, storage conditions) and affinity binding (activated substrate interacts non-covalently with a specific group of the protein sequence) are types of this method.11 In reversible immobilization of enzymes on graphene, the hexagonal lattice, hydrophobic, and hydrophilic domains of graphene and its derivatives contribute to the efficient immobilization. Hexagonal lattice of graphene composed of carbon aromatic rings can attach to aromatic residues of an enzyme via π–π stacking. Hydrophobic domains in the structure of biomolecules like enzymes interact non-covalently with the hydrophobic surface of graphene family members. Charged residues of enzymes interact electrostatically with negative functional groups of graphene derivatives.20
Immobilization of Biomolecules
Peptide
The binding affinity of different amino acids and their position in the peptide sequence are the factors that contribute to the non-covalent bonding of peptide to G and GO.69 A GO nanosheet can incubate peptides due to π–π interaction; in addition, charged and polar residues of peptides interact electrostatically with ionizable edges of GO.53,70 The strength of bonds between some residues and GO follows this order: Arg > His > Lys > Trp > Tyr > Phe. Among these amino acids, His interacts with GO by both electrostatic and π–π interactions.71 Woo et al investigated the molecular interaction between GO and Tyr and Trp and found that GO strongly quenches the fluorescence of Tyr and Trp because of several non-covalent intermolecular interactions like π–π interactions and hydrogen bonding between GO and these amino acids.72
Zuo et al applied sum frequency generation (SFG) vibrational spectroscopy and molecular dynamics (MD) simulation to investigate the interactions between two peptides (cecropin P1 and MSI-78(C1)) and graphene in real-time and in situ. It was found that both planar and hydrophilic residues in the peptide are responsible for the interaction with graphene.69 That end of peptide with enough planar side chains attaches strongly to the graphene surface, whereas the other end with hydrophilic residues abandons the graphene surface and is accessible for reaction in the aqueous solution. Either hydrophilic residues or π–π stacking help peptides to orient in the appropriate direction for interaction.69 Wang and Lin designed a glassy carbon electrode modified by GO–peptide–AgNPs nanohybrids to produce a non-enzymatic electrochemical hydrogen peroxide (H2O2) sensor. The π–π interaction between peptide and GO builds a compact network-like structure of peptide on both sides of GO.73
Protein/Enzyme
Immobilization of protein can be done through methods like mixing, sonication, ultrasound, and cyclic voltammetry to provide suitable conditions for adsorption via covalent bonding, non-covalent bonding, physical entrapment, and functionalization via EDC/NHS chemistry.9
Non-covalent attachments between protein and matrix happen through various forces such as intermolecular, dipole–dipole, and forces between similar surface residues and opposing surface charges.20 In addition, the surface density of the functional groups is affected by preparation technique and storage conditions. These factors influence the loading and stability of the proteins that interact with GO electrostatically.66 Aromatic residues of protein and backbone of graphene can bond by π–π stacking. The hydrophobic aromatic structure of graphene is in favor of hydrophobic interaction with protein, as protein molecules prefer to agglomerate on a hydrophobic surface.74 The polar groups such as hydroxyls, epoxides, and carboxyl groups promote adsorption of proteins mainly through electrostatic interactions. As an instance, at human physiological pH, the hexagonal aromatic structure of GO and the negatively charged functional groups can provide non-covalent interactions and high loading of serum protein on the surface of GO.70
Covalent immobilization of proteins on GO is achieved through chemical reactions between OFGs of GO and free amine groups of protein.49,66 For example, amidation reaction between GO’s carboxyl groups and amine groups of bovine serum albumin (BSA) helps their covalent bonding.66 The crosslinker molecule from one side attaches to graphene oxide non-covalently through hydrophobic interaction or π–π stacking, and from the other side bonds covalently to the biomolecule due to an amide bond.49 Glutaraldehyde for instance, works as a crosslinker for covalent attachment of alkaline protease to GO sheets.66 In another experiment, accomplished by Kodali et al, the aromatic pyrenyl group in 1-pyrenebutyric acid N-hydroxy succinimide ester (PYR-NHS) interacted strongly with the basal plane of graphene via π–π stacking, without perturbing the sp2 bond structure or the π band which is responsible for electronic properties of graphene. From the other side, the PYR-NHS reaction with lysine residues facilitates the immobilization of protein on graphene.75
Functionalization of the graphene family with peptides or crosslinking agents assists enzyme attachment through their C-terminal or side chains, whereas epoxide groups attach covalently to the enzyme through their N-terminal. Although covalent bonding is very stable and prevents leaching, it affects enzyme structure, activity, and reusability (Figure 6).20
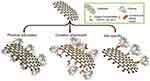 |
Figure 6 Illustration of enzyme immobilization methods onto graphene nanosheets. Reproduced from Karimi. Republished with permission of Royal Society of Chemistry (Great Britain), Karimi A, Othman A, Uzunoglu A, Stanciu L, Andreescu S. Graphene based enzymatic bioelectrodes and biofuel cells. Nanoscale. 2015;7(16):6909–6923. Permission conveyed through Copyright Clearance Center, Inc.111 |
Oxidoreductase, hydrolase, and transferase are some types of enzymes that are widely used in graphene-based biosensors due to the kind of reactions they catalyze (Table 3).
