Back to Journals » Infection and Drug Resistance » Volume 13
Ferritin Degradation by Pneumococcal HtrA, RadA and ClpP Serine Proteases : A Probable Way For Releasing and Acquisition Of Iron
Authors Kheirandish M , Motlagh B, Afshar D
Received 23 May 2020
Accepted for publication 7 August 2020
Published 11 September 2020 Volume 2020:13 Pages 3145—3152
DOI https://doi.org/10.2147/IDR.S264170
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mozhgan Kheirandish,1 Behrooz Motlagh,2 Davoud Afshar1
1Department of Microbiology and Virology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran; 2Department of Clinical Biochemistry, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
Correspondence: Davoud Afshar
Department of Microbiology and Virology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
Tel +98 2433140297
Email [email protected]
Purpose: Iron is a necessary element for the growth of bacteria; however, there are limited iron sources known for these microorganisms yet. Intracellular iron is stored as ferritin from, which releases iron in a gradual and controlled manner. The present study aimed to characterize ferritin-binding proteins (FBPs) of Streptococcus pneumoniae.
Material and Methods: S. pneumoniae species were cultured in BHI broth containing ferritin (1094 ng/mL) for 4h at 37°C. Ferritin level was measured using ELISA assay. Bacterial proteome was electrophoresed on SDS-PAGE and then transferred on PVDF nitrocellulose membrane. Afterward, the PVDF membranes were incubated with a ferritin solution. Identification of ferritin binding proteins was performed using anti-ferritin monoclonal antibody conjugated with HRP enzyme. Molecular docking was used to assess the interaction between pneumococcal proteases and FBPs applying phenylmethylsulfonyl fluoride (PMSF) as a protease inhibitor.
Results: No FBPs were identified in S. pneumoniae proteome. Moreover, ferritin levels have significantly (p< 0.05) decreased following the growth of S. pneumoniae in ferritin-rich BHI medium. Also, molecular docking showed that RadA protease, ClpP hydrolase, and HtrA protease can potentially interact with PMSF protease inhibitors. On the other hand, the addition of the PMSF to the culture of S. pneumoniae prevented the reduction of ferritin, which indicates a potential role of RadA, ClpP, and HtrA proteases in ferritin degradation.
Conclusion: Our results suggest that S. pneumoniae produces no FBPs and also cannot directly use ferritin as an iron source. However, ferritin may be degraded through a protease-mediated mode.
Keywords: Streptococcus pneumoniae, ferritin-binding proteins, proteome, iron
Introduction
Streptococcus pneumoniae is a pathogenic bacterium causing many infections such as meningitis, sinusitis, pneumonia, otitis media, and bacteremia.1 Although S. pneumoniae presents as the normal flora within sinuses and nasopharyngeal tract in some individuals, it may also cause disease in people with compromised immune system such as young children and elderly. In fact, S. pneumoniae is considered as an important cause of meningitis, pneumonia, and sepsis in patients with HIV.
Iron is essential for the growth of bacteria, and particularly for pneumococci. In human beings, iron exists in combinations of glycoproteins such as ferritin, transferrin, and lactoferrin.2 Therefore, iron is not easily available to bacteria, so they must recruit distinct strategies to earn this element.3–7
Iron uptake mechanisms of S. pneumoniae that have not been thoroughly understood. Although some bacteria produce siderophore, this strategy has not been reported in pneumococci.8 In this regard, some studies have shown that ABC transporters (eg, Piu, Pia and Pit) constitute the main iron acquisition systems in pneumococci.9
Ferritin, as the major iron storing protein, plays a significant role in iron homeostasis by preserving up to 4,500 iron atoms in its structure.10,11 In addition, it comprises 24 units from two H and L chains and presents in the cytoplasm of cells and in the serum.12 Iron is essential for S. pneumoniae to propagate within hosts; however, iron acquisition mechanisms of this bacterium are not fully understood yet. Therefore, in the present study, we aimed to characterize ferritin-binding proteins (FBPs) of S. pneumoniae.
