Back to Journals » Clinical Ophthalmology » Volume 11
Functional characterization of rs2229094 (T>C) polymorphism in the tumor necrosis factor locus and lymphotoxin alpha expression in human retina: the Retina 4 project
Authors Pastor-Idoate S , Rodríguez-Hernández I, Rojas J, Gonzalez-Buendia L, Delgado-Tirado S, Lopez JC, González-Sarmiento R , Pastor JC
Received 20 February 2017
Accepted for publication 19 April 2017
Published 22 May 2017 Volume 2017:11 Pages 973—981
DOI https://doi.org/10.2147/OPTH.S135170
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Salvador Pastor-Idoate,1,2 Irene Rodríguez-Hernández,2,3 Jimena Rojas,1 Lucia Gonzalez-Buendia,1 Santiago Delgado-Tirado,1,4 Jose Carlos López,1 Rogelio González-Sarmiento,2,3 Jose C Pastor1,4
1IOBA Eye Institute, University of Valladolid, Valladolid, 2Molecular Medicine Unit, Department of Medicine, 3Molecular and Cellular Cancer Biology Institute, High Council of Scientific Research, Biomedical Research Institute of Salamanca, University of Salamanca, Salamanca, 4Department of Ophthalmology, Hospital Clínico Universitario, Valladolid, Spain
Purpose: The objective of this study is to determine the expression and localization of lymphotoxin alpha (LTA) in human retinas and the functionality of one of its polymorphisms rs2229094 (C13R) (T>C), previously associated with proliferative vitreoretinopathy (PVR) development.
Materials and methods: Total RNA from three healthy human retinas were extracted and subjected to reverse transcription-polymerase chain reaction (RT-PCR) analysis, using flanking primers of LTA cDNA. In addition, three human eyes with retinal detachment (RD) and three healthy control eyes were subjected to immunohistochemistry (IHC) with a specific antibody against LTA. The functionality of T and C alleles was assessed by using pCEFL-Flag expression vector and transient transfection assays in COS-1 cell line. In addition, expression analysis by RT-PCR, Western blot and subcellular localization of both alleles and by immunofluorescence assay was performed.
Results: RT-PCR analysis revealed no significant levels of messenger RNA (mRNA) LTA in healthy human retinas. Sequential IHC staining showed differences between healthy human and RD retinas. No differences in mRNA and protein expression levels and in subcellular localization between both alleles were found. Both alleles were located in the cytoplasm of COS-1 cells.
Conclusion: Although results suggest lack of functionality, the differences found in IHC study and its strong association with PVR and its relationship with tumor necrosis factor locus, warrant further studies and could justify the use of this polymorphism as a valid biomarker to identify high-risk patients to develop PVR after RD.
Keywords: proliferative vitreoretinopathy, lymphotoxin alpha, tumor necrosis factor alpha, inflammation, cytokines, polymorphism
Introduction
Proliferative vitreoretinopathy (PVR) is the major cause of failure in retinal detachment (RD) surgery,1 affecting 5%–10% of RD and accounting for ~75% of all primary failures after RD surgery.1–3 PVR is a multifactorial disease4–9 considered as an abnormal wound-healing process induced by a retinal break, whereupon the levels of certain inflammatory cytokines (eg, tumor necrosis factor-alpha [TNFα]) and growth factors (eg, transforming growth factor beta [TGFβ], connective tissue growth factor or platelet-derived growth factor) are increased and may play an important role in its pathogenesis.2,4,5,10–13
The identification of single-nucleotide polymorphisms (SNPs) has an important implication in human diseases. They may help to assess the genetic susceptibility for certain diseases, either as a causative factor, a protective factor, or as a biomarker of the disease. Furthermore, the severity of inflammatory conditions and the circulating levels of some cytokines could be partially determined by related SNP.14 Thus, research focused on predictive molecular risk factors (biomarkers) of PVR susceptibility has been intensified in the recent years.