 |
Table 3 Different Types of Enzyme Immobilization on GO and Their Applications |
DNA
Biocompatibility, renewability, flexibility, and high specificity of DNA because of the unique sequences of its nucleotides make it an ideal candidate for biosensing purposes.9 DNA attachment to graphene is mostly done through non-covalent bindings such as hydrogen bonding, π–π stacking, and electrostatic interaction. Positively charged functionalized graphene interacts electrostatically with negatively charged nucleotide of DNA. Edges of graphene and functional groups of GO are involved in the covalent interaction with DNA.9,74
Bo et al immobilized DNA on a bioelectrode for designing an electrochemical DNA biosensor. They modified the glassy carbon electrode by layers of oxidized graphene and polyaniline nanowires (PANIw) with the aid of their specific synergetic effect. The immobilization is done through the phosphonamidite bonds between the phosphate group of oligonucleotides and the amino group of polyanilines. The ssDNA/PANIw/graphene/GCE bioelectrode exhibited high sensitivity and selectivity towards complementary DNA sequence.76,77
Aptamer
Short, single-stranded DNA or RNA (ssDNA or ssRNA) are called aptamers that can attach selectively to a specific target such as proteins. Graphene adsorbs unfolded flexible aptamer non-covalently but repulses the folded aptamer which has a rigid 3D structure.78 Pyramidic and puric bases of aptamers attach to the graphene lattice via π–π stacking interactions which is known as a plain immobilization technique for graphene-based aptasensors.9,78 Dye-labeled ssDNA attaches to GO by π–π stacking interaction, and GO quenches the fluorescence of the dye which is used in fluorescent biosensors.1
Negatively charged aptamers are adsorbed to positively charged graphene materials via electrostatic interaction in neutral buffers. This property is used for development of label-free electrochemical aptasensors in which graphene is functionalized with charged materials such as Fe2O3 and poly sodium 4-styrenesulfonate.78 Synthesis of graphene by oxidation methods or reduction of GO causes some carbonyl and carboxyl groups to remain in the structure which assists in the covalent interaction of graphene with amine groups of biomolecules.78
Loo et al examined three thrombin aptasensors with different immobilization techniques to detect thrombin (THR). They discovered that the aptasensor based on a physical attachment of aptamer is remarkably selective towards THR compared to targets like BSA, IgG (immunoglobulin G), and avidin. In comparison with physical immobilization, covalent immobilization exhibits similar selectivity towards analyte. In contrast, affinity immobilization does not show any considerable influence on selectivity of THR.68
Graphene and Graphene Oxide-Based Biosensors
Biosensors can be categorized into different types due to their transducing mechanisms. The most common types are: i) electrochemical; ii) field effect transistor (FET); iii) plasmonic resonance; iv) optical; and v) fluorescent biosensors. Some examples of these biosensors and their applications are exhibited in Table 4.
 |
Table 4 Types of Graphene and Graphene Oxide Biosensors and Their Applications |
Electrochemical Biosensors
Electrochemical sensors measure the resistance, current, or potential. The change in the electrical signal due to the interactions is detected by the transducer electrode.9,79 Defects in graphene structure are the active sites for transferring electrons to biomolecules, and sp2-like planes increase carbon materials’ activities towards oxidation or reduction of small biomolecules such as NADH, dopamine, and H2O2.80
Hydrophilicity of GO enables it to assemble with quantum dots, semiconducting nanoparticles, polymers, and metals (nanoparticles and oxides) to promote the performance of electrochemical biosensors by enhancing the electron transfer between the bioreceptor and transducer, which is an advantage for electrochemical sensors with high sensitivity.8,9
Figure 7 illustrates the principle of an electrochemical cholesterol biosensor. At first, cholesterol esterase (CHER) and cholesterol oxidase (CHOD) interact with GO via an amide bond by applying EDC/NHS as a crosslinker (Figure 7A). Then, gold nanoparticles electrodeposit on SPE. After that, the mixture of GO, CHOD, and CHER electrostatically attaches to the surface of the electrode. CHER and CHOD catalyze the oxidation of cholesterol into cholest-4-en-3-one and H2O2. Due to the oxidation of cholesterol, Ag+ is reduced to Ag and deposits on the modified electrode which produces a stripping anodic signal, determined by linear sweep voltammetry (LSV). The current of LSV is directly proportional to the concentration of H2O2. Therefore, detection of cholesterol with high sensitivity can be achieved by measuring the current of LSV (Figure 7B).81
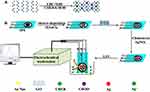 |
Figure 7 Principle of the electrochemical detection of cholesterol based on GO and AuNPs co-mediated enzymatic Ag deposition: (A) Attachment of CHOD/CHER to GO via EDC/NHS chemistry; (B) The stages to electrochemically detection of glucose by measuring LSV. Reprinted from Biosens Bioelectron, 102, Huang Y, Tan J, Cui L, et al. Graphene and Au NPs co-mediated enzymatic silver deposition for the ultrasensitive electrochemical detection of cholesterol, 560–567, Copyright (2018), with permission from Elsevier.81 |
Field Effect Transistor (FET) Biosensors
Bare graphene-FET shows only the charge of the biomolecule and has no detection ability; therefore, the surface of graphene must be modified with biomolecules. An illustration of a probe-modified G-FET biosensor with an Ag/AgCl reference probe is depicted in Figure 8. A solution-gated G-FET (Figure 8B) has typically three electrodes: source, drain, and gate. Possible leakage of current is prevented by insulating source and drain electrodes. In these G-FET biosensors (Figure 8A and C), antibody and aptamer work as bioreceptors for detecting E. coli (Escherichia coli).82 The charge and discharge status of the graphene layer changes by regulating the gate voltage, which transfers the Fermi levels of the nanomaterials. Therefore, the applied gate voltage can modify the conductance of the channel material.83 Matsumoto et al designed a FET biosensor in which the graphene surface was modified by aptamer for detection of antibody and immobilized fragment antibody on the graphene surface for detection of antigen. They used aptamer and fragment antibody as detection probes because their lengths are shorter than the Debye length (the required distance to screen the surplus charge by carriers).84,85
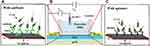 |