Materials and Methods
Evaluation of Ferritin, Transferrin and Iron Levels in Pneumococcal Culture
Bacterial Growth in BHI Medium Containing Human Serum
S. pneumoniae ATCC49619 was cultured in BHI broth for 24h at 37 ° C. Afterward, the culture medium was centrifuged at 9690 g for 3 min, and the pellet was re-cultured in BHI broth medium containing concentrated serum (1094 ng/mL ferritin, 135 µg/mL transferrin, and 88 µg/mL iron). Subsequently, the culture was further incubated in the candle jar for 4 hours at 37 ° C and finally centrifuged for 3 minutes at 9690 g. The supernatant was then separated to determine the levels of ferritin, transferrin, and iron using enzyme-linked immunosorbent assay (ELISA), immunoturbidometric, and photometric assays, respectively. The protocols were conducted in terms of the manufacturer’s instructions (Pishtazteb & Pars Azmoon, Iran).
Evaluation of FBPs
Purification of Proteins by the Acetone and Ammonium Sulfate Methods
In this study, acetone was used to precipitate pneumococcal proteins in the culture supernatant. 13 The following steps were performed to precipitate proteins:
- The growth medium supernatant and cold acetone (−20 ° C) (the ratio of 1 to 4) were mixed into a falcon. After mixing, the falcon was incubated for 60 minutes at −20 ° C.
- The sample was centrifuged for 10 min at 13000 g at −4 ° C.
- The supernatant was cautiously discarded, and the Falcon was kept for 30 minutes at room temperature until the acetone was completely vaporized.
- Distilled water was added to the falcon, and after 10 minutes of shaking, 2% SDS solution was added to the mixture.The proteins into pellets of S. pneumoniae culture were also purified by ammonium sulfate method following cell lysis by sonication. Protein concentration was then measured using Nanodrop, and the precipitated proteins were stored at −70 ° C.
Western Blotting
The purified proteins and pneumococcus pellet lysate were boiled in sample buffer and electrophoresed on 10% SDS-PAGE. Another sample mixed with 2ME-free sample buffer and without boiling, and subsequently transferred on polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, USA). The membrane was blocked by 3% blocking buffer (PBS-1X containing 3% nonfat skim milk and 0.05% Tween 20) overnight at 4 °C and rinsed three times with washing buffer (PBS-1X containing 0.05% Tween 20). The PVDF membrane was incubated with ferritin (62 ng/mL) solution at room temperature for 3 h and washed three times with the washing buffer. After that, the membrane was incubated (25 °C, 1 h) with diluted (1:1000) anti-ferritin antibody conjugated to Horseradish peroxidase (HRP) (Pishtazteb, Iran) and then rinsed three times with washing buffer. Finally, the membrane was treated with 3, 3-diaminobenzidine (DAB) solution (Sigma-Aldrich, USA) for ~3 min. Plasma sample was also used as a positive control.
The Activity of Pneumococcal Proteases Against Ferritin and Transferrin
S. pneumoniae was cultured in BHI broth for 24 h at 37 ° C. Subsequently, the bacterial suspension was then centrifuged at 9690 g for 3 min, and the supernatant discarded. Accordingly, the pellet was transferred into a sterile tube containing transferrin (135 µg/mL) and ferritin (1094 ng/mL), and the tube was then incubated for 4 hours at 37 ° C. Tube without bacterium culture (Serum and BHI (SBHI)) and another containing 1mM PMSF (SBHI+PMSF) were also used as controls 1 and 2, respectively. After incubation, the tubes were centrifuged at 9690 g for 3 min. Finally, the concentrations of ferritin, transferrin, and iron were measured using protocols mentioned earlier in section 1. All of the tests were repeated five times.
Docking of Pneumococcal Proteases with PMSF
A list of pneumococcal proteases was first obtained from the protein data bank (PDP) database and imported into Molegro virtual docker software (MVD 2013.6.0). Protease cavities were detected based on expanded Van der Waals on molecule surface and the following settings; minimum cavity volume: 10, maximum cavity volume: 10000, probe size: 1.20, max number of ray hits: 16, min number of hits: 12 and grid resolution: 0.80.
