Back to Journals » International Journal of Nanomedicine » Volume 12
Extracellular biosynthesis of silver nanoparticles using the cell-free filtrate of nematophagous fungus Duddingtonia flagrans
Authors Costa Silva LP, Oliveira JP, Keijok WJ, da Silva AR, Aguiar AR, Guimarães MCC, Ferraz CM , Araújo JV, Tobias FL, Braga FR
Received 22 March 2017
Accepted for publication 9 May 2017
Published 31 August 2017 Volume 2017:12 Pages 6373—6381
DOI https://doi.org/10.2147/IJN.S137703
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Laryssa Pinheiro Costa Silva,1 Jairo Pinto Oliveira,2 Wanderson Juvencio Keijok,2 André Romero da Silva,3 Anderson Rocha Aguiar,1 Marco Cesar Cunegundes Guimarães,2 Carolina Magri Ferraz,1 Jackson Victor Araújo,4 Fernando Luiz Tobias,5 Fábio Ribeiro Braga1
1Department of Parasitology, University Vila Velha, Vila Velha, Espírito Santo, Brazil; 2Morphology Department, Federal University of Espírito Santo, Vitória, Espírito Santo, Brazil; 3Federal Institute of Education, Science and Technology of Espírito Santo, Aracruz, Espírito Santo, Brazil; 4Department of Veterinary Medicine, Federal University of Viçosa, Viçosa, Minas Gerais, Brazil; 5Department of Microbiology, University Vila Velha, Vila Velha, Espírito Santo, Brazil
Abstract: The biosynthesis of metallic nanoparticles (NPs) using biological systems such as fungi has evolved to become an important area of nanobiotechnology. Herein, we report for the first time the extracellular synthesis of highly stable silver NPs (AgNPs) using the nematophagous fungus Duddingtonia flagrans (AC001). The fungal cell-free filtrate was analyzed by the Bradford method and 3,5-dinitrosalicylic acid assay and used to synthesize the AgNPs in the presence of a 1 mM AgNO3 solution. They have been characterized by UV–Vis spectroscopy, X-ray diffraction, transmission electron microscopy, dynamic light scattering, Zeta potential measurements, Fourier-transform infrared, and Raman spectroscopes. UV–Vis spectroscopy confirmed bioreduction, while X-ray diffractometry established the crystalline nature of the AgNPs. Dynamic light scattering and transmission electron microscopy images showed approximately 11, 38 nm monodisperse and quasispherical AgNPs. Zeta potential analysis was able to show a considerable stability of AgNPs. The N–H stretches in Fourier-transform infrared spectroscopy indicate the presence of protein molecules. The Raman bands suggest that chitinase was involved in the growth and stabilization of AgNPs, through the coating of the particles. Our results show that the NPs we synthesized have good stability, high yield, and monodispersion.
Keywords: silver nanoparticles, green synthesis, nematophagous fungi, Duddingtonia flagrans
Introduction
Myconanotechnology has been touted as an emerging science. Fungi have a promising role in the fabrication of nanoparticles (NPs) due to their ability to grow at nutrient-poor substrates and, consequently, low cost.1 Moreover, fungi are remarkable organisms that produce a wide range of metabolites. It has long been suggested that these metabolites can induce metal ions reduction by two mechanisms: intra- and extracellular.2,3 Furthermore, fungi are good candidates for large-scale production owing their resistance to flow pressure, agitation, and other bioreactor cultivation conditions.4,5
More recently, many authors have used fungi in biosynthetic routes to produce metallic NPs,6,7 which generally involves reduction and functionalization of metallic NPs. However, major challenges still exist at the surface level that prevent their biological applications.7–9 These challenges include molecular vibrations analysis on the silver NPs (AgNPs) surface and the synthesis of monodispersed NPs, crucial parameters for success in their application as antimicrobial,10–12 anticancer,13,14 and drug delivery.15,16
The synthesis and characterization of AgNPs using Duddingtonia flagrans have been reported here for the first time. D. flagrans is considered to be a suitable species for the biological control of gastrointestinal parasites, because its predatory capacity and the production of fungal structures of resistance ensure the propagation and survival of these organisms in environmental conditions. In addition, it is harmless to animal and human health and to the environment.17,18 In the crude extract of D. flagrans, extracellular enzymes can be identified, such as proteases and chitinases, capable of hydrolyzing the nematode cuticle, which facilitate predation of the parasite. Therefore, its applicability has been studied extensively.19–22
This study evaluates a new approach to the application of these organisms, involving the biosynthesis of AgNPs, since to date, there are few reports on the use of nematophagous fungi in the synthesis of NPs.23 However, it is worth noting that there is no scientific report of the production of AgNPs using D. flagrans.