Figure 8 The general schematic of the probe-modified G-FET biosensor: (A) linker/antibody-functionalized G-FET for E. coli detection; (B) general structure of the G-FET biosensor; (C) aptamer-functionalized G-FET for E. coli detection. Reproduced from Wu G, Meyyappan M, Lai KWC. Simulation of graphene field-effect transistor biosensors for bacterial detection. Sensors. 2018;18(6):1715.82 |
Mid-Infrared Plasmonic Biosensors
The massive light capture ability of graphene provides high overlaps with biomolecules and subsequently exhibits exceptional detection of vibrational fingerprints and refractive index of biomolecules.86 Figure 9 illustrates a graphene-based surface plasmon resonance (SPR) biosensor in which dielectric top layers are graphene. The sensing region is composed of a four-layer Fresnel structure (prism, Au, graphene, and PBS). A graphene monolayer as an effective light absorption medium is inserted between the sensing dielectric and the gold film. The immobilized DNA probes on the graphene surface receive and transduce signals which in turn increase the refractive index (RI) of graphene for sensing medium inter surface. For the next cycle of application, the surface of the graphene is cleaned by a suitable chemical to repel the target DNA samples without affecting the immobilized DNA probe.87–91
 |
Figure 9 Illustration of the four-layered model for fiber optic SPR biosensor: prism, Au (50 nm), graphene (0.34nm × £ (£ is the total of graphene coating)), and phosphate buffer solution (PBS) contains p-DNA (probe) and complementary target mr-DNA (mutated type) or wt-DNA (wild type) samples. Reproduced from Hossain MB, Akib TBA, Abdulrazak LF, Rana MM. Numerical modeling of graphene-coated fiber optic surface plasmon resonance biosensor for BRCA1 and BRCA2 genetic breast cancer detection. Optic Eng. 2019;58(03):1.87 |
Singh et al employed the superior optical properties of graphene monolayer to boost the SPR signals for immunosensing cholera as a disease model. The functional groups of nitrilotriacetic acid (NTA) attached to graphene via electropolymerization of a polypyrrole film or π–π interactions of pyrene derivatives, which were additionally secured by electropolymerization. NTA anchors the biotinylated bioreceptor cholera toxin and controls its immobilization.92
Optical Biosensors
The bandgap of GO can be modified by adjusting the sp2/sp3 ratio of carbon atoms which causes the enhancement of GO’s conductivity and absorption peak, and, consequently, the photoluminescence of GO alters.93,94 Zhang et al discovered that the specific sensitivity of peptides combined with optoelectronic properties of GO can sense TNT through high-throughput absorption spectra.94 A rGO-based optical biosensor is demonstrated in Figure 10 in which immobilized antigens on the surface of graphene detect antibodies in the solution. The polarized light reflects at the interface and splits into two polarized TE (transverse electric) and TM (transverse magnetic) waves. TE is more favorable for graphene than TM. The power difference of the separated light can be registered by a balanced photodetector. The inset of Figure 10 exhibits the attachment of antigen–antibody to the surface of rGO.95
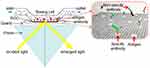 |
Figure 10 Schematic of reduced graphene oxide (rGO)-based optical sensor. Reproduced from Li Z, Zhang W, Xing F. Graphene optical biosensors. Int J Mol Sci. 2019;20(10):2461.95 |
Fluorescence Biosensors
Tagged targets absorb external light and can release fluorescent light. This is a mechanism for sensing molecules in biological systems. Graphene and its derivatives have high distance-dependent fluorescence-quenching ability based on fluorescence resonance energy transfer (FRET) which enables them to quench various adsorbed quantum dots, fluorophores, and fluorescent metal nanoclusters, then transduce them to fluorescent biosensors.9,96
Lu et al for the first time in 2009 established GO-based fluorescent sensors for selective and sensitive detection of DNA and protein.97 A natural DNA probe can detect its complementary target in fluorescent biosensors; however, high affinity of other biomolecules to GO causes desorption of DNA with the noise or high background signal.24 In comparison to ssDNA, the nucleotide bases of dsDNA are buried in the helical structure, which impacts the interaction between dsDNA and GO (Figure 11).1
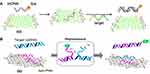 |
Figure 11 (A) Scheme of the ssDNA target-induced fluorescence change of the DNA probe-FAM-GO complex. Reprinted from Lu CH, Yang HH, Zhu CL, Chen X, Chen GN, Graphene A. Platform for sensing biomolecules. Angewandte Chemie Int. 2009;48(26):4785–4787, Copyright © 2009 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.97 reproduced with permission from Wiley Online Library, (B) Scheme of the direct detection of dsDNA using peptide nucleic acid (PNA) and GO. Reprinted from Biosens Bioelectron, 62, Lee J, Park I-S, Jung E, Lee Y, Min D-H. Direct, sequence-specific detection of dsDNA based on peptide nucleic acid and graphene oxide without requiring denaturation. 140–144, Copyright (2014), with permission from Elsevier.112 |
Chung et al designed a fluorescent biosensor for recognition of dsDNA by employing a GO–organic dye ionic complex. The detection principle relies on the exchange of ions adsorbed to the carboxyl groups which are placed at the GO edges. Li et al claimed that detecting label-free Hg+2 by quenching the fluorescence emission of aptamer-functionalized GO is possible. Without Hg2+, the aptamer molecules rest on the surface of GO, while Hg2+ is added, a stiff hairpin-shape dsDNA structure is composed because of the arrangement of the thymine–Hg2+–thymine complex, which quenches the GO fluorescence.93 Figure 12 shows a label-free fluorescent Cu(II) biosensor composed of a graphene/DNAzyme complex in which the fluorophore (weak fluorescent GelRed) is inserted into the folded structure of DNAzyme. Graphene self assembles to DNAzyme through π–π stacking, which increases the quenching of fluorescence. After adding Cu(II), DNAzyme splits, and quenched fluorophore is released.98
 |
Figure 12 Schematic illustration of label-free fluorescent Cu(II) biosensor based on graphene-quenched DNAzymes. Reproduced from Liu M, Zhao H, Chen S, Yu H, Zhang Y, Quan X. Label-free fluorescent detection of Cu(ii) ions based on DNA cleavage-dependent graphene-quenched DNAzymes. Chem Commun. 2011;47(27):7749–7751.98 |
Future Research Prospects
The exceptional properties of graphene family materials such as large surface area and high conductivity are the requirements of a suitable support for biomolecule immobilization. However, these characteristics need to be suitably tailored for effective immobilization and graphene-biomolecule conjugate for biosensing. Nevertheless, there are also challenges in this task.
