Back to Journals » Infection and Drug Resistance » Volume 10
Exploring bacterial outer membrane barrier to combat bad bugs
Received 19 June 2017
Accepted for publication 18 July 2017
Published 30 August 2017 Volume 2017:10 Pages 261—273
DOI https://doi.org/10.2147/IDR.S144299
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Ishan Ghai,1 Shashank Ghai2
1School of Engineering and Life Sciences, Jacobs University, Bremen, 2Leibniz University, Hannover, Germany
Abstract: One of the main fundamental mechanisms of antibiotic resistance in Gram-negative bacteria comprises an effective change in the membrane permeability to antibiotics. The Gram-negative bacterial complex cell envelope comprises an outer membrane that delimits the periplasm from the exterior environment. The outer membrane contains numerous protein channels, termed as porins or nanopores, which are mainly involved in the influx of hydrophilic compounds, including antibiotics. Bacterial adaptation to reduce influx through these outer membrane proteins (Omps) is one of the crucial mechanisms behind antibiotic resistance. Thus to interpret the molecular basis of the outer membrane permeability is the current challenge. This review attempts to develop a state of knowledge pertinent to Omps and their effective role in antibiotic influx. Further, it aims to study the bacterial response to antibiotic membrane permeability and hopefully provoke a discussion toward understanding and further exploration of prospects to improve our knowledge on physicochemical parameters that direct the translocation of antibiotics through the bacterial membrane protein channels.
Keywords: antibiotics, Gram-negative bacteria, cell envelope, protein channels, nanopores, influx, antibiotic resistance
Introduction
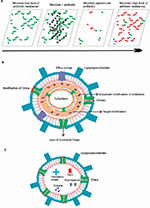
Antibiotic resistance can be defined as the capability of any microbial organism to counterattack effects of antimicrobial drugs (antibiotics) (Figure 1A) used against them.1,2 This phenomenon has become a global communal health threat due to an enormous increase in annual death rate.2 The emergence of highly resistant organisms has led to the requirement of new antibacterial drugs.1 Due to the slow progress of the current antibiotic research, there exists an enormous gap between bacterial evolution and the rate of development of novel antibiotic drugs.1,3,4 Only about two new classes of antibiotics have been brought to the market in the last three decades. On the technical front, there is an urgent need for a greater understanding of how antibiotics work, how bacteria progress with resistance against these antibiotics, and what molecular machinery could be exploited to get around bacterial defense mechanisms.1–4 The current innovative way of improving the potential of antibiotics is to effectively introduce them into bacteria and further prevent them from degradation by bacterial enzymes before they reach their targets. There is an extreme necessity for counteracting the problem of multi-antibiotic resistance.1,4 The important mechanism (Figure 1B) of resistance toward antibiotics known till date includes the enzymes-mediated deactivation of antibiotics for example, β-lactamase enzymes which hydrolyze and confer resistance against a diverse variety of antibiotics including penicillins, cephalosporins, carbapenems, and many more.4–7 The outer membrane vesicles (Figure 1C), these native vesicles released by Gram-negative bacteria, are mainly composed of periplasmic and outer membrane components including lipopolysaccharides, proteins, lipids, and other molecules.8–11 They help the producer cells while communicating with other cells concerning pathogenesis, secretion, nutrients acquisition, and self-defense.5,8–10 These moieties protect bacteria from various environmental stress factors including antibiotics, for example, gentamicin, imipenem, ampicillin, melittin, colistin, and many more.8–14 Further, resistance mechanism is also mediated by reducing the entry of antibiotics into the target site of bacteria which is mainly effected by specific alteration of outer membrane permeability (Figure 2). Efflux pumps effectively contribute towards resistance mechanism by antibiotic expulsion. In addition, antibiotic target proteins, for example, penicillin-binding proteins, are altered inside the bacterial cells, leading to antibiotic resistance.2,3,5,6,15–21
In this review, we present a systemic overview of the role of different membrane protein transporters responsible for antibiotic transport, present in the outer membrane of Gram-negative bacteria.4–6,22 We highlight the different achievements of the scientific community in understanding the uptake of different solutes including antibiotics.7,17,22 This active knowledge of the role of outer membrane influx in antibiotic transport in Gram-negative bacteria can be useful for antibiotic drug development in the future, where the computed data can be employed toward understanding the detailed mechanism of bacterial membrane transport, and to further design novel antibiotics with an effective permeability profile.