PMSF as a ligand was then imported into MVD software and molecular docking with tested proteases performed under the following parameters:
- Scoring function: Score: MolDock, Resolution Grade: 0.30A
- Binding site properties: radius 15, X axis (−1.95), Y axis (−0.97), and Z axis (−44.6).
- Search algorithm: MolDock SE
All of the predicted poses were separately analyzed for hydrogen bond interaction, bond length (Å) and other types of interactions between protease and PMSF.
Statistical Analysis
Statistical analysis was performed using SPSS V22.0 software. Differences in values of Iron, transferrin, and ferritin between test and control groups were analyzed by one-way analysis of variance (ANOVA). Also, a P value of <0.05 was considered as statistically significant. All the graphs were drawn using GraphPad Prism 6.0 (San Diego, Calif.)
Results
Changes of Ferritin and Transferrin Levels Following Growth of S. pneumoniae
In the present study, the BHI medium enriched with serum (SBHI) containing ferritin (1094.71 ng/mL), transferrin (135 µg/dl), and iron (88 µg/dl) was used to grow S. pneumoniae. Notably, the levels of ferritin (P=0.001) and transferrin (P=0.047) have significantly decreased after a 4-hour growth of S. pneumoniae. On the other hand, free iron concentration has significantly increased after a 4-hour incubation (P= 0.006).
Exploring FBPs in Bacterial Proteome
Western blotting revealed no FBPs in the proteome of S. pneumoniae. Also, serum and plasma samples were used as positive controls (Figure 1).
Evaluation of Ferritin-Degrading Proteases
Considering the lack of FBPs in pneumococcus proteome and significant reductions in ferritin and transferrin levels in the culture medium of S. pneumoniae, there was a possibility of the degradation of ferritin and transferrin by pneumococcal proteases. Therefore, to test this hypothesis, ferritin and transferrin degradation tests were performed using PMSF serine protease inhibitors. S. pneumoniae was grown in SBHI medium containing ferritin, transferrin, and iron with the concentrations of 172.18 ng/mL, 182 µg/dl, and 91 µg/dl, respectively. In this regard, the obtained results showed that there were significant reductions in the levels of ferritin compared to the control group1 (p: 0.000) and control group 2(p: 0.000), while there were no significant differences in the levels of ferritin between control groups1 and 2(p: 0.710). There were also significant reductions in the levels of transferrin compared to the control group1 (p: 0.000) and 2(p: 0.000). A significant difference was also observed in the levels of transferrin between the control group 1 and 2(p: 0.000), which show that transferrin has been degraded by proteases other than those inhibited by PMSF (Figures 2–4).
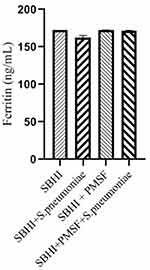 |
Figure 2 The levels of ferritin in the SBHI, SBHI + S. pneumoniae, SBHI + PMSF and SBHI + S. pneumoniae + PMSF groups. |
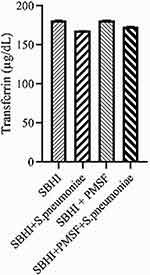 |
Figure 3 The levels of transferrin in the in the SBHI, SBHI + S. pneumoniae, SBHI + PMSF and SBHI + S. pneumoniae + PMSF groups. |
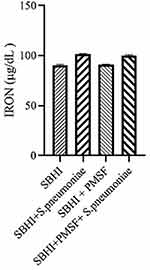 |
Figure 4 The levels of iron in the in the SBHI, SBHI + S. pneumoniae, SBHI + PMSF and SBHI + S. pneumoniae + PMSF groups. |
Identification of PMSF Sensitive Proteases
Since PMSF is effective only on serine proteases, molecular docking (Molegro software) was used to confirm and also to identify PMSF sensitive proteases in the S. pneumoniae proteome.
