Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Genetic Association Study Revealed Three Loci Were Associated Risk of Myopia Among Minors
Authors Zhou Z, Li S, Yang Q, Yang X, Hao K, Liu Y, Xu S
Received 11 December 2020
Accepted for publication 8 February 2021
Published 11 May 2021 Volume 2021:14 Pages 547—551
DOI https://doi.org/10.2147/PGPM.S296444
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin Bluth
Zixiu Zhou, Sizhen Li, Qingsong Yang, Xiaodong Yang, Kuanxiao Hao, Yating Liu, Shanshan Xu
Nanjing Tongren Hospital, School of Medicine, Southeast University, Nanjing, 211102, People’s Republic of China
Correspondence: Sizhen Li
Nanjing Tongren Hospital, School of Medicine, Southeast University, No. 2007, Jinyindadao Street, Jiangning District, Nanjing, 211102, People’s Republic of China
Tel +86-25-66987318
Email [email protected]
Background: Myopia has raised a predominant public concern among minors. A recent genome-wide association study (GWAS) identified six novel loci in Asian adults. Whether these genetic loci works for myopia in minors remains unknown and worthy of exploration.
Methods: In order to validate the findings, here we performed a case-control study (600 myopia minors, 110 high myopia (HM) minors, and 800 non-myopia minors as controls) utilizing the TaqMan single nucleotide polymorphism (SNP) genotyping assays. Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) was adopted.
Results: The median ages in controls, myopia, and HM were 15.1, 15.0, and 15.1, respectively, while the means ± standard deviations for them were 0.32± 0.41, - 3.2 ± 1.6, and − 9.8± 2.2, respectively. We found rs2246661 (allelic OR: 1.29; 95% CI: 1.09– 1.52; P =0.003), rs74633073 (allelic OR: 1.41; 95% CI: 1.12– 1.78; P =0.004), and rs76903431 (allelic OR: 1.42; 95% CI: 1.11– 1.81; P =0.005) were significantly associated with increased risk of myopia. Rs2246661 was also significantly associated with increased risk of HM in minors (OR: 1.37; 95% CI: 1.02– 1.84; P =0.035).
Conclusion: We identified three loci contributed to myopia in minors and these findings gave new insight into the genetic susceptibility mechanisms of myopia at the molecular level.
Keywords: myopia, genome-wide association study; GWAS, SNP, minors
Introduction
Myopia, the most common refractive error, results in a significant threat on global public health worldwide.1 As the myopic population increases globally, the severity of its impact is predicted.2 Children with early onset are particularly susceptible to myopia-related complications, like high myopia (HM) and myopic macular degeneration.3 According to a recent school-based epidemiology study of myopia in China with 14,551 participants (ages ranging from 5 to 16 years), the overall prevalence of myopia is 78.2%.4 Myopia is a complex disease which is contributed by various environmental and genetic factors. Environmental factors includes low outdoor time and near work, dim light exposure, the use of LED lamps for homework, low sleeping hours, and short reading distance.3,5 Meanwhile, there is growing evidence that susceptibility genes play a crucial role in the risk of myopia and single nucleotide polymorphisms (SNPs) may contribute to the risk of myopia.6–10
Recently, a genome-wide association study (GWAS) identified six novel loci (rs2246661, rs74633073, rs76903431, rs698047, rs17029206, and rs72748160) in Asian adults, and revealed the important role of genes in the nervous system in the pathogenesis of myopia.11 The findings highlighted a nervous system role in pathogenesis of myopia. Minors are better suited to studying the genetic factors of myopia. Whether these genetic loci works for myopia in minors remains unknown and worthy to be explored. Thus, here we aimed to evaluate the potential role of these GWAS identified loci in occurrence of myopia in this case-control study including 600 myopia minors, 110 HM minors and 800 non-myopia minors.