Gram-negative bacteria
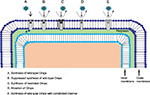
Gram-negative bacteria have a multifaceted cell envelope comprising an outer membrane that restricts the access to the periplasm by acting as a molecular filter, thus forming an efficient selective permeation barrier.4–6,23,24 This outer membrane, like other biological membranes, is fundamentally built up of a bilayer of lipids.6,18,25,26 As such, this lipid bilayer membrane is mostly impermeable to hydrophilic molecules including nutrients.22,25,27 The effective intake of hydrophilic molecules is mainly controlled by specific water-filled open channels termed as outer membrane proteins (Omps) or porins.22,27–29 These Omps are intensively characterized in Gram-negative bacteria and are further distinguished as nonspecific and specific Omps in accordance with their functional structure (monomeric or trimeric),6,7,22,24–26,28 substrate specificity, regulation, and expression.15,18,29,30 These membrane proteins do not show any hydrophobic stretches in their amino acid sequences and majorly form hollow β-barrel structures with a hydrophobic outer surface.28,31 The barrel structure encompasses the transmembranous pore-type structure with a crucial function of facilitating the passive flux of hydrophilic substances22,28 and further acting as a functional diffusional barrier for nonpolar solutes.6,28 These proteins might show specific selectivity in general for either cations or anions.5,22,28
Bacterial adaptation to reduce influx through these Omps is an increasing problem that contributes, together with efflux systems, to antibiotic resistance.3–5,20,23,32–34 An existing challenge for drug design is to interpret membrane permeability at molecular level to get a better insight into the role of membrane transport (Figure 2) in bacterial resistance mechanism.4–7,20,35 Like other hydrophilic molecules, polar antibiotics including β-lactam antibiotics and fluoroquinolones majorly sneak into Gram-negative bacteria using these Omps.5,31,33 Any slight modification by the bacteria in the responsible Omps can significantly affect the antibiotic drug therapy.33 Many clinically pertinent bacterial species including Enterobacter aerogenes, Escherichia coli, Enterobacter cloacae, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii have been sequenced for determining the effective key Omps (Table 1) present in the outer membrane.3–6,23,28,31–33,36,37 Further, bacterial bugs including Pseudomonas aeruginosa, and Acinetobacter baumannii possess an innate low vulnerability toward β-lactams, through reduced outer membrane permeability.5,6,20,22,38 For instance, reduced membrane permeability in Pseudomonas aeruginosa as compared to Enterobacteriaceae mainly occurs due to less number of Omps present in the outer membrane and their distinct physicochemical properties.22,38–41 In other Gram-negative bugs, for example, Escherichia coli, Enterobacter and Klebsiella pneumoniae, susceptibility toward β-lactam molecules is closely related to the presence of nonspecific diffusion Omps, for example, OmpF and OmpC.5,6,22
  | Table 1 Crucial Omps studied in different Gram-negative bacterial species Notes: Studies by Nikaido,5,6 Pages et al,22,42 and Schulz28 provide further insight. Abbreviation: Omps, outer membrane proteins. |
Previous works showing the effective role of different Omps (Table 1) in molecular influx of different antibiotics are shown in Table 2. We discuss the achievements of the scientific community in this area by studying the role of different Omps in outer membrane permeability, using separate set of theoretical and experimental techniques including molecular simulation (MS), electrophysiology, minimum inhibitory concentration assay, liposome swelling assay, X-ray crystallography, and fluorescence resonance energy transfer.
Discussion
Computing influx
Typical antibiotic activity toward bacterial cell occurs in micromolar concentration range, thereby representing values that are approximately limited to a thousand molecules inflowing the cell in few minutes to hours.7,22 Such numbers are considerably beneath the detection limit of most of the techniques and thus require significant amplification of the signal.4,7,22,110 Measuring the flux of small molecules across the outer cell membrane can be possibly achieved by different approaches including whole-cell assays, which require computation of flux using genetically engineered bacterial cell.7,111,112 These methods involve soaking bacteria in antibiotics for a fixed time followed by a separation process to remove the external media from the internalized antibiotics.7 However, the quality of the separation method is crucial for improving permeability.7,111,112 There are several published studies employing whole-cell assays to quantify the uptake, and their quality has been intensively compared.7,110–116 Once the separation technique allows collecting sufficient amounts of internalized antibiotics, several biophysical methods can be used to quantify the intracellular antibiotics.7,113–118 One of the promising tools for studying intracellular accumulation is mass spectrometry. The technique was successfully applied in measuring the uptake of antibiotics;117,118 for example, a work demonstrated cellular uptake of linezolid by E. coli using liquid chromatography–mass spectrometry.118