Lymphotoxin alpha (LTA), a member of the TNF family of cytokines, was initially isolated on the basis of an antitumor activity. Later, this cytokine was shown to have inflammatory and immunologic activities.15
A candidate gene association study, in which ~200 SNPs in 30 genes were analyzed, found a strong association between TNF locus and PVR.16 These results have been confirmed recently in an independent sample.17 Among those SNPs significantly associated with PVR, the r2229094 was particularly of interest. First, within the LTA gene, located in the TNF locus (6p21.3), the LTA rs2229094 SNP encodes a cysteine to arginine change in the signal peptide (both amino acids with a remarkable different hydropathy profile). Second, it showed a single significant association with PVR and also it was present in every haplotypic significant association.5 In addition, it was not present in any nonsignificant association.5 Third, when predictive models of PVR based on genetic variables were analyzed, the rs2229094 showed the highest individual predictor value.16 Finally, each allelic variant showed different hydropathy profile, different prediction of the transmembrane region and different quality of the splicing probability.5
Despite its shown association, the functionality of this SNP and the role of LTA in human retinas and in PVR pathogenesis remain to be determined. Hence, the purpose of this study was to characterize the functionality of rs2229094 SNP and determine the expression and localization of LTA in human retinas as part of the project named Retina 4.
Materials and methods
All tissue samples were used in accordance with applicable laws for research involving human tissues and samples (14/2007 of Biomedical Research) and the Declaration of Helsinki. The study was approved by the research committee of the IOBA, Eye Institute (University of Valladolid, Valladolid, Spain) and by the ethics committee of the Valladolid Clinic University Hospital (Valladolid, Spain).
Human samples
Three postmortem eyeballs from the Regional tissue bank (Castilla y León, Spain) from subjects with no reported history of eye disease within the age range of 40–60 years and two peripheral blood samples from healthy donors were obtained for RNA extraction. Eyes were transported on dry ice to the laboratory where, under aseptic conditions, each eyeball was bisected with scissors, dividing the ocular globe into anterior and posterior eye-cups. The vitreous was removed, and the posterior eyecup was placed into a dish with clean transport medium. A paintbrush was used to mechanically detach the neuroretina from the retinal pigment epithelium layer. In addition, three human eye globe specimens diagnosed with RD during the period 1990–2013 selected from the collection of the IOBA’s Ocular Pathology Laboratory (University of Valladolid, Spain) and three healthy eyes from the eye tissue bank were fixed in formalin and embedded in paraffin for immunohistochemistry (IHC) analysis. Normal adult human brain sections from the hippocampal area, provided by the Fundación Centro de Investigación en Enfermedades Neurológicas (CIEN) tissue bank (Madrid, Spain), were used as a positive control for the IHC study.
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA from three postmortem neuroretinas and two peripheral blood samples of human subjects was isolated using Trizol reagent (Invitrogen™, Carlsbad, CA, USA) according to manufacturer protocol. RNA concentration was determined by spectrophotometry on a Nanodrop (Thermo Scientific, Waltham, MA, USA). cDNA was synthesized with the Improm-II™ Reverse Transcription System (Promega, Madison, WI, USA) according to manufacturer specifications, with 1 μg total RNA per reaction. LTA cDNA was amplified from these samples with GoTaq Hot Start Polymerase (Promega) using flanking primers 5′-ATGACACCACCTGAACGTCTC-3′ (forward) and 5′-CTACAGAGCGAAGGCTCCAA-3′ (reverse) following the protocol: denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds and polymerization at 72°C for 1 minute. The glyceraldehyde-3-phosphate dehydrogenase expression levels in each sample were used as internal control using 5′-CCACCCATGGCAAATTCCATGGCA-3′ (forward) and 5′-TCTAGACGGCAGGTCAGGTCCACC-3′ (reverse) primers. PCR products were separated by electrophoresis in TBE agarose gels, and LTA RT-PCR products were sequenced to identify the different LTA transcripts obtained, and the LTA rs2229094 genotype of the LTA full-length of each sample in an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Generation of LTA expressing vectors
Complete cDNA carrying LTA rs2229094 allele C was amplified from a peripheral blood sample with primers containing EcoRI and NotI sites: 5′-GAATTCACACCACCTGAACG-3′ (forward) and 5′-GCGGCCGCCTACAGAGCGAAGG-3′ (reverse) (restriction sites are underlined) following the program described previously. The product was cloned in frame with an aminoterminal Flag epitope into the EcoRI-NotI sites of mammalian vector pCEFL-Flag to generate pCEFL-Flag-LTA-C. The polymorphic substitution rs2229094 was introduced in pCEFL-Flag-LTA-C with the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, San Diego, CA, USA) using the following primers: 5′-CTCCCAAGGGTGTGTGGCACCACCC-3′ (forward) and 5′-GGGTGGTGCCACACACCCTTGGGAG-3′ (reverse) to generate pCEFL-Flag-LTA-T. All vectors were transformed in Escherichia coli DH5α, and plasmid DNA was purified using the Rapid DNA plasmid miniprep kit (Genedan, S.L., Barcelona, Spain). All constructs were verified by automated sequencing.