Methods
Obtaining the fungal filtrate
An isolate of the nematophagous fungus, D. flagrans (AC001), from the mycology repository of the Parasitology Laboratory of the Veterinary Department of the Federal University of Viçosa, Viçosa, Minas Gerais was used. The isolate was kept at 4°C, protected from light in test tubes containing 2% corn flour agar. To prepare the biomass for the biosynthesis studies, fungal mycelia was obtained by transferring culture disks (around 5 mm in diameter) of fungal isolates kept in 2% water agar (WA 2%) to Erlenmeyer flasks (250 mL) containing 200 mL of liquid nutrient-poor medium composed of bacteriological peptone, 2 g/L; yeast extract, 3 g/L; bibasic potassium phosphate, 0.1 g/L; magnesium sulfate hexahydrate, 0.05 g/L; lactic acid, 100 μL, pH =9. The flasks were incubated in an orbital shaker (120 rpm) at 25°C. The biomass formed was removed after 10 days, using a previously autoclaved stainless steel strainer, followed by extensive washing with ultrapure water to remove any component of the medium from the biomass. Two reactive conditions were tested: the first contained only 100 mL of ultrapure water and 10 g (wet weight) of the biomass (Filtrate 1), and the second was enriched with 0.1 g of tick shells, a natural source of chitin (Filtrate 2). The flasks were once again incubated in an orbital shaker under the same conditions, mentioned above, for 15 days. After incubation, the fungal filtration technique was carried out based on the techniques of Fatima et al24 and Kar et al25 with modification. Briefly, cellular filtrate was obtained using 0.22 nm membranes.
Analysis of the fungal filtrate – protein content and chitinase activity
The two fungal filtrates were analyzed for total protein content, using the Bradford method.26 Bovine serum albumin (98%, Sigma, Rio de Janeiro, Brazil) was used for the standard curve, and the readings were performed in a UV–Vis spectrophotometer (Spectrophotometer Spectramax 190, Molecular Devices, Sunnyvale, CA, USA) at 595 nm.
Chitinase activity was assessed through the measure of the reducing sugars using 3,5-dinitrosalicylic acid assay.27 One unit of chitinase was defined as the amount of enzyme required to liberate 1.0 μmol of N-acetylglucosamine per minute under the assay conditions.
Biosynthesis of the AgNPs from the fungal filtrate
To determine the necessary condition for the formation of the NPs, the fungal filtrates 1 and 2 were mixed with 100 mL of 1 mM AgNO3 solution at different temperatures (30°C and 60°C) and at different pH (5 and 10) and kept in a shaker at 120 rpm in the dark. The formation of silver NPs was first visualized by a change in the color of the solution and was then confirmed by optical absorption (UV–Vis spectroscopy).