Docking of HtrA Serine Protease with PMSF
Docking of HtrA serine protease (PDB accession number: 2L97) with PMSF indicated that PMSF was likely to bind to the protease at 5 positions. Moreover, high-grade protease analysis predicted two potential stearic bonds between PMSF and HtrA serine protease at His 73 and Ile 109 positions (Figure 5).
 |
Figure 5 Interactions between PMSF and HtrA protease. Stearic bonds between HtrA protease and PMSF at histidine 73 and isoleucine 109 positions were predicted. |
Docking of ClpP Serine Protease with PMSF
Docking of ClpP hydrolase (PDB accession number: 1Y7O) with PMSF indicated some possible interactions at 5 positions. In addition, high-grade protease analysis showed the interactions between two hydrogen atoms of PMSF and Arg 14 and Met 1 of the hydrolase (Figure 6).
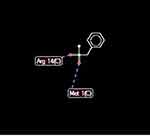 |
Figure 6 Docking between PMSF and ClpP protease indicated possible hydrogen bonds (length of 2.6 A) at arginine 14 and methionine 1 positions. |
Docking of RadA Protease with PMSF
Docking of the RadA (protease domain) hydrolase (PDB accession number: 5LKQ) with PMSF indicated some probable interactions at 5 positions. Moreover, high-grade protease analysis showed hydrogen bonds at the Gly 358 and Arg 330, as well as stearic interactions at the Gly 325, Lys 354, and Gly 357 positions (Figure 7).
Discussion
S. pneumoniae is known as an important cause of pneumonia, meningitis, and otitis media. Besides, it was shown that, the polysaccharide capsule and various virulence factors of S. pneumoniae play major roles in its pathogenesis and also facilitate its dissemination from the upper respiratory tract to the deeper regions of the lungs.
In the present study, we investigated the presence of FBPs in S. pneumoniae proteome. However, the results showed that S. pneumoniae had no FBPs. Moreover, it has been previously reported that S. pneumoniae lacks siderophores, and its iron uptake mechanisms are not well clear yet.13
Iron plays several important roles in the electron transport chain, energy metabolism, and many other biological functions.14 Accordingly, bacteria exploit different glycoproteins (eg, siderophores) to absorb iron from environmental sources and their hosts. Notably, many of these systems have been shown to play important roles in the pathogenesis of bacterial infections.15
Although hosts’ tissues are relatively iron-rich, this iron is not easily accessible to bacteria, due to the actions of various iron chelating glycoproteins. Therefore, bacteria need efficient iron absorption systems to compete with the host’s iron-chelating glycoproteins.16
The ability of pathogens in obtaining iron from transferrin, ferritin, hemoglobin, and other iron-containing proteins is considered as one of the most important factors ensuring their survival. Some pathogenic bacteria also acquire the host’s iron either through the production of siderophores or through a direct contact with iron-containing proteins. Correspondingly, bacteria that lack these abilities are slowly growing and effectively eliminated by host’s defense mechanisms.17
In human beings, iron is located within erythrocytes (ie, hemoglobin), ferritin, as well as in the serum and secretions (eg, glycoproteins such as transferrin and lactoferrin).2,18
Pneumococcal bacteria do not produce siderophore.19 In fact, the presence of a large capsule layer negates the presence of siderophore binding proteins on the bacteria surface. Therefore, siderophore binding proteins have no access to siderophore, which limits the applicability of these proteins for pneumococci. Lactoferrin is a transferrin-like protein present in secretions such as milk, tears, nasal secretions, saliva, bronchial mucosa, gastrointestinal fluid, and the bile. Also, iron-binding glycoproteins are associated with the pathogenesis of some bacterial species. In this regard, by producing lactoferrin-binding proteins; for example, Neisseria pathogenic strains use human lactoferrin as a source of iron.20 Nevertheless, most of the non-pathogenic strains are unable to exploit iron-binding proteins.21,22
In S. pneumoniae, PspA protein plays a key role in binding to lactoferrin at pneumococcal surface.23 Moreover, lactoferrin is the only source of iron in epithelial secretions. Accordingly, it is essential for S. pneumoniae to bind to lactoferrin and to absorb iron via PspA for ensuring its proliferation and colonization. The expression of PspA has been shown to be associated with the reduced concentration of free iron in secretions.24