Cell culture and DNA transfections
COS-1 cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 1% L-glutamine and 1% penicillin–streptomycin at 37°C in a humidified 5% CO2 atmosphere. Exponentially, growing COS-1 cells were transiently co-transfected with 1 μg total plasmid DNA (pCEFL-Flag-LTA-C and pCEFL-Flag-LTA-T) using X-tremeGENE HP DNA Transfection Reagent (Roche, Switzerland) at ratio 3:1. After 24–48 hours, cells were harvested to assess expression at RNA and protein level of pCEFL-Flag-LTA-C and pCEFL-Flag-LTA-T vectors by RT-PCR and Western blot. RT-PCR was performed as described previously.
Western blot
COS-1 cells were harvested 24–48 hours after transfection and resuspended in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA and 1% NP40) supplemented with Complete Mini (Roche, Switzerland) for protein extraction. Protein concentrations were determined using the BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Protein extracts (200 μg) were fractionated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis, transferred to an Immobilon-P membrane (Millipore, Billerica, MA, USA) and incubated with primary specific antibodies anti-Flag M2 and anti-β-actin (Sigma-Aldrich Co., St Louis, MO, USA). The ECL Plus detection System (GE Healthcare, Buckinghamshire, UK) with HRP-conjugated anti-mouse secondary antibodies (Sigma-Aldrich Co.) was used for detection.
Confocal immunofluorescence assay
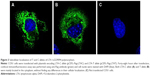
COS-1 cells were grown on glass cover-slips and transfected as indicated earlier. At 48 hours after transfection, cells were fixed with ice-cold 3.7% formaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.5% Triton X-100 in PBS and incubated with the primary antibody anti-Flag M2 (Sigma-Aldrich Co.) and the secondary antibody Alexa Fluor 488 Donkey Anti-Goat IgG conjugated to fluorescein isothiocyanate (FITC; Invitrogen). Finally, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich Co.), and cover-slips were mounted onto microscope slides in the presence of Mowiol mounting medium. Fluorescence images were captured with a LEICA TCS SP5 DMI-6000B confocal microscope (Leica Microsystems Inc., Wetzlar, Germany) using excitation wavelength of 488 nm (for FITC) and were analyzed with LEICA LAS AF (Leica Microsystems Inc.). Final processing of images was done with the Adobe Photoshop program 6.0 (Adobe Systems Incorporated, San Jose, CA, USA).
Immunohistochemical study
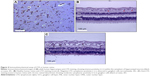
The aforementioned samples were used for the IHC study. The 3 μm sections were mounted on FLEX IHC Microscope slides (Dako, Glostrup, Denmark) and subjected to deparaffinization and hydration followed by heat-induced epitope retrieval in citrate buffer (pH 6.0). IHC staining was performed on these sections using a specific antibody against LTA (dilution 1:10; following manufacturer’s instructions; reference HPA007729; Sigma-Aldrich Co.), and the reaction was visualized by the EnVision (TM FLEX detection system kit (Dako). Normal adult human brain sections from the hippocampal area were used as a positive control. Images were taken using a Leica DM4000 B light microscope equipped with a Leica DFC490 digital camera (Leica Microsystems Inc.), and final processing of images was done with the Adobe Photoshop (Adobe Systems) program. The slides were evaluated semiquantitatively in a blind fashion by a pathologist using the following scoring system: −, negative; +, weakly positive; ++, moderately positive and +++, strongly positive.