Characterization of the AgNPs
Confirmation of AgNP formation after the color change of the solution and determination of AgNPs by localized surface plasmon resonance (LSPR) were performed on a UV–Vis spectrophotometer (Spectrophotometer Spectramax 190) at a resolution of 1 nm and scanning of 200–600 nm. The morphology of the NPs was confirmed by transmission electron microscopy (TEM). AgNPs were placed on a Formvar-coated copper grid (Ted Pella Inc., Redding, CA, USA) and examined by TEM using a JEOL microscope (JEOL, Inc., Peabody, MA, USA), model JEM1400, operating at 120 V with lathanum hexaboride filament (LAB6). The size distribution of the particles and the zeta potential were determined using dynamic light scattering (DLS) combined with the interaction of random Brownian motion and the electrical field movement of the particle suspensions (NPA152 Zetatrac, Microtrac Instruments, York, PA, USA). The spherical shape of AgNPs was determined based on the aspect ratio (greatest diameter/smallest diameter) using the software ImageJ 1.51i (n=500 NPs). For the analysis of the difference of sphericity of the AgNPs synthesized in different reaction conditions, analysis of variance (BioEstat 5.3) was used. After the synthesis of the AgNPs, the samples were centrifuged at 15,000 rpm for 30 minutes and the pellet formed was used for the Fourier-transform infrared analysis (FTIR). FTIR was performed in attenuated total reflection mode (FT-MIR FTLA 2000 Bomem, CEM Specialties Inc., London, ON, Canada) to investigate which organic compounds could be associated with AgNPs. AgNPs were characterized using Raman scattering (Raman Confocal Spectrometer ALPHA 300R, WITec, Ulm, Germany) in a region of 640–4,000 cm−1 at a laser strength of 532 nm.
Finally, the crystal structures of NPs were determined using X-ray diffraction (XRD). The AgNP samples were prepared by drop-coating the pelletized AgNPs on a glass slide and scanning in a 2θ region from 30° to 80° at 0.01° per minute with a time constant of 2 seconds using a D8 Advance (Bruker-axs, Billerica, MA, USA) X-ray diffractometer.
Results and discussion
Analysis of cell-free fungal filtered
Microorganisms have great potential for the production of AgNPs for several applications. This study showed that it is possible to biosynthesize AgNPs from compounds secreted by the nematophagous fungus D. flagrans. Wang et al23 described the possibility of the natural formation of NPs from the nematophagous fungus Arthrobotrys oligospora. However, the presence of possible reducing agents in the fungus or in its filtrate capable of reducing metals, and consequently forming metallic NPs, has not been described. In this study, we report a simple biological way to synthesize extracellular AgNPs using a cell-free filtrate of the fungus D. flagrans.
Based on the results obtained using the Bradford method, total proteins present in the fungal filtrate was influenced by the type of medium in which the biomass was held, given that the concentration of total proteins in Filtrate 2 (TP =1.16 mg/mL) was around triple that in Filtrate 1 (TP =0.41 mg/mL), as shown in Figure 1A. This occurs because the D. flagrans fungus can be stimulated to produce hydrolytic enzymes such as protease, pectinase, phospholipase, and chitinase according to the constituents of the medium.20,22,28 In this study, the medium was enriched with a natural source of chitin (tick shells) in order to increase the production of chitinase. Braga et al22 reported that the chitinase produced by the D. flagrans fungus has a weight of 34 kDa and maximum activity at pH 8–10 and 60°C. However, on that occasion, the authors already envisioned larger applicability studies around this chitinase. In the present work, total chitinase activity was measured in order to confirm the presence of the enzyme.29 Activity was measured (0.22 U/mL), which demonstrated chitinase presence in Filtrate 2.
Biosynthesis and characterization of as-prepared samples
UV–Vis spectroscopy analysis
AgNPs exhibit strong absorption of electromagnetic waves in the visible range due to LSPR. This phenomenon occurs due to collective oscillations of the NP conduction electrons by visible light irradiation.30 The color of the colloidal silver is attributed to the specific LSPR; therefore, the formation of NPs was established by UV–Vis spectroscopy. The results shown in Figure 1B and C indicate that the condition necessary for this synthesis of AgNPs is pH 10 at 60°C, since there was a peak between 380 and 450 nm in the UV, a wavelength range characteristic of the LSPR of AgNPs.31 Initially, the formation of NPs was observed due to the change in the color of the solution, which turned yellowish brown 24 hours after the addition of the AgNO3 solution to the fungal filtrate. Of the two filtrates used in the synthesis, the one that showed the best NP spectrum was the filtrate of the biomass kept in the chitin-enriched medium (Figure 1C).