Although methods for synthesis of G and GO are continuously improving, there are still challenges such as in achieving precision with respect to the position and number of defects and distribution of OFGs. Designing synthesis strategies to obtain an even spread of functional groups on the surface with defined edges is an aspect that has received limited research attention.3,23,33 Furthermore, there is a need to develop a more comprehensive understanding of the influence of various synthesis parameters such as reaction time, pH, and temperature on the morphology and properties of the functional groups. Regarding successful attachment of biomacromolecules such as proteins to the substrate (G or GO), the influence of distribution, charge, and geometry of functional groups needs to be understood. For example, Asal et al encountered some fundamental issues such as choice of suitable support material, proper immobilization of biomolecule, and their stability after interacting with graphene.11
The variety of amino acid sequences and their folding create an extensive number of proteins with complex structures that perform distinct functions. The unique structure and characteristics of each protein, together with the influence of the environment, can profoundly affect their interaction with G and GO. In addition, there are still lots of biomolecules whose applications in graphene-based biosensing have not been explored. These numerous factors open opportunities for the development of a new generation of biosensors. These possibilities can trigger further research in biomedical science, recognition and treatment of diseases, drug delivery, imaging, and a lot more.
A few of the prospects for biosensing investigations are described below:
- Developing new functionalization methods that can preserve the usability and functionality of the biomolecule, while providing a strong bond with the substrate (G or GO).
- Designing procedures for controlling the amount, location, and geometry of the oxygen functional groups which play major roles in interaction with other biomolecules and their immobilization on the surface of substrate.
- Diagnosing the immobilization process of the biomolecule on the support surface (G or GO).
- Determining the preferred attachment site and position of the biomolecule on the substrate and availability of the active interaction sites for analyte.
Acknowledgments
We gratefully acknowledge Dr. Katherine Nairn’s assistance in editing the manuscript.
Disclosure
The authors report no conflicts of interests in this work.
References
1. Sharma D, Kanchi S, Sabela MI, Bisetty K. Insight into the biosensing of graphene oxide: present and future prospects. Arab J Chem. 2016;9(2):238–261. doi:10.1016/j.arabjc.2015.07.015
2. Nazari M, Kashanian S, Maleki N, Shahabadi N. Laccase immobilized onto graphene oxide nanosheets and electrodeposited gold–cetyltrimethylammonium bromide complex to fabricate a novel catechol biosensor. Indian Acad Sci. 2019;42(2):1–10.
3. Skoda M, Dudek I, Jarosz A, Szukiewicz D. Graphene: one material, many possibilities–application difficulties in biological systems. J Nanomater. 2014;2014:1–11. doi:10.1155/2014/890246
4. Yang L, Wang G, Liu Y, Wang M. Development of a biosensor based on immobilization of acetylcholinesterase on NiO nanoparticles-carboxylic graphene-nafion modified electrode for detection of pesticides. Talanta. 2013;113:135. doi:10.1016/j.talanta.2013.03.025
5. Li D, Zhang W, Yu X, Wang Z, Su Z, Wei G. When biomolecules meet graphene: from molecular level interactions to material design and applications. Nanoscale. 2016;8(47):19491–19509. doi:10.1039/C6NR07249F
6. Keeley GP, O’Neill A, Holzinger M, Cosnier S, Coleman JN, Duesberg GS. DMF-exfoliated graphene for electrochemical NADH detection. Phys Chem Chem Phys. 2011;13(17):7747–7750. doi:10.1039/c1cp20060g
7. Jimenez-Cervantes E, Lopez Barroso J, Martínez-Hernández A, Velasco-Santos C. Graphene‐based materials functionalization with natural polymeric biomolecules. Recent Adv Graphene Res. 2016;1:257–298.
8. Thangamuthu M, Hsieh KY, Kumar PV, Chen GY. Graphene- and graphene oxide-based nanocomposite platforms for electrochemical biosensing applications. Int J Mol Sci. 2019;20(12):2975. doi:10.3390/ijms20122975
9. Peea-Bahamonde J, Nguyen HN, Fanourakis SK, Rodrigues DF. Recent advances in graphene-based biosensor technology with applications in life sciences. J Nanobiotechnology. 2018;16(1):1–7.
10. Gs S, Cv A, Mathew BB. Biosensors: a modern day achievement. J Instrument Technol. 2014;2(1):26–39.
11. Asal M, Ozen O, Sahinler M, Baysal HT, Polatoglu I. An overview of biomolecules, immobilization methods and support materials of biosensors. Sensor Rev. 2019;39(3):377–386. doi:10.1108/SR-04-2018-0084
12. Karaman C, Aktas Z, Bayram E, Karaman O, Kizil C. Correlation between the molecular structure of reducing agent and pH of graphene oxide dispersion on the formation of 3D-graphene networks. ECS J Solid State Sci Technol. 2020;9(7):071003. doi:10.1149/2162-8777/abb192
13. Karaman C, Karaman O, Atar N, Yola ML. Tailoring of cobalt phosphide anchored nitrogen and sulfur co-doped three dimensional graphene hybrid: boosted electrocatalytic performance towards hydrogen evolution reaction. Electrochim Acta. 2021;380:138262. doi:10.1016/j.electacta.2021.138262
14. Liu X, Dai L. Carbon-based metal-free catalysts. Nat Rev Mater. 2016;1(11):16064. doi:10.1038/natrevmats.2016.64
15. Karaman C, Karaman O, Atar N, Yola ML. Sustainable electrode material for high-energy supercapacitor: biomass-derived graphene-like porous carbon with three-dimensional hierarchically ordered ion highways. PCCP. 2021;23(22):12807–12821. doi:10.1039/D1CP01726H
16. Karaman C, Karaman O, Yola BB, Ulker I, Atar N, Yola ML. A novel electrochemical aflatoxin B1 immunosensor based on gold nanoparticle-decorated porous graphene nanoribbon and Ag nanocube-incorporated MoS2 nanosheets. N J Chem. 2021;45(25):11222–11233. doi:10.1039/D1NJ02293H
17. Gizem Güneştekin B, Medetalibeyoglu H, Atar N, Lütfi Yola M. Efficient direct‐methanol fuel cell based on graphene quantum dots/multi‐walled carbon nanotubes composite. Electroanalysis (New York, NY). 2020;32(9):1977–1982.