Results
LTA messenger RNA (mRNA) expression was detected in peripheral blood but not in healthy human retinas
LTA expression was evaluated both at mRNA and protein levels by RT-PCR. LTA mRNA expression was below the significance level of the used method, and none of the three healthy human neuroretinas showed expression of LTA mRNA.
However, mRNA expression of LTA was detected in both DNA peripheral blood samples analyzed. Interestingly, RT-PCR analysis revealed different alternative LTA transcripts (Figure 1), but the analysis by sequencing of these new transcripts revealed premature stop codons, and consequently, these LTA isoforms may not have any function.
Functional characterization revealed no differences in either mRNA or protein expression levels between T and C alleles
The functionality of the T and C alleles of LTA rs2229094 polymorphism was investigated by cloning these alleles in the pCEFL-Flag expressing vector and by analyzing their expression (pCEFL-Flag-LTA-T and pCEFL-Flag-LTA-C) both at RNA and protein levels as well as their subcellular localization in COS-1 cells using a specific anti-Flag antibody.
mRNA expression of LTA-T and LTA-C evaluated by RT-PCR at 7, 24 and 48 hours after transfection (Figure 2A) showed no differences between the two alleles. In addition, no differences were observed in the protein expression level of LTA-T and LTA-C by Western blot analysis (Figure 2B).
To investigate whether the T to C substitution in the signal peptide of LTA polymorphism may affect the corresponding protein localization, an immunofluorescence assay to study the subcellular location of the T and C LTA alleles (pCEFL-Flag-LTA-T and pCEFL-Flag-LTA-C) was performed in the COS-1 cell line at 48 hours after transfection. Both LTA alleles were located mainly in the cellular cytoplasm; however, no differences in the subcellular location between the two alleles were observed (Figure 3A and B).
LTA protein expression was detected in retinas with RD but not in healthy human retinas
Constitutive LTA protein expression was evaluated by IHC in healthy human eyes, chronic RD donor eyes and n adult human brain sections from the hippocampal area (used as a positive control). Strong positive staining for LTA was detected in control brain sections, which was confined to hippocampal neuronal cells and their cytoplasms (Figure 4A).
The intensity of the immunostaining was negative in retinal sections from healthy eyes (Figure 4B), whereas the intensity of the immunostaining and the number of stained cells were moderately positive (++) in retinal sections from chronic RD eyes (Figure 4C). The immunostaining was confined to ganglion neuron cells and their cytoplasms (Figure 4C).
Discussion
Currently, PVR is considered a multifactorial disease and thus the result of interaction between genetic and environmental factors.4–9 Despite the fact that the exact mechanisms responsible of PVR are not completely understood, it is widely accepted that inflammation plays an important role in its pathogenesis.8,18
LTA expression and localization
TNF superfamily of cytokines regulates many physiological processes, including inflammation, proliferation, differentiation and cell death.19
The implication of proinflammatory cytokines, especially TNFα, in RD and PVR pathogenesis has been suggested in many reports. Previous studies reported high vitreous levels of TNFα and its receptors (TNFR 1 and 2) in eyes with PVR,20 and local production of TNFα has been proposed to occur in these eyes.21 Moreover, it has been reported that TNFα is able to mediate photoreceptor cell death after RD,22–24 and the addition of exogenous TNFα to organotypic culture of porcine neuroretinas has demonstrated to result in morphologic changes that resemble some of those found in human PVR.10,25,26
TNFα and LTA are two molecules that play an important role in inflammation. Previous studies demonstrate that LTA, with a 40% of homology to TNFα, shares the same membrane receptors as TNFα, exerting its biological effect largely via TNF receptor 1 (TNFR1) and TNFR2 activation.27 However, retinal expression and localization of LTA have not been previously described in either normal or RD eyes.
In this study, a positive immunostaining of LTA predominantly localized in the cytoplasm of ganglion cells in chronic RD eyes was observed. This particular localization (inside of the cell) could be due probably because LTA is internalized after its receptor binding as TNFα.28–31 But the reason why it was only visualized in ganglion cells remains unclear.