Iron is also available in the structure of the “heme” within erythrocytes (ie, hemoglobin) and partially in the serum. In a study by Tai et al, it has been shown that pneumococcus produces a hemoglobin-binding protein (ie, PiaA) on the surface of its cytoplasmic membrane.19 Furthermore, Yang et al showed that the PiaA in a complex with ferrochrome siderophore is involved in iron transportation.25 Moreover, in another study, Brown et al reported that ABC transporter-like proteins, may be involved in iron absorption by S. pneumoniae.13 In this regard, other possible iron transporters of S. pneumoniae have been identified as Pit, Pia, and Piu.25
Ferritin is the main intracellular iron storage, which is also present in small amounts in the serum. Therefore, ferritin is considered as an important source of iron for microorganisms growing. In the present study, the results of Western blot analysis showed that S. pneumoniae had no FBPs. Actually, the thick polysaccharide capsule of S. pneumoniae can interfere with both binding of FBPs to ferritin and transporting of the complex into the bacterium. Despite this, ferritin concentration has significantly decreased in the culture medium of growing S. pneumoniae. Compared to the control group, ferritin level has significantly preserved after adding a protease inhibitor (ie, PMSF) to the culture medium of S. pneumoniae (p<0.05). Molecular docking results also showed that, pneumococcal proteases such as HtrA, ClpP, and RadA can be inhibited by the PMSF. Accordingly, these observations indicated that S. pneumoniae may recruit protease-dependent pathways to obtain iron. In fact, such pathways provide several effective strategies obviating the need for specific receptors and transporters. In addition, iron is a necessary element for the growth of bacteria; however, there are limited iron sources known for these microorganisms yet. Intracellular iron is stored as ferritin from, which releases iron in a gradual and controlled manner. The present study aimed to characterize ferritin-binding proteins (FBPs) of S. pneumoniae.
Conclusion
In the present study, we observed that S. pneumoniae had no FBPs, so it is unable to use ferritin as a direct source of iron. However, reduction of ferritin concentration in the culture medium of S. pneumoniae growing suggested FBPs-independent mechanisms for iron absorption. Moreover, preservation of ferritin level after using the PMSF protease inhibitor suggested a protease-dependent pathway for degradation of ferritin, probably as a source of iron.
Abbreviations
FBPs, ferritin-binding proteins; ELISA, enzyme-linked immunosorbent assay; PVDF, polyvinylidene difluoride; PDP, protein data bank; PMSF, phenylmethylsulfonyl fluoride.
Data Sharing Statement
The data and materials used during the current study are available from the corresponding and first authors on reasonable request.
Ethics Approval
The ethical code for the present study was IR.ZUMS.REC.1397.117.
Acknowledgments
We thank all the staff of the Microbiology department of ZUMS for their technical assistance.
Author Information
Dr. Davoud Afshar; Assistant professor in Medical Bacteriology, Department of Microbiology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran.
Dr. Behrooz Motlagh; Assistant professor in Clinical Biochemistry, Department of Clinical Biochemistry, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran.
Ms. Mozhgan Kheirandish; MSc student in Medical microbiology, Department of Microbiology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Pfaller MA, Sader HS, Shortridge D, Castanheira M, Flamm RK, Mendes RE. Activity of tedizolid against gram-positive clinical isolates causing infections in Europe and surrounding areas (2014–2015). J Chemother. 2019;31(4):188–194. doi:10.1080/1120009X.2019.1609740
2. Kawabata H, Sakamoto S, Masuda T, et al. Roles of transferrin receptors in erythropoiesis. [Rinsho Ketsueki] Jpn J Clin Hematol. 2016;57(7):951–958. doi:10.11406/rinketsu.57.951
3. Caza M, Kronstad JW. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol. 2013;3:80. doi:10.3389/fcimb.2013.00080
4. Barber MF, Elde NC. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346(6215):1362–1366. doi:10.1126/science.1259329
5. Carver PL. The battle for iron between humans and microbes. Curr Med Chem. 2018;25(1):85–96. doi:10.2174/0929867324666170720110049
6. Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6(8):e1000949. doi:10.1371/journal.ppat.1000949
7. Xia W. Competition for iron between host and pathogen: a structural case study on Helicobacter pylori. Methods Mol Biol. 2017;1535:65–75.