18. Tolga Colak A, Eren T, Yola ML, Besli E, Sahin O, Atar N. 3D polyoxometalate-functionalized graphene quantum dots with mono-metallic and bi-metallic nanoparticles for application in direct methanol fuel cells. J Electrochem Soc. 2016;163(10):F1237–F1244. doi:10.1149/2.0911610jes
19. Atar N, Yola ML. A novel QCM immunosensor development based on gold nanoparticles functionalized sulfur-doped graphene quantum dot and h-ZnS-CdS NC for Interleukin-6 detection. Anal Chim Acta. 2021;1148:338202. doi:10.1016/j.aca.2021.338202
20. Atta N, El-Ads E, Galal A. Graphene — a platform for sensor and biosensor applications. Biosensors Micro Nanoscale Appl. 2015;9:38–84.
21. Fang Y, Wang E. Electrochemical biosensors on platforms of graphene. Chem Commun (Camb). 2013;49(83):9526–9539. doi:10.1039/c3cc44735a
22. Seo HM, Park JH, Dao TD, Jeong HM. Compatibility of functionalized graphene with polyethylene and its copolymers. (Research Article). J Nanomater. 2013;2013(2013):1–8. doi:10.1155/2013/805201
23. Shin DS, Kim HG, Ahn HS, et al. Distribution of oxygen functional groups of graphene oxide obtained from low-temperature atomic layer deposition of titanium oxide. RSC Adv. 2017;7(23):13979–13984. doi:10.1039/C7RA00114B
24. Lee J, Kim J, Kim S, Min D-H. Biosensors based on graphene oxide and its biomedical application. Adv Drug Deliv Rev. 2016;105(Pt B):275–287. doi:10.1016/j.addr.2016.06.001
25. Dideikin AT, Vul AY. Graphene oxide and derivatives: the place in graphene family. (report). Front Phys. 2019;6. doi:10.3389/fphy.2018.00149
26. Adeel M, Bilal M, Rasheed T, Sharma A, Iqbal HMN. Graphene and graphene oxide: functionalization and nano-bio-catalytic system for enzyme immobilization and biotechnological perspective. Int J Biol Macromol. 2018;120(Pt B):1430–1440. doi:10.1016/j.ijbiomac.2018.09.144
27. Luo H. A Theoretical Study of Graphene Oxide Chemical Structure. Australia: The School of Mechanical and Mining Engineering. The University of Queensland; 2017.
28. Zhang X, Yuan Q, Shu H, Ding F. Mechanisms of graphene chemical vapor deposition (CVD) growth. In: Chen DE, editor. Graphene Chemistry: Theoretical Perspectives. Chichester, United Kingdom: Wiley; 2013:255–290.
29. Sun L. Structure and synthesis of graphene oxide. Chin J Chem Eng. 2019;27(10):2251–2260. doi:10.1016/j.cjche.2019.05.003
30. Rana S, Jonnalagadda S. Covalently Functionalized Nano-Graphene Oxide for Fine Chemical Synthesis. IntechOpen; 2016.
31. Hermanov S, Zarevck M, Boua D, Pumera M, Sofer Z. Graphene oxide immobilized enzymes show high thermal and solvent stability. Nanoscale. 2015;7(13):5852–5858. doi:10.1039/C5NR00438A
32. Huitao Y, Bangwen Z, Chaoke B, Ruihong L, Ruiguang X. High-efficient synthesis of graphene oxide based on improved hummers method. Sci Rep. 2016;6(1):1–7.
33. Gao W. Graphene Oxide: Reduction Recipes, Spectroscopy, and Applications.
34. Liu Z, Norgaard K, Overgaard MH, et al. Direct observation of oxygen configuration on individual graphene oxide sheets. Carbon (New York). 2018;127:141–148.
35. Aliyev E, Khan M, Abetz C, Abetz V, Abetz C, Abetz V. Structural characterization of graphene oxide: surface functional groups and fractionated oxidative debris. Nanomaterials. 2019;9(8):1180. doi:10.3390/nano9081180
36. Liu Z, Rios-Carvajal T, Ceccato M, Hassenkam T. Nanoscale chemical mapping of oxygen functional groups on graphene oxide using atomic force microscopy-coupled infrared spectroscopy. J Colloid Interface Sci. 2019;556:458–465. doi:10.1016/j.jcis.2019.08.089
37. Wu H, Lu W, Shao -J-J, et al. pH-dependent size, surface chemistry and electrochemical properties of graphene oxide. Carbon (New York). 2014;67:795.
38. Luo L, Peng T, Yuan M, Sun H, Dai S, Wang L. Preparation of graphite oxide containing different oxygen-containing functional groups and the study of ammonia gas sensitivity. Sensors (Basel, Switzerland). 2018;18(11):3745. doi:10.3390/s18113745
39. Guobin Jia JP, Dellith J, Dellith A, Wahyuono RA, Andra G. Large area graphene deposition on hydrophobic surfaces, flexible textiles, glass fibers and 3d structures. J Technol. 2019;9:3579.