LTA mediates its proinflammatory effects in part through the induction of adhesion molecules as vascular cell-adhesion molecule 1 and chemokine expression as monocyte chemotactic protein-1 (MCP-1).32 Increased levels of TNFα in outer nuclear layer, inner nuclear layer and MCP-1 in ganglion cell layer after a experimental RD have been reported.33
LTA is able to act through LTA–TNFR1 complex as TNFα, but it seems to exist differences in their signal strength or capabilities.34 In particular, it has been reported that LTA is less able to promote TNFR1-induced cell death and nuclear factor-κB activation,35 expression of cell surface markers36 and cytokine production37,38 than TNFα. Alternatively, TNFα and LTA have shown protective effects in neuronal cells when acting via TNFR2, especially in ischemic conditions both in brain and in retina.39,40 Thus, despite LTA is able to induce signals via TNFR1 and could have a crucial role in the initiation and development of some inflammatory diseases,41,42 due to its diverse bioactivities, it is presently not clear under which conditions LTA promotes beneficial or deleterious effects on neuronal tissues. Therefore, further studies will be critical to better understand the role of LTA in retinal neuronal cells after RD.
In this study, only three retinas suffering chronic RD and three healthy retinas could be analyzed due to the scarcity of this material, from human origin. Collecting a greater number of samples would be extremely challenging, especially samples from PVR patients. In addition, quantification of positive LTA staining was based on the impression of the pathologist, and a formal recount with consecutive serial sections was not performed, which is one of the major limitations of this study. However, to the best of our knowledge, this is the first report of LTA expression in human neuroretina.
The intensity of the immunostaining was considered moderately positive compared with positive brain section controls. RD samples used in this study presented chronic RD and intraocular tissues were relatively disorganized, which is another limitation of this study. For this reason, they were enucleated. Thus, it is possible that in these eyes immediate proinflammatory mediators may not be present as it occurs in acute RD. In addition, the increase of neuronal cell death could be one of the possible causes leading to a smaller number of positive cells and weaker immunostaining in chronic RD eyes.
Functionality of the T and C alleles of LTA rs2229094 polymorphism
LTA gene is located on chromosome 6, between the HLA class II and class I loci. LTA gene SNPs are also shown to be associated with the inflammatory and immunomodulatory diseases including cancer.43–48 In addition, the contribution of TNF genes to PVR susceptibility has been previously discussed. A positive association of TNFα and LTA SNPs with PVR has been reported.5,17 However, the functionality of most of these SNPs remains unknown.
LTA rs2229094 SNP is a nonsynonymous polymorphism located in the signal peptide of LTA gene, which results in a change from cysteine to arginine in codon 13 (Cys13Arg). While cysteine is a neutral hydrophobic amino acid, arginine is a hydrophilic, positively charged amino acid. Regarding the results of bioinformatic tool analysis,5 the hydropathy profile changed for each allelic variant could affect the function of LTA protein or modify its production or its localization, due to the hydrophobicity of the signal peptide and the asymmetric distribution of the hydrophobic and hydrophilic potentials, which determine the translocation specificity of proteins.49–51
However, the functional analysis performed in this study did not show any differences in the expression levels between the LTA-T and C alleles (cysteine and arginine alleles, respectively). Furthermore, the subcellular location of C allele did not vary on the T allele, both being located in the cytoplasm, suggesting that the C allele has no significant effect in the LTA gene transcriptional regulation and subcellular location.
In this study, the times adopted for the expression analysis could not have been sufficient to prove the existence of these differences between T and C alleles. Besides, LTA gene is located within TNF locus, and it is closely linked to TNFα and lymphotoxin beta genes. Therefore, modifications derived from functional SNPs in any of these could lead to collateral alterations in the neighboring genes.48 In addition, only three normal retinas and two peripheral blood samples could be analyzed to assess the mRNA and protein expression of LTA, due to the scarcity of this material and strict inclusion criteria in the obtention of samples.
Conclusion
Although the results suggest a lack of functionality of the LTA rs2229094 SNP, this polymorphism could remain as a valid biomarker to identify high-risk patients to develop PVR after RD, due to the strong association between PVR and LTA rs2229094 SNP previously shown by our group. In addition, we believe that differences found in the IHC studies, its strong association with PVR and its relation to TNF locus warrant further studies to clarify its potential role in RD and PVR pathogenesis.