The discussed methods allow quantifying the total turnover of a cell uptake which represents the relevant actual effective concentration seen by the bacteria. On the contrary, the comprehensive flux depends on a multitude of parameters and renders the molecular understanding difficult.7,22 To understand the molecular origin of the antibiotic uptake, we need information on the role of each individual involved component. For example, the so-called liposome swelling assay provides information on a model system.35,52,55,60,80,97,105 The method involves reconstitution of batches of purified Omps into (multilamellar) liposomes.7,22 Under isosmotic addition, the diffusion of substrate inside the liposome results in alteration of the light-scattering pattern. The effective change in light-scattering signal is then correlated with the relative permeability of the molecules. The main disadvantage of this method is that it requires a large quantity of material and is only effective for uncharged molecules, whereas for charged molecules, the effect of counterion flow affects the quality of the measurement. Moreover, the assay can only determine average turnover numbers and often does not provide conclusive values.7
Moreover, using conventional electrophysiology, computation of rate of flux of discrete small molecules across Omps present in bacterial outer cell membrane involves measurement of flux values at single molecular level.7,36,45,52,56,66,67 Here, electrophysiological measurement using single Omps provides the best high-resolution (Figure 3) signal-to-noise ratio,7,18,40,73,74,83 thereby suggesting the higher efficacy of this method in sensing and understanding uptake at molecular level.7,15,22 The method includes reconstitution of a single or multiple Omps into an artificial planar lipid bilayer and further uses transmembrane potential-driven ion current across the channel as a detection probe.7,67 Using ion current as a probe specifically demonstrates very well-characterized electrophysiological properties of the Omps,15,34,45,65,66,84,106,119–121 including size,122,123 single-channel conductance, channel ion selectivity,58,75,76,90,99–101 channel gating dynamics, and more.47,95,109 Likewise, the size of Omps is a key factor defining transport through the channel.107,108 This factor plays a key role in antibiotic susceptibility.72–74 Determination of the size of Omps using electrophysiology provides a crucial insight into the maximum size of molecule they can transport.122,123 This, further, helps in evaluating the inner structure including constriction site.122–125 Further, single-channel conductance of Omps, ion selectivity,58,75,76,84,89 and gating dynamics35,47,94,95,109 give an insight into the channel–substrate binding and channel–substrate interactions.35,71,83,85,97,99,101 An insight into the channel conductance can be obtained, specifically using staircase electrophysiology (Figure 3A and B), where real-time insertions of single channels at constant voltage can be attained.59,123 The conductance of any channel can be termed as its unique characteristic. This allows a better understanding of the open/close states of the channel and its gating dynamics which can then be employed in studying channel structure–activity relationship.35,71,107,108
Using these functions, a proper insight into the channel interaction with different substrates can be obtained including substrate-induced partial or full blockage (Figure 3C) of channel52,53,67 and substrate-induced gating.67 The function of these pores has been well documented on the basis of pore characteristics, chemical modification, and genetic mutations.15 These parameters were further used to elaborate transport of the following antibiotics: meropenem,52 imipenem,52 cefotaxime,48 cefpirome,44 ceftriaxone,44,45 cefepime,45 ceftazidime,44,45 ciprofloxacin,45 norfloxacin,45 and enrofloxacin45 through OmpC; imipenem,74 meropenem,53 ceftazidime,44,45,74 cefepime,45,55,74 ceftriaxone,44,45 cefpirome,44 ampicillin,57,60,61,67 benzylpenicillin,60 amoxicillin,57 carbenicillin,57 azlocillin,57 piperacillin,57 ciprofloxacin,45 norfloxacin,45,56,126 enrofloxacin,7,45,54,66,72 moxifloxacin,65 different poly arginines,64 polyamines,106 and antimicrobial peptides51 through OmpF; ceftriaxone80 through Omp35; cefepime16 through Omp36; imipenem91 and meropenem91 through OccD3; imipenem86 through OccD1; and meropenem,104 glutamic acid,104 arginine,104 and imipenem104 through CarO Omp (Table 2).