40. Ding J, Liu Y, Yuan N, Ding G, Fan Y, Yu C. The influence of temperature, time and concentration on the dispersion of reduced graphene oxide prepared by hydrothermal reduction. Diam Relat Mater. 2012;21:11–15. doi:10.1016/j.diamond.2011.08.004
41. Nazri SRB, Liu -W-W, Khe C-S, et al. Synthesis, characterization and study of graphene oxide. AIP Conf Proc. 2018;2045:020033.
42. Soozanipour A, Taheri-Kafrani A. Enzyme immobilization on functionalized graphene oxide nanosheets: efficient and robust biocatalysts. Methods Enzymol. 2018;609:371–403.
43. Yu W, Sisi L, Haiyan Y, Jie L. Progress in the functional modification of graphene/graphene oxide: a review. RSC Adv. 2020;10(26):15328–15345. doi:10.1039/D0RA01068E
44. Liu J, Tang J, Gooding JJ. Strategies for chemical modification of graphene and applications of chemically modified graphene. J Mater Chem. 2012;22(25):12435. doi:10.1039/c2jm31218b
45. Guo Y, Yang W, He F, et al. Electrostatic interaction-based self-assembly of paraffin@graphene microcapsules with remarkable thermal conductivity for thermal energy storage. Fullerenes Nanotubes Carbon Nanostruct. 2019;27(2):120–127. doi:10.1080/1536383X.2018.1517754
46. Duan X, Indrawirawan S, Sun H, Wang S. Effects of nitrogen-, boron-, and phosphorus-doping or codoping on metal-free graphene catalysis. Catalys Today. 2015;249(C):184–191. doi:10.1016/j.cattod.2014.10.005
47. Wang X, Shi G. An introduction to the chemistry of graphene. Phys Chem Chem Phys. 2015;17(43):28484–28504. doi:10.1039/C5CP05212B
48. Kasprzak A, Zuchowska A, Poplawska M. Functionalization of graphene: does the organic chemistry matter? Beilstein J Org Chem. 2018;14(1):2018. doi:10.3762/bjoc.14.177
49. Simsikova MST. Interaction of graphene oxide with proteins and applications of their conjugates. Nanomed Res. 2017;5(2):00109.
50. Wang F, Yu Z, Betegon M, et al. Amino and PEG-amino graphene oxide grids enrich and protect samples for high-resolution single particle cryo-electron microscopy. J Struct Biol. 2020;209(2):107437. doi:10.1016/j.jsb.2019.107437
51. Georgakilas V. Addition of organic groups through reactions with oxygen species of graphene oxide. In: Functionalization of Graphene. Wiley Online Library; 2014:59–94.
52. Niyogi S, Bekyarova E, Itkis ME, McWilliams JL, Hamon MA, Haddon RC. Solution properties of graphite and graphene. J Am Chem Soc. 2006;128(24):7720–7721. doi:10.1021/ja060680r
53. Zhang H, Li Z-F, Snyder A, Xie J, Stanciu LA. Functionalized graphene oxide for the fabrication of paraoxon biosensors. Anal Chim Acta. 2014;827:86–94. doi:10.1016/j.aca.2014.04.014
54. Li Q, Fan F, Wang Y, Feng W, Ji P. Enzyme immobilization on carboxyl-functionalized graphene oxide for catalysis in organic solvent. Ind Eng Chem Res. 2013;52(19):6343–6348. doi:10.1021/ie400558u
55. Li W, Wen H, Shi Q, Zheng G. Study on immobilization of (+) gamma-lactamase using a new type of epoxy graphene oxide carrier. Process Biochem. 2016;51(2):270–276. doi:10.1016/j.procbio.2015.11.030
56. Akkaya B, Cakiroglu B, Ozacar M. Tannic acid-reduced graphene oxide deposited with Pt nanoparticles for switchable bioelectronics and biosensors based on direct electrochemistry. ACS Sustain Chem Eng. 2018;6(3):3805–3814. doi:10.1021/acssuschemeng.7b04164
57. Bekir C, Ozacar M. Tannic acid modified electrochemical biosensor for glucose sensing based on direct electrochemistry. Electroanalysis (New York, NY). 2017;29(12):2719–2726.
58. Mao K, Wu D, Li Y, et al. Label-free electrochemical immunosensor based on graphene/methylene blue nanocomposite. Anal Biochem. 2012;422:22–27. doi:10.1016/j.ab.2011.12.047
59. Hu L, Hu X-R, Wu X, Du C, Dai Y, Deng J. Density functional calculation of transition metal adatom adsorption on graphene. Physica B Condens Matter. 2010;405:3337–3341. doi:10.1016/j.physb.2010.05.001
60. Szunerits S, Boukherroub R. Graphene-based bioelectrochemistry and bioelectronics: a concept for the future? Curr Opin Electrochem. 2018;12:141–147. doi:10.1016/j.coelec.2018.03.028
61. Song W, Li D-W, Li Y, Li Y, Long Y-T. Disposable biosensor based on graphene oxide conjugated with tyrosinase assembled gold nanoparticles. Biosens Bioelectron. 2011;26:3181–3186. doi:10.1016/j.bios.2010.12.022
62. Cakıroglu B, Ozacar M. Photoelectrochemical and non-enzymatic glucose sensor based on modified Fehling’s test by using Ti/TiO2 NTs-rGO-Cu2O electrode. J Electrochem Soc. 2019;166:B728–B734. doi:10.1149/2.1201908jes
63. Laaksonen P, Kainlauri M, Laaksonen T, et al. Interfacial engineering by proteins: exfoliation and functionalization of graphene by hydrophobins. Angewandte Chemie Int. 2010;49(29):4946–4949. doi:10.1002/anie.201001806
64. Liebana Girona S, Drago G, Estrela P. Bioconjugation and stabilisation of biomolecules in biosensors. Essays Biochem. 2016;60:59–68. doi:10.1042/EBC20150007
65. Prieto-Simín B, Campàs M, Marty JL. Biomolecule immobilization in biosensor development: tailored strategies based on affinity interactions. Protein Pept Lett. 2008;15:757–763. doi:10.2174/092986608785203791
66. Zhang Y, Wu C, Guo S, Zhang J. Interactions of graphene and graphene oxide with proteins and peptides. Nanotechnol Rev. 2013;2:27–45. doi:10.1515/ntrev-2012-0078
67. Srivastava G, Singh K, Talat M, Srivastava ON, Kayastha AM. Functionalized graphene sheets as immobilization matrix for Fenugreek β-amylase: enzyme kinetics and stability studies. PLoS One. 2014;9(11):e113408–e113408. doi:10.1371/journal.pone.0113408
68. Loo AH, Bonanni A, Pumera M. Biorecognition on graphene: physical, covalent, and affinity immobilization methods exhibiting dramatic differences. Chemistry. 2013;8(1):198–203. doi:10.1002/asia.201200756
69. Zou X, Wei S, Jasensky J, et al. Molecular interactions between graphene and biological molecules. J Am Chem Soc. 2017;139(5):1928–1936. doi:10.1021/jacs.6b11226
70. Wahab RA, Elias N, Abdullah F, Ghoshal SK. On the taught new tricks of enzymes immobilization: an all-inclusive overview. React Funct Polym. 2020;152:104613.