Acknowledgments
The authors would like to thank Nieves Fernández (IOBA-University of Valladolid, Spain) for her assistance with the pictures that illustrate this work and for performing the laboratory experiments. Also, we would like to thank Fundación Centro de Investigación en Enfermedades Neurológicas (CIEN) tissue bank (Madrid, Spain) and the Hospital Clínico Universitario (Valladolid, Spain) for providing normal adult human brain sections and human eyes from donors. This article was presented in ARVO Annual Meeting, Denver, CO, May 2014, as a poster and EVER Annual meeting, Nice, France, October 2014, as a Rapid Fire. RGS is supported by SAF 2007-66394, FIS PI13/01741 and Group of Excellence Grant (GR15) from Junta de Castilla y León. The funding organization had no role in the design or conduct of this research.
Disclosure
The authors report no conflicts of interest in this work.
References
Machemer R, Aaberg TM, Freeman HM, Irvine AR, Lean JS, Michels RM. An updated classification of retinal detachment with proliferative vitreoretinopathy. Am J Ophthalmol. 1991;112(2):159–165. | ||
Pastor JC, de la Rúa ER, Martín F. Proliferative vitreoretinopathy: risk factors and pathobiology. Prog Retin Eye Res. 2002;21(1):127–144. | ||
de la Rua ER, Pastor JC, Fernandez I, et al. Non-complicated retinal detachment management: variations in 4 years. Retina 1 project; report 1. Br J Ophthalmol. 2008;92(4):523–525. | ||
Sanabria Ruiz-Colmenares MR, Pastor Jimeno JC, Garrote Adrados JA, Telleria Orriols JJ, Yugueros Fernández MI. Cytokine gene polymorphisms in retinal detachment patients with and without proliferative vitreoretinopathy: a preliminary study. Acta Ophthalmol Scand. 2006;84(3):309–313. | ||
Rojas J, Fernandez I, Pastor JC, et al. A strong genetic association between the tumor necrosis factor locus and proliferative vitreoretinopathy: the Retina 4 project. Ophthalmology. 2010;117(12):2417.e1–e2–2423.e1–e2. | ||
Pastor-Idoate S, Rodriguez-Hernández I, Rojas J, et al. The p53 codon 72 polymorphism (rs1042522) is associated with proliferative vitreoretinopathy: the Retina 4 project. Ophthalmology. 2013;120(3):623–628. | ||
Pastor-Idoate S, Rodríguez-Hernández I, Rojas J, et al. The T309G MDM2 gene polymorphism is a novel risk factor for proliferative vitreoretinopathy. PLoS One. 2013;8(12):e82283. | ||
Pastor JC, Rojas J, Pastor-Idoate S, Di Lauro S, Gonzalez-Buendia L, Delgado-Tirado S. Proliferative vitreoretinopathy: a new concept of disease pathogenesis and practical consequences. Prog Retin Eye Res. 2016;51:125–155. Review. | ||
Pastor-Idoate S, Rodríguez-Hernández I, Rojas J, et al. BAX and BCL-2 polymorphisms, as predictors of proliferative vitreoretinopathy development in patients suffering retinal detachment: the Retina 4 project. Acta Ophthalmol. 2015;93(7):e541–e549. | ||
Pastor JC, Méndez MC, de la Fuente MA, et al. Intraretinal immunohistochemistry findings in proliferative vitreoretinopathy with retinal shortening. Ophthalmic Res. 2006;38(4):193–200. | ||
Pennock S, Rheaume MA, Mukai S, Kazlauskas A. A novel strategy to develop therapeutic approaches to prevent proliferative vitreoretinopathy. Am J Pathol. 2011;179(6):2931–2940. | ||
Moysidis SN, Thanos A, Vavvas DG. Mechanisms of inflammation in proliferative vitreoretinopathy: from bench to bedside. Mediators Inflamm. 2012;2012:815937. | ||
Pennock S, Haddock LJ, Eliott D, Mukai S, Kazlauskas A. Is neutralizing vitreal growth factors a viable strategy to prevent proliferative vitreoretinopathy? Prog Retin Eye Res. 2014;40:16–34. | ||
Clark AG. The role of haplotypes in candidate gene studies. Genet Epidemiol. 2004;27(4):321–333. | ||
Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. | ||
Rojas J, Fernandez I, Pastor JC, et al. Development of predictive models of proliferative vitreoretinopathy based on genetic variables: the Retina 4 project. Invest Ophthalmol Vis Sci. 2009;50(5):2384–2390. | ||
Rojas J, Fernandez I, Pastor JC, et al. A genetic case-control study confirms the implication of SMAD7 and TNF locus in the development of proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2013;54(3):1665–1678. | ||
Delyfer MN, Raffelsberger W, Mercier D, et al. Transcriptomic analysis of human retinal detachment reveals both inflammatory response and photoreceptor death. PLoS One. 2011;6(12):e28791. | ||
Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104(4):487–501. | ||
Limb GA, Hollifield RD, Webster L, Charteris DG, Chignell AH. Soluble TNF receptors in vitreoretinal proliferative disease. Invest Ophthalmol Vis Sci. 2001;42(7):1586–1591. | ||
El-Ghrably IA, Dua HS, Orr GM, Fischer D, Tighe PJ. Detection of cytokine mRNA production in infiltrating cells in proliferative vitreoretinopathy using reverse transcription polymerase chain reaction. Br J Ophthalmol. 1999;83(11):1296–1299. | ||
Nakazawa T, Kayama M, Ryu M, et al. Tumor necrosis factor-alpha mediates photoreceptor death in a rodent model of retinal detachment. Invest Ophthalmol Vis Sci. 2011;52(3):1384–1391. | ||
Murakami Y, Notomi S, Hisatomi T, et al. Photoreceptor cell death and rescue in retinal detachment and degenerations. Prog Retin Eye Res. 2013;37:114–140. | ||
Kataoka K, Matsumoto H, Kaneko H, et al. Macrophage-and RIP3-dependent inflammasome activation exacerbates retinal detachment-induced photoreceptor cell death. Cell Death Dis. 2015;6:e1731. | ||
Sethi CS, Lewis GP, Fisher SK, et al. Glial remodeling and neural plasticity in human retinal detachment with proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2005;46(1):329–342. | ||
Fernandez-Bueno I, Fernandez-Bueno I, Garcia-Gutierrez MT, Gayoso MJ, Gonzalo-Orden JM, Pastor JC. Adalimumab (tumor necrosis factor-blocker) reduces the expression of glial fibrillary acidic protein immunoreactivity increased by exogenous tumor necrosis factor alpha in an organotypic culture of porcine neuroretina. Mol Vis. 2013;19:894–903. | ||
Naoum JJ, Chai H, Lin PH, Lumsden AB, Yao Q, Chen C. Lymphotoxin-alpha and cardiovascular disease: clinical association and pathogenic mechanisms. Med Sci Monit. 2006;12(7):RA121–RA124. | ||
Bradley JR, Thiru S, Pober JS. Disparate localization of 55-kd and 75-kd tumor necrosis factor receptors in human endothelial cells. Am J Pathol. 1995;146(1):27–32. | ||
Botchkina GI, Meistrell ME 3rd, Botchkina IL, Tracey KJ. Expression of TNF and TNF receptors (p55 and p75) in the rat brain after focal cerebral ischemia. Mol Med. 1997;3(11):765–781. | ||
Schutze S, Machleidt T, Adam D, et al. Inhibition of receptor internalization by monodansylcadaverine selectively blocks p55 tumor necrosis factor receptor death domain signaling. J Biol Chem. 1999;274(15):10203–10212. | ||
Tezel G, Li LY, Patil RV, Wax MB. TNF-alpha and TNF-alpha receptor-1 in the retina of normal and glaucomatous eyes. Invest Ophthalmol Vis Sci. 2001;42(8):1787–1794. | ||
Cuff CA, Schwartz J, Bergman CM, Russell KS, Bender JR, Ruddle NH. Lymphotoxin alpha3 induces chemokines and adhesion molecules: insight into the role of LT alpha in inflammation and lymphoid organ development. J Immunol. 1998;161(12):6853–6860. | ||
Nakazawa T, Matsubara A, Noda K, et al. Characterization of cytokine responses to retinal detachment in rats. Mol Vis. 2006;12:867–878. | ||