To determine the best conditions for synthesis of the NPs, three reaction mixtures were studied: 1:100, 1:50, and 1:25 (fungal filtrate in 1 mM AgNO3 solution). These solutions were monitored at time intervals of 1, 2, 3, 4, 5, 10, 20, and 30 days, using a UV–Vis spectroscope. The synthesized AgNPs had a mean maximum peak of absorbency of 414, 406, and 413 nm for the 1:100, 1:50, and 1:25 solutions, respectively, and the yield was analyzed for 30 days (Figure 2A–C). The 1:50 sample showed the minor regularity of the peaks, suggesting a variation in the yield of the reaction (Figure 2B). Peak intensity in the UV–Vis is directly proportional to NP synthesis yield. Thus, in Figure 2D, it can be observed that the best yield of the 1:100 solutions is in 20 days, of the 1:50 solution in 1 day, and of the 1:25 solution in 30 days.
DLS and zeta potential
Meléndrez et al32 reported that increasing the quantity of NPs in suspension reduced the distance between the particles. This enables the attractive forces responsible for colloidal stability to prevail over other the repulsive forces. These attractive forces diminish zeta potential and electrophoretic mobility, allowing the interaction between the NPs and the consequent formation of aggregates. Thus, the colloidal stability of the AgNPs can be confirmed by measuring the zeta potential to estimate the load on the surface of the NPs.14 In general, NPs with zeta potential greater than +25 mV or less than −25 mV have sufficient electrostatic repulsion to remain stable in solution.33 The zeta potential obtained for 1:100, 1:50, and 1:25 samples was −49,205 mV, −28,655 mV, and −61,935 mV, respectively (Figure 2E). These values indicate good stability of the colloids. The zeta potential values for the AgNPs produced in this study indicate good stability of the colloids, which can be attributed to the presence of fungal proteins that functionalize the NPs. The 1:25 solution had the highest negative value of the zeta potential, and so was the most stable because the repulsive forces between negatively charged particles prevent agglomeration.32–34
DLS is used to determine the size of the nanoparticle considering the metallic nucleus of the NP and the cloud of electrons that surrounds it as well as defining the zeta potential.35,36 The results of the DLS analysis demonstrated that the size of the biosynthesized 1:100 AgNPs was between 36 and 486 nm, of the 1:50 AgNPs between 30 and 409 nm, and of the 1:25 AgNPs between 12 and 409 nm (Figure 3). The 1:50 solution was the one that presented the highest monodispersion.
  | Figure 3 Distribution of particle size of AgNPs 1:100 (A), 1:50 (B), and 1:25 (C). |
XRD analysis
The crystalline nature of the biosynthesized silver NPs was further confirmed by intense XRD analysis. The XRD patterns of the synthesized silver NPs are shown in Figure 4. Three diffraction peaks were observed at 2θ values of 39°, 43.45°, and 64.5°, corresponding to the (111), (200), and (220) reflections of metallic crystalline silver, respectively.24,37,38 The data obtained were matched with the database of Joint Committee on Powder Diffraction Standards (JCPDS file No 04-0783).
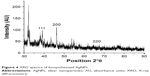  | Figure 4 XRD spectra of biosynthesized AgNPs. |
Structural characterization with TEM
Using the images obtained by the TEM (Figure 5) in the ImageJ 1.51i program, it was possible to calculate the diameter of the NPs (n=500) and thus assess the size, monodispersion, and spherical shape of the AgNPs. The synthesized AgNPs in the 1:100 condition revealed smaller size and dispersed particles (mean diameter =10.29±7.18) (Figure 5B–D), while the NPs of the 1:50 sample were larger and more agglomerated (mean diameter =12.17±5.94) (Figure 5E–G). The most dispersed AgNPs were those synthesized in the 1:25 condition (mean diameter =11.69±8.49) (Figure 5H–J). The NPs of the 1:50 solution, because they are more agglomerated, indicated lower stability, which lead to the obtained zeta potential results (Figure 5E).