71. Zhang M, Yin B-C, Wang X-F, Ye B-C. Interaction of peptides with graphene oxide and its application for real-time monitoring of protease activity. Chem Commun. 2011;47(8):2399–2401. doi:10.1039/C0CC04887A
72. Woo J, Tran L-H, Jang S-H, Lee C, Kang T. Molecular interactions of graphene oxide with aromatic amino acids tyrosine and tryptophan. Bull Korean Chem Soc. 2015;36:
73. Wang L, Lin J. Phenylalanine-rich peptide mediated binding with graphene oxide and bioinspired synthesis of silver nanoparticles for electrochemical sensing. Appl Sci. 2017;7(2):160. doi:10.3390/app7020160
74. Ramakrishna TRB, Nalder TD, Yang W, Marshall SN, Barrow CJ. Controlling enzyme function through immobilisation on graphene, graphene derivatives and other two dimensional nanomaterials. J Mater Chem B. 2018;6(20):3200–3218. doi:10.1039/C8TB00313K
75. Kodali VK, Scrimgeour J, Kim S, et al. Nonperturbative chemical modification of graphene for protein micropatterning. Langmuir. 2011;27(3):863–865. doi:10.1021/la1033178
76. Bo Y, Yang H, Hu Y, Yao T, Huang S. A novel electrochemical DNA biosensor based on graphene and polyaniline nanowires. Electrochim Acta. 2011;56(6):2676–2681. doi:10.1016/j.electacta.2010.12.034
77. Noll JD, Nicholson MA, Van Patten PG, Chung C-W, Myrick ML. Template electropolymerization of polypyrrole nanostructures on highly ordered pyrolytic graphite step and pit defects. J Electrochem Soc. 1998;145(10):3320. doi:10.1149/1.1838806
78. Ping J, Zhou Y, Wu Y, et al. Recent advances in aptasensors based on graphene and graphene-like nanomaterials. Biosens Bioelectron. 2015;64:373–385. doi:10.1016/j.bios.2014.08.090
79. Cho I-H, Lee J, Kim J, et al. Current technologies of electrochemical immunosensors: perspective on signal amplification. Sensors (Basel, Switzerland). 2018;18(1):207. doi:10.3390/s18010207
80. Shao Y, Wang J, Wu H, Liu J, Aksay IA, Lin Y. Graphene based electrochemical sensors and biosensors: a review. Electroanalysis. 2010;22(10):1027–1036. doi:10.1002/elan.200900571
81. Huang Y, Tan J, Cui L, et al. Graphene and Au NPs co-mediated enzymatic silver deposition for the ultrasensitive electrochemical detection of cholesterol. Biosens Bioelectron. 2018;102:560–567. doi:10.1016/j.bios.2017.11.037
82. Wu G, Meyyappan M, Lai KWC. Simulation of graphene field-effect transistor biosensors for bacterial detection. Sensors. 2018;18(6):1715. doi:10.3390/s18061715
83. Pengfei L, Nan L, Jie X, Wei X. High-yield fabrication of graphene chemiresistors with dielectrophoresis. IEEE Trans Nanotechnol. 2012;11(4):751–759. doi:10.1109/TNANO.2012.2196524
84. Matsumoto K, Maehashi K, Ohno Y, Inoue K. Recent advances in functional graphene biosensors. J Phys D: Appl Phys. 2014;47(9):094005. doi:10.1088/0022-3727/47/9/094005
85. Ohno Y, Maehashi K, Matsumoto K. Label-free biosensors based on aptamer-modified graphene field-effect transistors. J Am Chem Soc. 2010;132(51):18012–18013. doi:10.1021/ja108127r
86. Rodrigo D, Limaj O, Janner D, et al. APPLIED PHYSICS. Mid-infrared plasmonic biosensing with graphene. Science (New York, NY). 2015;349(6244):165. doi:10.1126/science.aab2051
87. Hossain MB, Akib TBA, Abdulrazak LF, Rana MM. Numerical modeling of graphene-coated fiber optic surface plasmon resonance biosensor for BRCA1 and BRCA2 genetic breast cancer detection. Optic Eng. 2019;58(03):1. doi:10.1117/1.OE.58.3.037104
88. Jiri Homola MP. Surface Plasmon Resonance (SPR) sensors. In: Springer Series on Chemical Sensors and Biosensors. Vol. 4. Berlin, Heidelberg: Springer; 2006:45–67.