Etemadi N, Holien JK, Chau D, et al. Lymphotoxin alpha induces apoptosis, necroptosis and inflammatory signals with the same potency as tumour necrosis factor. FEBS J. 2013;280(21):5283–5297. | ||
Chaturvedi MM, LaPushin R, Aggarwal BB. Tumor necrosis factor and lymphotoxin. Qualitative and quantitative differences in the mediation of early and late cellular response. J Biol Chem. 1994;269(20):14575–14583. | ||
Andrews JS, Berger AE, Ware CF. Characterization of the receptor for tumor necrosis factor (TNF) and lymphotoxin (LT) on human T lymphocytes. TNF and LT differ in their receptor binding properties and the induction of MHC class I proteins on a human CD4+ T cell hybridoma. J Immunol. 1990;144(7):2582–2591. | ||
Broudy VC, Harlan JM, Adamson JW. Disparate effects of tumor necrosis factor-alpha/cachectin and tumor necrosis factor-beta/lymphotoxin on hematopoietic growth factor production and neutrophil adhesion molecule expression by cultured human endothelial cells. J Immunol. 1987;138(12):4298–4302. | ||
Oster W, Lindemann A, Horn S, Mertelsmann R, Herrmann F. Tumor necrosis factor (TNF)-alpha but not TNF-beta induces secretion of colony stimulating factor for macrophages (CSF-1) by human monocytes. Blood. 1987;70(5):1700–1703. | ||
Shohami E, Ginis I, Hallenbeck JM. Dual role of tumor necrosis factor alpha in brain injury. Cytokine Growth Factor Rev. 1999;10(2):119–130. | ||
Fontaine V, Mohand-Said S, Hanoteau N, Fuchs C, Pfizenmaier K, Eisel U. Neurodegenerative and neuroprotective effects of tumor Necrosis factor (TNF) in retinal ischemia: opposite roles of TNF receptor 1 and TNF receptor 2. J Neurosci. 2002;22(7):RC216. | ||
Buch MH, Conaghan PG, Quinn MA, Bingham SJ, Veale D, Emery P. True infliximab resistance in rheumatoid arthritis: a role for lymphotoxin alpha? Ann Rheum Dis. 2004;63(10):1344–1346. | ||
Calmon-Hamaty F, Combe B, Hahne M, Morel J. Lymphotoxin alpha stimulates proliferation and pro-inflammatory cytokine secretion of rheumatoid arthritis synovial fibroblasts. Cytokine. 2011;53(2):207–214. | ||
Garrote JA, Arranz E, Telleria JJ, Castro J, Calvo C, Blanco-Quirós A. TNF alpha and LT alpha gene polymorphisms as additional markers of celiac disease susceptibility in a DQ2-positive population. Immunogenetics. 2002;54(8):551–555. | ||
Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. | ||
Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9(5):361–371. | ||
Drutskaya MS, Efimov GA, Kruglov AA, Kuprash DV, Nedospasov SA. Tumor necrosis factor, lymphotoxin and cancer. IUBMB Life. 2010;62(4):283–289. | ||
Lu R, Dou X, Gao X, Zhang J, Ni J, Guo L. A functional polymorphism of lymphotoxin-alpha (LTA) gene rs909253 is associated with gastric cancer risk in an Asian population. Cancer Epidemiol. 2012;36(6):e380–e386. | ||
Huang Y, Yu X, Wang L, et al. Four genetic polymorphisms of lymphotoxin-alpha gene and cancer risk: a systematic review and meta-analysis. PLoS One. 2013;8(12):e82519. | ||
Goldstein J, Lehnhardt S, Inouye M. Enhancement of protein translocation across the membrane by specific mutations in the hydrophobic region of the signal peptide. J Bacteriol. 1990;172(3):1225–1231. | ||
Hoyt DW, Gierasch LM. Hydrophobic content and lipid interactions of wild-type and mutant OmpA signal peptides correlate with their in vivo function. Biochemistry. 1991;30(42):10155–10163. | ||
Talmud P, Lins L, Brasseur R. Prediction of signal peptide functional properties: a study of the orientation and angle of insertion of yeast invertase mutants and human apolipoprotein B signal peptide variants. Protein Eng. 1996;9(4):317–321. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.