In the histogram elaborated from the NP values, it was possible to conclude that the NPs of the 1:25 solution have a larger size variation and those of the 1:50 solution a smaller size variation, and thus higher monodispersity (Figure 6A), which confirms the results obtained by DLS (Figure 3B). The AgNPs were not completely spherical; therefore, it was necessary to calculate the aspect ratio, the ratio between the greatest diameter and the smallest diameter of each metallic NP. From the aspect ratio of the AgNPs, it was possible to determine that the particles produced were spherical particles and that there was no statistical difference regarding the sphericity of the AgNPs produced by the solutions 1:100, 1:50, and 1:25 (analysis of variance, P>0.005) (Figure 6B).
  | Figure 6 Histogram of the diameter (A) and AR (B) of the AgNPs. |
FTIR and Raman signature
For a better understanding of the biosynthesized NPs, techniques like FTIR and Raman scattering were used to characterize the molecules that can be absorbed on the surface of these AgNPs.
According to Stuart, the FTIR protein spectra exhibit absorption bands with their characteristic amide group. The bands of the protein amide group are similar to the absorption bands exhibited by the secondary amides (amide A, amide B, and amide I–VII). All the amides show a carbonyl absorption band, better known as an amide I band, which occurs between 1,680 and 1,640 cm−1. Their position depends on the degree of hydrogen bonding and the physical state of the compound. The number of N–H bonds in an amide is equal to the number of N–H deformation bands observed in the spectrum; thus, secondary amides have only one axial deformation band between 1,560 and 1,530 cm−1, called amide band II.39,40 FTIR spectrum of the AgNPs (Figure 7) showed the presence of two bands: 1,640 and 1,540 cm−1, identified as amide I and amide II, respectively, which emerged due to the C=O axial deformation vibrations and the vibrations of the angular deformation of N–H in the amide bonds of the proteins. It was also possible to observe the presence of carboxylic acid OH deformation band at 2,665 cm−1 39 and stretch bands at 730 cm−1 (CH2-S-S, possibly referring to disulfide cross-links between the amino acid cysteine),41 1,000 cm−1 (C-O),29 1,811 cm−1 (C=O anhydride),26 2,100 cm−1 (C =O),42 2,326 cm−1 (NH2+),41 and 2,665, 3,600, and 3,751 cm−1 (O-H).35,43
  | Figure 7 FTIR spectrum of the biosynthesized AgNPs. |
Basavaraja et al30 confirmed by IR spectroscopy that the carbonyl group present in the amino acid and peptide protein residues has a greater capacity to bond metal such that the proteins can form a coating that covers the metal NPs to prevent agglomeration of the particles and stabilize them in the medium. This evidence suggests that the biological molecules may play a role in the formation and stabilization of silver NPs in aqueous medium.
Generally in the FTIR, the NH2 and N–H vibration bands are fundamental in clarifying the type of amide present, whereas in the Raman spectrum, the vibratory bands are very useful in conformational studies to determine the secondary structure of the biological molecules.41 Therefore, it was possible to confirm, from the results obtained by Raman scattering (Figure 8), that the molecule –C(=O)-NHR, which is adsorbed on the metallic surface of the NP, has a cis confirmation due to the presence of the peak at 1,440 cm−1. In both the AgNP samples and the filtrate, the presence of the 1,600 cm−1 band was observed. Stuart39 reports that vibrations of the benzene ring in 1,600 and 1,602 cm−1 may indicate the presence of essential amino acids with aromatic side chains, such as phenylalanine and tyrosine, respectively. Yamagami et al44 reported the presence of amino acids such as aspartate, alanine, cysteine, phenylalanine, tyrosine, and arginine, among others, in the composition of chitinase.