In contrast, single-channel recording provides the best signal-to-noise ratio and intrinsic data on Omp–substrate interaction.40,45,65–67 But the interpretation of molecule translocation cannot be made directly as the chances of molecule exit on the entry side are almost identical when compared to the transport of the molecule across the pore.7 Whereas in the case of charged molecules, direct conclusion of translocation can be made as the increasing voltage will reduce the residence time of the molecules inside the Omp, which might provide some evidence of transport across the Omp. In addition, using channel selectivity, that is, channel inherent selection of either anion or cation, a quantitative flux assessment of the charged molecules can be made using electrophysiological reversal potential measurements.36,59 Using this approach, flux of β-lactamase inhibitors across OmpF and OmpC was estimated, showing the role of Omps in their transport across bacterial biobarrier.36,59 However, most of the molecules did not carry a net charge or show low intrinsic solubility which makes them trivial to measure and thus excludes them from screening via this method. Furthermore, the finite time resolution of electrophysiology also makes the method limited in screening of antibiotics uptake.7,45,66,67
Molecular simulation
In the current scenario, MS is well suited to obtain a particular information at an atomic scale.121 Thus far, knowledge of the antibiotic translocation problem has pointed essentially toward three mechanisms including diffusion with molecule binding, a mechanism based on pore dehydration induced by the permeating molecule, and slow diffusion with molecule binding.50,61,62,70,71,97,99,121 Further, to discriminate among these mechanisms, and to attain a better description of the Omps behavior and their role in substrate transport, understanding the communication between pore and substrate is essential.119–121,127,128 Thanks to the high-resolution, molecular modeling simulations, detailed characterization is possible in terms of energetics (Figure 4 from Ghai et al)36 and bond formation including hydrogen bonds, hydrophobic contacts, and more.50,62,71,121
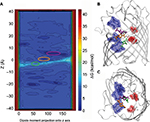  | Figure 4 (A) Intrinsic depiction of the two-dimensional free energy of translocation of β-lactamase inhibitor (avibactam), reassembled from metadynamic simulations. (B) Lateral view and (C) topmost view of the avibactam inside OmpF pore in the two lowest minima near the constriction region and at the subsequent transition state. Reprinted with permission from Ghai I, Pira A, Scorciapino MA, et al. General method to determine the flux of charged molecules through nanopores applied to beta-lactamase inhibitors and OmpF. J Phys Chem Lett. 2017;8(6):1295–1301.36 Copyright (2017) American Chemical Society. |
The complete control over the characteristics of the system allows MS to explain the impact of pinpoint mutations and the effects that arise due to different domains of the same proteins.95,100,101 Further, MS significantly allows understanding and interpreting available experimental data.50,61,62,70,121 When combined with experimental approach, MS proves to be a complementary method. For instance, together with electrophysiology,36,48,54,55,57,58,60,61,65–67,71–73,95,97,99–101 MS was used for understanding the transport of β-lactamase inhibitors (Figure 4), interaction of substrates with Omps (enrofloxacin, moxifloxacin, ampicillin, benzylpenicillin, carbenicillin, amoxicillin, azlocillin, piperacillin, ertapenem, imipenem, meropenem, cefepime, cefpirome, cefotaxime, ceftazidime, cefoxitin, and cefepime with OmpF;7,19,21,36,50,54,55,57,58, 60–63,65–67,70–73 cefotaxime, imipenem, and meropenem with OmpC;19,48 natural amino acids with OccD1),87 ion transport including transport of phosphate potassium and chloride ion via OprP90,100–102 and OprO,99 and interaction of glycine ornithine, glucose, and imipenem with CarO isoforms.105 Further, for liposome swelling55,60,97,105 and minimum inhibitory concentration assay48,60,73 (not described), MS was helpful for understanding and interpreting the experimental results.
Rationalizing the process of permeation of antibiotics into Gram-negative bacteria via MS requires an accurate and exhaustive description of some key molecular properties of the antibiotic molecule.121 MS is the best alternative tool to obtain homogenously derived physical–chemical descriptors for molecules with or without experimental approach.121,127,128 MS based on all-atom empirical force fields with the resolution in microsecond time range and beyond could potentially provide a good level of description of the structural and dynamical properties of biological systems.119,121,127,128