Patients and Methods
Study Population
A total of 600 consecutive myopia minors, 110 HM minors, and 800 non-myopia minors, which were frequency-matched by age and gender, were recruited in this case-control study. All subjects were Chinese Han population. Myopia was defined as mean spherical equivalent (MSE) of both eyes ≤ −0.5 diopters (D), while HM was defined as MSE less than or equal to −6.0 D.12 Patients with a predisposition to myopic eye disease, other known ocular or systemic diseases were excluded. Controls were selected from subjects coming for routine vision screening. The criteria for the control group were as follows: minors with MSE between −0.5 D and +1.0 D, best unaided visual acuity ≥ 0.8, and no other known ocular or systemic diseases.13 Patients are tested for refractive errors using an automated refractometer (Topcon RM-8000B, Topcon Co., Tokyo, Japan). The refraction was taken under cycloplegia. Information of all participants, including age, gender, body mass index (BMI, calculated using weight/height2), self-reported outdoors time, self-reported time using electronic equipment, and parental myopia, was collected through questionnaire responses, and all subjects donated 5 mL peripheral venous blood. The study protocol was approved by the ethics committee of Nanjing Tongren Hospital. All subjects gave their written informed consent, and the study complied with the Declaration of Helsinki.
DNA Extraction and Genotyping
Genomic DNA was extracted by blood DNA extraction kit (Promega, Madison, Wisconsin, USA) and stored in TE buffer. Genotyping was performed by TaqMan assay in 384-well ABI 7900HT Real-Time PCR system (Applied Biosystems [ABI], Foster City, CA). The qPCR reactions proceeded in a final volume of 10 μL mix including 5 μL TaqMan Genotyping Master Mix (Thermo Fisher Scientific), 0.5 μL pre-designed TaqMan probe (Thermo Fisher Scientific), 20 ng genomic DNA and ultrapure water. Each plate included blank samples as negative controls to verify genotyping quality. Genotype data were analyzed using their System SDS Allelic Discrimination Software version 2.3 (Applied Biosystems). For quality control, about 5% of the samples were genotyped repeatedly with Sanger sequencing and the results of both methods were in good agreement.
Statistical Analysis
SPSS 22.0 (SPSS, Chicago, IL) was used for statistical analysis, and a two-sided P-value of less than 0.05 was used as statistical significance. Chi -square goodness-of-fit test was adopted to derive the Hardy-Weinberg equilibrium (HWE). In the case-control study, Student t-test and/or Chi -square test were used to demonstrate how demographic and clinical characteristics and frequency of genotypes differ between case and control groups. Using unconditional logistic regression model, adjusted odds ratios (ORs) and 95% confidence intervals (CIs) were adopted (only significant variables in Table 1 were included for adjustment) to evaluate the effects of SNPs and to quantify the association between the SNPs and myopia in minors.
 |
Table 1 Characteristics of Participating Minors |
Results
Characteristics of Study Population
Table 1 presented the 600 myopia minors, 110 HM minors, and 800 non- myopia minors in this case-control study. The groups were comparable in age, and gender (P>0.05). While the highly myopic spent more time using electronic devices (P<0.001), less time outdoors (P<0.001) and had more myopic parents than non-myopic ones (P=0.015). The median ages in controls, myopia, and HM were 15.1, 15.0, and 15.1, respectively, while the means ± standard deviation for them were 0.32±0.41, −3.2 ±1.6, and −9.8±2.2, respectively.
Genetic Association Study of Myopia
All six SNPs analyzed were in HWE in non-myopia controls, indicating that the sampled subjects were representative of the population and did not show any bias in genotype frequency (p>0.05). Subsequently, we evaluated the associations between the selected SNPs and the risk of myopia adjusting for BMI, self-reported outdoors time, and self-reported time using electronic equipment. Table 2 showed the results of genotypic frequency analysis for selected loci. SNP rs2246661 (allelic OR: 1.29; 95% CI: 1.09–1.52; P =0.003), rs74633073 (allelic OR: 1.41; 95% CI: 1.12–1.78; P =0.004), and rs76903431 (allelic OR: 1.42; 95% CI: 1.11–1.81; P =0.005) significantly contributed to elevated susceptibility of myopia. Under additive genetic model, all of three SNPs showed statistically significant associations. For rs2246661, the CT genotype was associated with a 1.42-fold increased risk (95% CI = 1.12–1.81; P = 0.004), while the TT genotype conferred 1.64-fold increased risk of myopia (95% CI = 1.1–2.43; P = 0.014), compared with the TT genotype. For rs74633073, the CT genotype was associated with a 1.39-fold increased risk (95% CI = 1.05–1.85; P = 0.022), while the TT genotype conferred 2.61-fold increased risk of myopia (95% CI = 1.12–6.08; P = 0.026). For rs76903431, genotype GG was associated with a 1.48-fold increased risk (95% CI = 1.11–1.97; P = 0.007), while the GG genotype conferred 2.57-fold increased risk of myopia (95% CI = 1.04–6.37; P = 0.041), compared with the CC genotype.