  | Figure 8 Raman spectrum of the biosynthesized AgNPs. |
The results obtained by FTIR and Raman scattering corroborate the hypothesis that the molecule responsible for the reduction and stability of the silver nanoparticle is chitinase, given the presence of amino acids that belong to the amino acid of this protein on the surface of the AgNPs. Finally, the reduction of the silver was only possible under ideal conditions for activity of the enzyme, having the best yield in the synthesis that used the filtrate of the chitin-enriched biomass. Tests with different concentrations of the filtrate were conducted to establish the best conditions for the formation of NPs. Based on the data obtained, it was possible to determine that the 1:50 solution was the condition that provided AgNPs that were more stable and more monodisperse.
Thus, in this study, we report a simple biological process for the synthesis of AgNPs using the nematophagous fungus D. flagrans. In addition to biological synthesis being an inexpensive, ecologically correct, and high-yield process, it was possible to perform extracellular synthesis, which makes the process easier since no extra processing is required to separate the particles from the live cells. Biosynthesized and functionalized AgNPs have good stability and high yield, in addition to antibacterial, antifungal, antiviral, and anticancer properties, making them a promising strategy for therapeutic applications, which potentiates new experimental designs using the fungus D. flagrans.
Acknowledgments
We thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for providing the author Fabio Ribeiro Braga’s Productivity Grant; the FAPES (Fundação de Amparo a Pesquisa e Inovação do Espírito Santo) for the scholarship grant; LabPetro and Luccar Ufes (Laboratório de Ultraestrutura Celular Carlos Alberto Redins); and Fapemig (Fundação de Amparo a Pesquisa de Minas Gerais). The authors also thank Professors Emy Hiura and Filippe Elias de Freitas Soares for their contribution.
Disclosure
The authors report no conflicts of interest in this work.
References
Alghuthaymi MA, Almoammar H, Rai M, Said-Galiev E, Abd-Elsalam KA. Myconanoparticles: synthesis and their role in phytopathogens management. Biotechnol Biotechnol Equip. 2015;29(2):221–236. | ||
Kulkarni N, Muddapur U. Biosynthesis of metal nanoparticles: a review. J Nanotechnol. 2014;2014:1–8. | ||
Zhang XF, Liu ZG, Shen W, Gurunathan S. Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci. 2016;17:1534–1568. | ||
Chowdhury S, Basu A, Kundu S. Green synthesis of protein capped silver nanoparticles from phytopathogenic fungus Macrophomina phaseolina (Tassi) Goid with antimicrobial properties against multidrug-resistant bacteria. Nanoscale Res Lett. 2014;9(1):365–376. | ||
Balakumaran MD, Ramachandran R, Balashanmugam P, Mukeshkumar DJ, Kalaichelvan PT. Mycosynthesis of silver and gold nanoparticles: optimization, characterization and antimicrobial activity against human pathogens. Microbiol Res. 2016;182:8–20. | ||
Moazeni M, Rashidi N, Shahverdi AR, Noorbakhsh F, Rezaie S. Extracellular production of silver nanoparticles by using three common species of dermatophytes: Trichophyton rubrum, Trichophyton mentagrophytes and Microsporum canis. Iran Biomed J. 2012;16(1):52–58. | ||
Ottoni CA, Simões MF, Fernandes S, et al. Screening of filamentous fungi for antimicrobial silver nanoparticles synthesis. AMB Express. 2017;7:31–41. | ||