Toward translational research
Translational research on understanding antimicrobial resistance has led to implausible development in recent years4,129 together with the expansion of novel techniques including proteomic analyses, high-sensitivity mass spectrometry, computational bioinformatics, and many more approaches.4 For the most part, the discovery of novel technologies, the development of new infrastructures, along with the training of budding scientists have reinforced this evolution.1,4,129,130 But the transition is still not complete, and roadblocks still exist on the path to scientific progress, for example, combining different data into a shared database that can be intrinsically used to understand how Omps located in the outer membrane of Gram-negative bacteria are able to filter molecular influx.24 The imperative need for new, effective Gram-negative antibacterial drugs comes at a time when techniques needed for innovative assays can provide significant crucial data over understanding the effective bottleneck.4 Ideally, the overall penetration–efflux puzzle4 will form part of a larger understanding of the Gram-negative cell envelope as well as direction on how to create small molecules that can easily penetrate across the outer membranes.4 This information should move the antibacterial research community toward more rational approaches, which may enable the delivery of new agents to treat life-threatening infections.1,4,129,130
Conclusive remarks
This review summarizes the progressive scientific evidence explaining the role of Omps in membrane permeability of Gram-negative bacteria. The control of bacterial membrane permeability is a complex process that is strongly structured by an intricate network of arrangements that senses and retorts to pH, osmotic shock, temperature, and external chemical stress. Bacteria majorly make use of cultured regulated cascades that perceive and distinguish toxic compounds and respond through various resistance mechanisms including regulation of Omps.6,7,15,18,22 The information on the role of effective Omps in substrate uptake and their structural relationship associated with their role in transport highlights the efforts of the scientific upfront in the direction of understanding the bacterial resistance.6,7,15,18,22 Translocation across the Omps can be assumed as the first step in the journey of an antibiotic along the defined pathway toward its target. Consequently, interpretation of antibiotic translocation through porins at the molecular level is crucial for understanding the correlation between influx and antibiotic activities within bacteria. The function of the general diffusion pores has been well studied based on pore characteristics, chemical modification, and genetic mutations. Our understanding of the structure of the pore-forming complex has tremendously improved over the last decade with the emergence of MS, state-of-the-art X-ray data, mass spectrometry assay protocols, and novel high-resolution experimental approaches including electrophysiology. However, a better understanding of the transportation mechanism by outer membrane pores is required. The molecular basis of the antibiotic transport via specific porins is still completely open at present, and further rigorous studies are needed to give insight into the structure–activity relationship of pores associated with antibiotic transport. The data computed for these Omps can be further employed to elucidate the antibiotic uptake pathway through Omps at molecular level, which could possibly empower rational drug design to further enhance permeation and support novel strategies to dodge “impermeability”-mediated resistance mechanism.
Acknowledgments
The publication of this article was funded by the Open Access fund of Leibniz Universität Hannover. The authors would like to thank Prof. Dr Mathias Winterhalter and Prof. Dr Richard Wagner for their constructive comments.
Disclosure
The authors report no conflicts of interest in this work.
References
Kostyanev T, Bonten MJ, O’Brien S, et al. The Innovative Medicines Initiative’s New Drugs for Bad Bugs programme: European public-private partnerships for the development of new strategies to tackle antibiotic resistance. J Antimicrob Chemother. 2016;71(2):290–295. | ||
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–283. | ||
Gootz TD. The global problem of antibiotic resistance. Crit Rev Immunol. 2010;30(1):79–93. | ||
Stavenger RA, Winterhalter M. TRANSLOCATION project: how to get good drugs into bad bugs. Sci Transl Med. 2014;6(228):228ed7. | ||
Nikaido H. Role of permeability barriers in resistance to beta-lactam antibiotics. Pharmacol Ther. 1985;27(2):197–231. | ||
Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. | ||
Winterhalter M, Ceccarelli M. Physical methods to quantify small antibiotic molecules uptake into Gram-negative bacteria. Eur J Pharm Biopharm. 2015;95(Pt A):63–67. | ||
Chattopadhyay MK, Jaganandham MV. Vesicles-mediated resistance to antibiotics in bacteria. Front Microbiol. 2015;6:758. | ||
Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014;160(10):2109–2121. | ||
Chattopadhyay MK, Jagannadham MV. Corrigendum: vesicles-mediated resistance to antibiotics in bacteria. Front Microbiol. 2015;6:758. | ||
McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63(2):545–558. | ||
Maredia R, Devineni N, Lentz P, et al. Vesiculation from Pseudomonas aeruginosa under SOS. Scientific World J. 2012;2012:18. | ||
Kulkarni HM, Swamy ChV, Jagannadham MV. Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res. 2014;13(3):1345–1358. | ||
Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13(10):605–619. | ||
Benz R. Structure and function of porins from gram-negative bacteria. Annu Rev Microbiol. 1988;42:359–393. | ||
James CE, Mahendran KR, Molitor A, et al. How beta-lactam antibiotics enter bacteria: a dialogue with the porins. PLoS One. 2009;4(5):e5453. | ||
Schirmer T. General and specific porins from bacterial outer membranes. J Struct Biol. 1998;121(2):101–109. | ||
Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565(2):308–317. | ||
Scorciapino MA, D’Agostino T, Acosta-Gutierrez S, Malloci G, Bodrenko I, Ceccarelli M. Exploiting the porin pathway for polar compound delivery into Gram-negative bacteria. Future Med Chem. 2016;8(10):1047–1062. | ||
Singh H, Thangaraj P, Chakrabarti A. Acinetobacter baumannii: a brief account of mechanisms of multidrug resistance and current and future therapeutic management. J Clin Diagn Res. 2013;7(11):2602–2605. | ||
Ziervogel BK, Roux B. The binding of antibiotics in OmpF porin. Structure. 2013;21(1):76–87. | ||
Pages JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6(12):893–903. | ||
Dupont H, Choinier P, Roche D, et al. Structural alteration of OmpR as a source of ertapenem resistance in a CTX-M-15-producing Escherichia coli O25b:H4-ST131 clinical isolate. Antimicrob Agents Chemother. 2017;61(5):e00014–e00017. | ||
Chakradhar, S. Breaking through: How researchers are gaining entry into barricaded bacteria. Nat Med. 2017;23(8):907–910. | ||
Nakae T. Outer membrane of Salmonella. Isolation of protein complex that produces transmembrane channels. J Biol Chem. 1976;251(7):2176–2178. | ||
Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49(1):1–32. | ||
Nguyen-Disteche M, Pollock JJ, Ghuysen JM, et al. Sensitivity to ampicillin and cephalothin of enzymes involved in wall peptide crosslinking in Escherichia coli K12, strain 44. Eur J Biochem. 1974;41(3):457–463. | ||
Schulz GE. Porins: general to specific, native to engineered passive pores. Curr Opin Struct Biol. 1996;6(4):485–490. | ||
Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20(17):2338–2348. | ||
Fahie MA, Yang B, Mullis M, Holden MA, Chen M. Selective detection of protein homologues in serum using an OmpG nanopore. Anal Chem. 2015;87(21):11143–11149. | ||
Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol. 2012;302(2):63–68. | ||
Dupont H, Gaillot O, Goetgheluck AS, et al. Molecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother. 2015;60(1):215–221. | ||
Hancock RE, Woodruff WA. Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa. Rev Infect Dis. 1988;10(4):770–775. | ||
Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ. 2016;352:h6420. | ||
Zahn M, Bhamidimarri Satya P, Baslé A, Winterhalter M, van den Berg B. Structural insights into outer membrane permeability of Acinetobacter baumannii. Structure. 2016;24(2):221–231. | ||
Ghai I, Pira A, Scorciapino MA, et al. General method to determine the flux of charged molecules through nanopores applied to beta-lactamase inhibitors and OmpF. J Phys Chem Lett. 2017;8(6):1295–1301. | ||
Kretschmer N, Damianakos H, Chinou I, et al. Comparison of the cytotoxicity and antimicrobial activity of several isohexenylnaphthazarins. Planta Med. 2011;77(12):PM199. | ||
Hancock RE, Brinkman FS. Function of pseudomonas porins in uptake and efflux. Annu Rev Microbiol. 2002;56:17–38. | ||
Fito-Boncompte L, Chapalain A, Bouffartigues E, et al. Full virulence of Pseudomonas aeruginosa requires OprF. Infect Immun. 2011;79(3):1176–1186. | ||
Nestorovich EM, Sugawara E, Nikaido H, Bezrukov SM. Pseudomonas aeruginosa porin OprF: properties of the channel. J Biol Chem. 2006;281(24):16230–16237. | ||
Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem. 2006;281(24):16220–16229. | ||