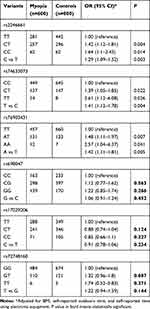 |
Table 2 Associations Between Candidate Loci and Myopia in Minors |
Genetic Association Study of HM
We further evaluated the associations of these six candidate SNPs with HM adjusting for self-reported outdoors time, self-reported time using electronic equipment, and parental myopia. We only found rs2246661 (OR: 1.37; 95% CI: 1.02–1.84; P =0.035), significantly contributed to elevated susceptibility of HM (Table 3). Under additive genetic model, the CT genotype was associated with a 1.55-fold increased risk (95% CI = 1.01–2.37; P = 0.044), while the TT genotype conferred 1.88-fold increased risk of myopia (95% CI = 1.03–3.43; P = 0.040), compared with the TT genotype.
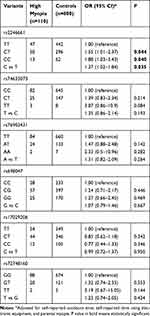 |
Table 3 Associations Between Candidate Loci and High Myopia in Minors |
Discussion
The current study investigated the potential function of six GWAS identified loci in occurrence of minors' myopia in a case-control study in Chinese population. We found three loci, including rs2246661, rs74633073, and rs76903431, significantly contributed to elevated risk of myopia. Besides, we also found rs2246661 significantly contributed to HM in minors. Our results confirm the GWAS findings in Asian adults and further provide a causal explanation for the occurrence of myopia at the molecular level.
The prevalence of myopia grew rapidly in minors.3,5,14 Finding the causes of the disease and taking effective preventive measures are vital to controlling the damage caused by myopia in young people. To date, a series of GWASs have been conducted to characterize the molecular mechanism responsible for myopia worldwide.11, 15–20 However, not all could be replicated. For example, Wang et al21 replicated findings of two Japanese GWAS in a Chinese population, and got null results. This was because myopia in adults was a genetically heterogeneous disease, which was influenced by inborn genetic factors and acquired environmental factors. On the contrary, minors are better suited to exploring the genetic factors of myopia. Thus, we attempted to classify the occurrence of myopia in minors was affected by GWAS loci identified in adults in this case-control study.
In the current study, rs2246661, rs74633073, and rs76903431 were identified to be associated risk of myopia in minors. Through searching Pubmed, we did not find any other genetic associations. According to RegulomeDB 2.0, rs2246661 and rs74633073 were located at the transcription factor (TF) binding site, which could affect the combination of TFs and their targets.22 HaploReg v4.1 revealed rs2246661 could cause Ets Motifs change, and rs74633073 could cause AP-2, RFX5 Motifs change, while rs76903431 could cause CDP, Pbx-1, RXRA Motifs change.23 This evidence supported the important role of these genetic loci.
Our study had several limitations. First, the selection bias of a case-control study design cannot be avoided. Second, early-onset myopia, which refers to myopia occurring before the age of 11 years, was not evaluated in current study, due to the limitations of sample size.24 Third, based on existing sample size, the associations might not have the strength to achieve real results, especially for HM. Fourth, the biological function of these SNPs and its detailed effect on occurrence of myopia need to be deep investigated by further biological studies. There are also several strengths in our research, including the detailed inspection and accurate diagnosis of cases, structured questionnaire by well-trained interviewers, and strict quality control of genotyping.
Conclusions
Conclusively, this study provides the evidence of the promotional role of rs2246661, rs74633073, and rs76903431 loci on the susceptibility of myopia. Replicated researches in independent ethnic samples and functional investigation are needed to confirm our findings.
Disclosure
The authors declare that they have no conflict of interest.
References
1. Foster PJ, Jiang Y. Epidemiology of myopia. Eye (Lond). 2014;28(2):202–208. doi:10.1038/eye.2013.280
2. Angle J, Wissmann DA. The epidemiology of myopia. Am J Epidemiol. 1980;111(2):220–228. doi:10.1093/oxfordjournals.aje.a112889
3. Grzybowski A, Kanclerz P, Tsubota K, Lanca C, Saw SM. A review on the epidemiology of myopia in school children worldwide. BMC Ophthalmol. 2020;20(1):27. doi:10.1186/s12886-019-1220-0
4. Wang J, Li Y, Zhao Z, et al. School-based epidemiology study of myopia in Tianjin, China. Int Ophthalmol. 2020;40(9):2213–2222. doi:10.1007/s10792-020-01400-w
5. Mak CY, Yam JC, Chen LJ, Lee SM, Young AL. Epidemiology of myopia and prevention of myopia progression in children in East Asia: a review. Hong Kong Med J. 2018;24(6):602–609.