8. Ebrahimi F, Kheirandish M, Jafarisani M, Afshar D. Evaluation of siderophore production in Streptococcus pneumoniae by chrome azurol S method. Shahroud Univ Med Sci. 2019;13(4):27–31.
9. Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. Characterization of pit, a Streptococcus pneumoniae iron uptake ABC Transporter. Infect Immun. 2004;72(11):6754. doi:10.1128/IAI.72.11.6754.2004
10. Anderson GJ, Frazer DM. Current understanding of iron homeostasis. Am J Clin Nutr. 2017;106(Suppl 6):1559S–1566S. doi:10.3945/ajcn.117.155804
11. MacKenzie EL, Iwasaki K, Tsuji Y. Intracellular iron transport and storage: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10(6):997–1030. doi:10.1089/ars.2007.1893
12. Zhuo Y, Chen F, Kong L, et al. Magnetic resonance imaging of the human ferritin heavy chain reporter gene carried by dendrimer-entrapped gold nanoparticles. J Biomed Nanotechnol. 2019;15(3):518–530. doi:10.1166/jbn.2019.2697
13. Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. Characterization of pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect Immun. 2002;70(8):4389–4398. doi:10.1128/IAI.70.8.4389-4398.2002
14. Cherayil BJ. The role of iron in the immune response to bacterial infection. Immunol Res. 2011;50(1):1–9. doi:10.1007/s12026-010-8199-1
15. Yamamoto S, Shinoda S. Iron uptake mechanisms of pathogenic bacteria. Nihon Saikingaku Zasshi. 1996;51(2):523–547. doi:10.3412/jsb.51.523
16. Wooldridge KG, Williams PH. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12(4):325–348. doi:10.1111/j.1574-6976.1993.tb00026.x
17. Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54(1):881–941. doi:10.1146/annurev.micro.54.1.881
18. Brock JH. The physiology of lactoferrin. Biochem Cell Biol. 2002;80(1):1–6. doi:10.1139/o01-212
19. Tai SS, Yu C, Lee JK. A solute binding protein of Streptococcus pneumoniae iron transport. FEMS Microbiol Lett. 2003;220(2):303–308. doi:10.1016/S0378-1097(03)00135-6
20. Schryvers AB, Morris LJ. Identification and characterization of the human lactoferrin-binding protein from Neisseria meningitidis. Infect Immun. 1988;56(5):1144–1149. doi:10.1128/IAI.56.5.1144-1149.1988
21. Mickelsen PA, Sparling PF. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from transferrin and iron compounds. Infect Immun. 1981;33(2):555–564. doi:10.1128/IAI.33.2.555-564.1981
22. Simonson C, Brener D, DeVoe I. Expression of a high-affinity mechanism for acquisition of transferrin iron by Neisseria meningitidis. Infect Immun. 1982;36(1):107–113. doi:10.1128/IAI.36.1.107-113.1982
23. Hammerschmidt S, Bethe G, Remane PH, Chhatwal GS. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect Immun. 1999;67(4):1683–1687. doi:10.1128/IAI.67.4.1683-1687.1999
24. Cheng W, Li Q, Jiang Y-L, Zhou C-Z, Chen Y. Structures of Streptococcus pneumoniae PiaA and its complex with ferrichrome reveal insights into the substrate binding and release of high affinity iron transporters. PLoS One. 2013;8(8):e71451. doi:10.1371/journal.pone.0071451
25. Yang X-Y, He K, Du G, et al. Integrated translatomics with proteomics to identify novel iron–transporting proteins in Streptococcus pneumoniae. Front Microbiol. 2016;7:78. doi:10.3389/fmicb.2016.00078
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