Pages JM, Peslier S, Keating TA, Lavigne JP, Nichols WW. Role of the outer membrane and porins in susceptibility of beta-lactamase-producing Enterobacteriaceae to ceftazidime-avibactam. Antimicrob Agents Chemother. 2015;60(3):1349–1359. | ||
Acosta Gutierrez S, Bodrenko I, Scorciapino MA, Ceccarelli M. Macroscopic electric field inside water-filled biological nanopores. Phys Chem Chem Phys. 2016;18(13):8855–8864. | ||
Lovelle M, Mach T, Mahendran KR, Weingart H, Winterhalter M, Gameiro P. Interaction of cephalosporins with outer membrane channels of Escherichia coli. Revealing binding by fluorescence quenching and ion conductance fluctuations. Phys Chem Chem Phys. 2011;13(4):1521–1530. | ||
Mahendran KR, Kreir M, Weingart H, Fertig N, Winterhalter M. Permeation of antibiotics through Escherichia coli OmpF and OmpC porins: screening for influx on a single-molecule level. J Biomol Screen. 2010;15(3):302–307. | ||
Pinet E, Franceschi C, Davin-Regli A, Zambardi G, Pages JM. Role of the culture medium in porin expression and piperacillin-tazobactam susceptibility in Escherichia coli. J Med Microbiol. 2015;64(11):1305–1314. | ||
Liu N, Samartzidou H, Lee KW, Briggs JM, Delcour AH. Effects of pore mutations and permeant ion concentration on the spontaneous gating activity of OmpC porin. Protein Eng. 2000;13(7):491–500. | ||
Lou H, Chen M, Black SS, et al. Altered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi-drug resistant E. coli. PLoS One. 2011;6(10):e25825. | ||
Sugawara E, Kojima S, Nikaido H. Klebsiella pneumoniae major porins OmpK35 and OmpK36 allow more efficient diffusion of beta-lactams than their Escherichia coli homologs OmpF and OmpC. J Bacteriol. 2016;198(23):3200–3208. | ||
Aguilella-Arzo M, Garcia-Celma JJ, Cervera J, Alcaraz A, Aguilella VM. Electrostatic properties and macroscopic electrodiffusion in OmpF porin and mutants. Bioelectrochemistry. 2007;70(2):320–327. | ||
Apetrei A, Asandei A, Park Y, Hahm KS, Winterhalter M, Luchian T. Unimolecular study of the interaction between the outer membrane protein OmpF from E. coli and an analogue of the HP(2–20) antimicrobial peptide. J Bioenerg Biomembr. 2010;42(2):173–180. | ||
Bajaj H, Scorciapino MA, Moynie L, et al. Molecular basis of filtering carbapenems by porins from beta-lactam-resistant clinical strains of Escherichia coli. J Biol Chem. 2016;291(6):2837–2847. | ||
Bodrenko I, Bajaj H, Ruggerone P, Winterhalter M, Ceccarelli M. Analysis of fast channel blockage: revealing substrate binding in the microsecond range. Analyst. 2015;140(14):4820–4827. | ||
Brauser A, Schroeder I, Gutsmann T, et al. Modulation of enrofloxacin binding in OmpF by Mg2+ as revealed by the analysis of fast flickering single-porin current. J Gen Physiol. 2012;140(1):69–82. | ||
Bredin J, Saint N, Mallea M, et al. Alteration of pore properties of Escherichia coli OmpF induced by mutation of key residues in anti-loop 3 region. Biochem J. 2002;363(Pt 3):521–528. | ||
Cama J, Bajaj H, Pagliara S, et al. Quantification of fluoroquinolone uptake through the outer membrane channel OmpF of Escherichia coli. J Am Chem Soc. 2015;137(43):13836–13843. | ||
Danelon C, Nestorovich EM, Winterhalter M, Ceccarelli M, Bezrukov SM. Interaction of zwitterionic penicillins with the OmpF channel facilitates their translocation. Biophys J. 2006;90(5):1617–1627. | ||
Danelon C, Suenaga A, Winterhalter M, Yamato I. Molecular origin of the cation selectivity in OmpF porin: single channel conductances vs. free energy calculation. Biophys Chem. 2003;104(3):591–603. | ||
Ghai I, Winterhalter M, Wagner R. Probing transport of charged beta-lactamase inhibitors through OmpC, a membrane channel from E. coli. Biochem Biophys Res Commun. 2017;484(1):51–55. | ||
Hajjar E, Bessonov A, Molitor A, et al. Toward screening for antibiotics with enhanced permeation properties through bacterial porins. Biochemistry. 2010;49(32):6928–6935. | ||
Hajjar E, Mahendran KR, Kumar A, et al. Bridging timescales and length scales: from macroscopic flux to the molecular mechanism of antibiotic diffusion through porins. Biophys J. 2010;98(4):569–575. | ||
Im W, Roux B. Ion permeation and selectivity of OmpF porin: a theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theory. J Mol Biol. 2002;322(4):851–869. | ||
Kumar A, Hajjar E, Ruggerone P, Ceccarelli M. Molecular simulations reveal the mechanism and the determinants for ampicillin translocation through OmpF. J Phys Chem B. 2010;114(29):9608–9616. | ||
Lamichhane U, Islam T, Prasad S, Weingart H, Mahendran KR, Winterhalter M. Peptide translocation through the mesoscopic channel: binding kinetics at the single molecule level. Eur Biophys J. 2013;42(5):363–369. | ||