6. Kunceviciene E, Liutkeviciene R, Budiene B, Sriubiene M, Smalinskiene A. Independent association of whole blood miR-328 expression and polymorphism at 3ʹUTR of the PAX6 gene with myopia. Gene. 2019;687:151–155. doi:10.1016/j.gene.2018.11.030
7. Zhang D, Zeng G, Hu J, McCormick K, Shi Y, Gong B. Association of IGF1 polymorphism rs6214 with high myopia: a systematic review and meta-analysis. Ophthalmic Genet. 2017;38(5):434–439. doi:10.1080/13816810.2016.1253105
8. Jin GM, Zhao XJ, Chen AM, Chen YX, Li Q. Association of COL1A1 polymorphism with high myopia: a Meta-analysis. Int J Ophthalmol. 2016;9(4):604–609.
9. Liang Y, Song Y, Zhang F, Sun M, Wang N. Effect of a single nucleotide polymorphism in the LAMA1 promoter region on Transcriptional activity: implication for pathological myopia. Curr Eye Res. 2016;41(10):1379–1386. doi:10.3109/02713683.2015.1118129
10. Chen T, Shan G, Ma J, Zhong Y. Polymorphism in the RASGRF1 gene with high myopia: a meta-analysis. Mol Vis. 2015;21:1272–1280.
11. Meguro A, Yamane T, Takeuchi M, et al. Genome-wide association study in asians identifies novel loci for high myopia and highlights a nervous system role in its pathogenesis. Ophthalmology. 2020;127(12):1612–1624. doi:10.1016/j.ophtha.2020.05.014
12. Luo HD, Gazzard G, Liang Y, Shankar A, Tan DT, Saw SM. Defining myopia using refractive error and uncorrected logMAR visual acuity >0.3 from 1334 Singapore school children ages 7–9 years. Br J Ophthalmol. 2006;90(3):362–366. doi:10.1136/bjo.2005.079657
13. Tideman JW, Polling JR, Voortman T, et al. Low serum vitamin D is associated with axial length and risk of myopia in young children. Eur J Epidemiol. 2016;31(5):491–499. doi:10.1007/s10654-016-0128-8
14. Recko M, Stahl ED. Childhood myopia: epidemiology, risk factors, and prevention. Mo Med. 2015;112(2):116–121.
15. Huang Y, Kee CS, Hocking PM, et al. A genome-wide association study for susceptibility to visual experience-induced myopia. Invest Ophthalmol Vis Sci. 2019;60(2):559–569. doi:10.1167/iovs.18-25597
16. Khor CC, Miyake M, Chen LJ, et al. Genome-wide association study identifies ZFHX1B as a susceptibility locus for severe myopia. Hum Mol Genet. 2013;22(25):5288–5294. doi:10.1093/hmg/ddt385
17. Meng W, Butterworth J, Bradley DT, et al. A genome-wide association study provides evidence for association of chromosome 8p23 (MYP10) and 10q21.1 (MYP15) with high myopia in the French Population. Invest Ophthalmol Vis Sci. 2012;53(13):7983–7988. doi:10.1167/iovs.12-10409
18. Li Z, Qu J, Xu X, et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20(14):2861–2868. doi:10.1093/hmg/ddr169
19. Solouki AM, Verhoeven VJ, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42(10):897–901. doi:10.1038/ng.663
20. Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42(10):902–905. doi:10.1038/ng.664
21. Wang Q, Gao Y, Wang P, et al. Replication study of significant single nucleotide polymorphisms associated with myopia from two genome-wide association studies. Mol Vis. 2011;17:3290–3299.
22. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using Regulome DB. Genome Res. 2012;22(9):1790–1797. doi:10.1101/gr.137323.112
23. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–881. doi:10.1093/nar/gkv1340
24. Baird PN, Saw SM, Lanca C, et al. Myopia. Nat Rev Dis Primers. 2020;6(1):99.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