Xue B, He D, Gao S, Wang D, Yokoyama K, Wang L. Biosynthesis of silver nanoparticles by the fungus Arthroderma fulvum and its antifungal activity against genera of Candida, Aspergillus and Fusarium. Int J Nanomedicine. 2016;11:1899–1906. | ||
Rathod V, Banu A, Ranganath E. Biosynthesis of highly stabilized silver nanoparticles by Rhizopus stolonifer and their Anti-fungal efficacy. Int J Cur Biomed Phar Res. 2012;2(1):241–245. | ||
Franci G, Falanga A, Galdiero S, et al. Silver nanoparticles as potential antibacterial agents. Molecules. 2015;20:8856–8874. | ||
Singh P, Kim YJ, Singh H, et al. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int J Nanomedicine. 2015;10:2567–2577. | ||
Mallmann EJ, Cunha FA, Castro BN, Maciel AM, Menezes EA, Fechine PB. Antifungal activity of silver nanoparticles obtained by green synthesis. Rev I Med Trop. 2015;57(2):165–167. | ||
Alsalhi MS, Devanesan S, Alfuraydi AA, et al. Green synthesis of silver nanoparticles using Pimpinella anisum seeds: antimicrobial activity and cytotoxicity on human neonatal skin stromal cells and colon cancer cells. Int J Nanomedicine. 2016;11:4439–4449. | ||
Kummara S, Patil MB, Uriah T. Synthesis, characterization, biocompatible and anticancer activity of green and chemically synthesized silver nanoparticles – a comparative study. Biomed Pharmacother. 2016;84:10–21. | ||
Abamor ES, Allahverdiyev AM. A nanotechnology based new approach for chemotherapy of cutaneous leishmaniasis: TIO2@AG nanoparticles – Nigella sativa oil combinations. Exp Parasitol. 2016; 166:150–163. | ||
Jahangirian H, Lemraski EG, Webster TJ, Rafiee-Moghaddam R, Abdollahi Y. A review of drug delivery systems based on nanotechnology and green chemistry: green nanomedicine. Int J Nanomedicine. 2017;12:2957–2978. | ||
Braga FR, Araújo JV. Nematophagous fungi for biological control of gastrointestinal nematodes in domestic animals. Appl Microbiol Biot. 2014;98:71–82. | ||
Hiura E, Lopes AD, Paz JS, et al. Fungi predatory activity on embryonated Toxocara canis eggs inoculated in domestic chickens (Gallus gallus domesticus) and destruction of second stage larvae. Paratisol Res. 2015;114:3301–3308. | ||
Braga FR, Araújo JV, Soares FE, et al. Proteolytic action of the crude extract of Duddingtonia flagrans on cyathostomins (Nematoda: Cyathostominae) in coprocultures. Rev Bras Parasitol Vet. 2013;22(1):143–146. | ||
Braga FR, Araújo JV, Soares FEF, et al. Interaction of the nematophagous fungus Duddingtonia flagrans on Amblyomma cajennense engorged females and enzymatic characterisation of its chitinase. Biocontrol Sci Technol. 2013;23:584–594. | ||
Braga FR, De Freitas Soares FE, Araujo JM, et al. Statistical experimental design to assess the influence of enzymes of nematophagous fungi versus helminths. Res Vet Sci. 2014;97:527–532. | ||
Braga FR, Soares FE, Giuberti TZ, et al. Nematocidal activity of extracellular enzymes produced by the nematophagous fungus Duddingtonia flagrans on cyathostomin infective larvae. Vet Parasitol. 2015;212:214–218. | ||
Wang Y, Sun L, Yi S, Huang Y, Lenaghan SC, Zhang M. Naturally occurring nanoparticles from arthrobotrys oligospora as a potential immunostimulatory and antitumor agent. Adv Funct Mater. 2013;23:2175–2184. | ||
Fatima F, Bajpai P, Pathak N, Singh S, Priya S, Verma SR. Antimicrobial and immunomodulatory efficacy of extracellularly synthesized silver and gold nanoparticles by a novel phosphate solubilizing fungus Bipolaris tetramera. BMC Microbiol. 2015;15:52–62. | ||
Kar PK, Murmu S, Saha S, Tandon V, Acharya K. Anthelmintic efficacy of gold nanoparticles derived from a phytopathogenic fungus, Nigrospora oryzae. PLoS One. 2014;9(1):1–9. | ||