89. Hossain MB, Rana MM. DNA hybridization detection based on resonance-frequency-readout in graphene on Au SPR biosensor. J Sensors. 2016;2015. doi:10.1155/2016/6070742
90. Shushama K, Rana M, Inum R, Hossain M. Sensitivity enhancement of graphene coated surface plasmon resonance biosensor. Optical Quant Electron. 2017;49(11):1–13. doi:10.1007/s11082-017-1216-z
91. Hossain MB, Rana MM. Graphene coated high sensitive surface plasmon resonance biosensor for sensing DNA hybridization. Sens Lett. 2016;14:145–152. doi:10.1166/sl.2016.3596
92. Singh M, Holzinger M, Tabrizian M, et al. Noncovalently functionalized monolayer graphene for sensitivity enhancement of surface plasmon resonance immunosensors. J Am Chem Soc. 2015;137(8):2800–2803. doi:10.1021/ja511512m
93. Li M, Zhou X, Ding W, Guo S, Wu N. Fluorescent aptamer-functionalized graphene oxide biosensor for label-free detection of mercury(II). Biosens Bioelectron. 2013;41(1):889–893. doi:10.1016/j.bios.2012.09.060
94. Zhang Q, Zhang D, Lu Y, Yao Y, Li S, Liu Q. Graphene oxide-based optical biosensor functionalized with peptides for explosive detection. Biosens Bioelectron. 2015;68:494. doi:10.1016/j.bios.2015.01.040
95. Li Z, Zhang W, Xing F. Graphene optical biosensors. Int J Mol Sci. 2019;20(10):2461.
96. Bai Y, Xu T, Zhang X. Graphene-based biosensors for detection of biomarkers. Micromachines. 2019;11:60.
97. Lu CH, Yang HH, Zhu CL, Chen X, Chen GN. A Graphene Platform for sensing biomolecules. Angewandte Chemie Int. 2009;48(26):4785–4787. doi:10.1002/anie.200901479
98. Liu M, Zhao H, Chen S, Yu H, Zhang Y, Quan X. Label-free fluorescent detection of Cu(ii) ions based on DNA cleavage-dependent graphene-quenched DNAzymes. Chem Commun. 2011;47(27):7749–7751. doi:10.1039/c1cc12006a
99. Filip J, Andicsova-Eckstein A, Vikartovska A, Tkac J. Immobilization of bilirubin oxidase on graphene oxide flakes with different negative charge density for oxygen reduction. The effect of GO charge density on enzyme coverage, electron transfer rate and current density. Biosens Bioelectron. 2017;89:384–389. doi:10.1016/j.bios.2016.06.006
100. Chu X. 2-hydroxy-4-amino-azobenzene modified graphene oxide with incorporation of bilirubin oxidase for photoelectrochemical catalysis of oxygen reduction reaction. Int J Electrochem Sci. 2020;11531–11554. doi:10.20964/2020.11.46
101. Zhang J, Zhang F, Yang H, et al. Graphene oxide as a matrix for enzyme immobilization. Langmuir. 2010;26(9):6083–6085. doi:10.1021/la904014z
102. Besharati Vineh M, Saboury AA, Poostchi AA, Rashidi AM, Parivar K. Stability and activity improvement of horseradish peroxidase by covalent immobilization on functionalized reduced graphene oxide and biodegradation of high phenol concentration. Int J Biol Macromol. 2018;106:1314–1322. doi:10.1016/j.ijbiomac.2017.08.133
103. Rouhani S, Azizi S, Mamba B, Msagati TAM. Covalent immobilization of laccase on Fe3O4-graphene oxide nanocomposite for biodegradation of phenolic compounds. Environ Protect Eng. 2020;47. doi:10.37190/epe210107
104. Han Z, Shaofeng H, Lei Z, et al. Characterization of immobilized α-amylase on functionalized graphene oxide surface. Indian J Biochem Biophys. 2020;57:411–419.
105. Singh A, Sinsinbar G, Choudhary M, et al. Graphene oxide-chitosan nanocomposite based electrochemical DNA biosensor for detection of typhoid. Sens Actuators B Chem. 2013;185:675–684. doi:10.1016/j.snb.2013.05.014
106. Sekhon SS, Kaur P, Kim Y-H, Sekhon SS. 2D graphene oxide–aptamer conjugate materials for cancer diagnosis. NPJ 2D Mater Appl. 2021;5(1):21. doi:10.1038/s41699-021-00202-7
107. Gupta R, Kaul S, Singh V, Kumar S, Singhal NK. Graphene Oxide and Fluorescent Aptamer Based Novel Biosensor for Detection of 25-Hydroxyvitamin D3. Research Square; 2021.
108. Yang L, Wang G, Liu Y, Wang M. Development of a biosensor based on immobilization of acetylcholinesterase on NiO nanoparticles–carboxylic graphene–nafion modified electrode for detection of pesticides. Talanta (Oxford). 2013;113:135–141.
109. Vajedi FS, Dehghani H. A high-sensitive electrochemical DNA biosensor based on a novel ZnAl/layered double hydroxide modified cobalt ferrite-graphene oxide nanocomposite electrophoretically deposited onto FTO substrate for electroanalytical studies of etoposide. Talanta (Oxford). 2020;208:120444.
110. Moshari M, Koirala D, Allen PB. Electrochemical biosensors based on divinyl sulfone conjugation of DNA to graphene oxide electrodes. J Solid State Electrochem. 2021;25(5):1667–1678. doi:10.1007/s10008-021-04930-0
111. Karimi A, Othman A, Uzunoglu A, Stanciu L, Andreescu S. Graphene based enzymatic bioelectrodes and biofuel cells. Nanoscale. 2015;7(16):6909–6923. doi:10.1039/C4NR07586B
112. Lee J, Park I-S, Jung E, Lee Y, Min D-H. Direct, sequence-specific detection of dsDNA based on peptide nucleic acid and graphene oxide without requiring denaturation. Biosens Bioelectron. 2014;62:140–144. doi:10.1016/j.bios.2014.06.028
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