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. | ||
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chem. 1959;31(3):426–428. | ||
Meyer WJ, Wiebe MG. Enzyme production by the nematode-trapping fungus, Duddingtonia flagrans. Biotechnol Lett. 2003;25:791–795. | ||
Gusakov AV, Kondratyeva EG, Sinitsyn AP. Comparison of two methods for assaying reducing sugars in the determination of carbohydrase activities. Int J Anal Chem. 2011;2011:1–4. | ||
Basavaraja S, Balajia SD, Arunkumar Lagashettyc, Rajasab AH, Venkataramana A. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium semitectum. Mater Res Bull. 2008;43:1164–1170. | ||
Ninganagouda S, Rathod V, Jyoti H, Singh D, Prema K, Haq MU. Extracellular biosynthesis of silver nanoparticles using Aspergillus flavus and their antimicrobial activity against gram negative MDR strains. Int J Pharma Bio Sci. 2013;4(2):222–229. | ||
Meléndrez MF, Cárdenasb G, Arbiol J. Synthesis and characterization of gallium colloidal nanoparticles. J Colloid Interface Sci. 2010;346:279–287. | ||
Milaneze BA, Oliveira JP, Augusto I, et al. Facile synthesis of monodisperse gold nanocrystals using Virola oleifera. Nanoscale Res Lett. 2016;11:465–474. | ||
Netala VR, Bethu MS, Pushpalatha B, et al. Biogenesis of silver nanoparticles using endophytic fungus Pestalotiopsis microspora and evaluation of their antioxidant and anticancer activities. Int J Nanomedicine. 2016;11:5683–5696. | ||
Zheng T, Bott S, Huo Q. Techniques for accurate sizing of gold nanoparticles using dynamic light scattering with particular application to chemical and biological sensing based on aggregate formation. ACS Appl Mater Interfaces. 2016;8(33):21585–21594. | ||
Elbeshehy EKF, Elazzazy AM, Aggelis G. Silver nanoparticles synthesis mediated by new isolates of Bacillus spp. nanoparticle characterization and their activity against bean yellow mosaic virus and human pathogens. Front Microbiol. 2015;6:453–466. | ||
Vigneshwaran N, Ashtaputre NM, Varadarajan PV, Nachane RP, Paralikar KM, Balasubramanya RH. Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus. Mater Lett. 2007;61:1413–1418. | ||
Chitra K, Annadurai G. Antibacterial activity of pH-dependent biosynthesized silver nanoparticles against clinical pathogen. Biomed Res Int. 2014;2014:725165. | ||
Stuart BH. Infrared Spectroscopy: Fundamentals and Applications. Chichester, UK: John Wiley & Sons; 2004. | ||
Silverstein RM, Webster FX, Kiemle DJ. Identificação Espectrométrica de Compostos Orgânicos. [Spectrometric identification of organic compounds]. 7th ed. Rio de Janeiro; LTC; 2007. Portuguese. | ||
Lin Vien D, Colthup NB, Fateley WG, Graselli JG, editor. The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules. San Diego, CA: San Diego Academic Press; 1991. | ||
Lopes WA, Fascio M. Esquema para interpretação de espectros de substâncias orgânicas na região do infravermelho. [Scheme for Interpreting Spectra of Organic Substances in the Infrared Region]. Quím Nova. 2004;27(4):670–673. Portuguese. | ||
Pal R, Panigrahi S, Bhattacharyya D, Chakraborti AS. Characterization of citrate capped gold nanoparticle-quercetin complex: experimental and quantum chemical approach. J Mol Structure. 2013;1046:153–163. | ||
Yamagami T, Tanigawa M, Masatsune I, Funatsu G. Complete amino acid sequence of chitinase-A from leaves of pokeweed. Biosci Biotechnol Biochem. 1998;62(4):825–828. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