Mach T, Neves P, Spiga E, et al. Facilitated permeation of antibiotics across membrane channels—interaction of the quinolone moxifloxacin with the OmpF channel. J Am Chem Soc. 2008;130(40):13301–13309. | ||
Mahendran KR, Hajjar E, Mach T, et al. Molecular basis of enrofloxacin translocation through OmpF, an outer membrane channel of Escherichia coli—when binding does not imply translocation. J Phys Chem B. 2010;114(15):5170–5179. | ||
Nestorovich EM, Danelon C, Winterhalter M, Bezrukov SM. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc Natl Acad Sci U S A. 2002;99(15):9789–9794. | ||
Neves P, Berkane E, Gameiro P, Winterhalter M, de Castro B. Interaction between quinolones antibiotics and bacterial outer membrane porin OmpF. Biophys Chem. 2005;113(2):123–128. | ||
Neves P, Sousa I, Winterhalter M, Gameiro P. Fluorescence quenching as a tool to investigate quinolone antibiotic interactions with bacterial protein OmpF. J Membr Biol. 2009;227(3):133–140. | ||
Pezeshki S, Chimerel C, Bessonov AN, Winterhalter M, Kleinekathofer U. Understanding ion conductance on a molecular level: an all-atom modeling of the bacterial porin OmpF. Biophys J. 2009;97(7):1898–1906. | ||
Phale PS, Philippsen A, Widmer C, Phale VP, Rosenbusch JP, Schirmer T. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry. 2001;40(21):6319–6325. | ||
Singh PR, Ceccarelli M, Lovelle M, Winterhalter M, Mahendran KR. Antibiotic permeation across the OmpF channel: modulation of the affinity site in the presence of magnesium. J Phys Chem B. 2012; 116(15):4433–4438. | ||
Vidal S, Bredin J, Pages JM, Barbe J. Beta-lactam screening by specific residues of the OmpF eyelet. J Med Chem. 2005;48(5):1395–1400. | ||
Weichbrodt C, Bajaj H, Baaken G, et al. Antibiotic translocation through porins studied in planar lipid bilayers using parallel platforms. Analyst. 2015;140(14):4874–4881. | ||
Benz R, Darveau RP, Hancock RE. Outer-membrane protein PhoE from Escherichia coli forms anion-selective pores in lipid-bilayer membranes. Eur J Biochem. 1984;140(2):319–324. | ||
Darveau RP, Hancock RE, Benz R. Chemical modification of the anion selectivity of the PhoE porin from the Escherichia coli outer membrane. Biochim Biophys Acta. 1984;774(1):67–74. | ||
Bornet C, Davin-Regli A, Bosi C, Pages JM, Bollet C. Imipenem resistance of Enterobacter aerogenes mediated by outer membrane permeability. J Clin Microbiol. 2000;38(3):1048–1052. | ||
Lavigne JP, Sotto A, Nicolas-Chanoine MH, Bouziges N, Pages JM, Davin-Regli A. An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int J Antimicrob Agents. 2013;41(2):130–136. | ||
Thiolas A, Bornet C, Davin-Regli A, Pages JM, Bollet C. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem Biophys Res Commun. 2004;317(3):851–856. | ||
Bornet C, Saint N, Fetnaci L, et al. Omp35, a new Enterobacter aerogenes porin involved in selective susceptibility to cephalosporins. Antimicrob Agents Chemother. 2004;48(6):2153–2158. | ||
Arunmanee W, Pathania M, Solovyova AS, et al. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proc Natl Acad Sci U S A. 2016;113(34):E5034–E5043. | ||
Castanheira M, Mendes RE, Sader HS. Low frequency of ceftazidime-avibactam resistance among Enterobacteriaceae isolates carrying blaKPC collected in U.S. hospitals from 2012 to 2015. Antimicrob Agents Chemother. 2017;61(3). | ||
Eren E, Parkin J, Adelanwa A, et al. Toward understanding the outer membrane uptake of small molecules by Pseudomonas aeruginosa. J Biol Chem. 2013;288(17):12042–12053. | ||
Liu J, Wolfe AJ, Eren E, et al. Cation selectivity is a conserved feature in the OccD subfamily of Pseudomonas aeruginosa. Biochim Biophys Acta. 2012;1818(11):2908–2916. | ||
Biswas S, Mohammad MM, Patel DR, Movileanu L, van den Berg B. Structural insight into OprD substrate specificity. Nat Struct Mol Biol. 2007;14(11):1108–1109. | ||
Huang H, Hancock RE. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1996;178(11):3085–3090. | ||
Samanta S, Scorciapino MA, Ceccarelli M. Molecular basis of substrate translocation through the outer membrane channel OprD of Pseudomonas aeruginosa. Phys Chem Chem Phys. 2015;17(37):23867–23876. | ||
Eren E, Vijayaraghavan J, Liu J, et al. Substrate specificity within a family of outer membrane carboxylate channels. PLoS Biol. 2012;10(1): e1001242. | ||
Liu J, Eren E, Vijayaraghavan J, et al. OccK channels from Pseudomonas aeruginosa exhibit diverse single-channel electrical signatures but conserved anion selectivity. Biochemistry. 2012;51(11):2319–2330. | ||
Modi N, Benz R, Hancock RE, Kleinekathofer U. Modeling the ion selectivity of the phosphate specific channel OprP. J Phys Chem Lett. 2012;3(23):3639–3645. | ||
Soundararajan G, Bhamidimarri SP, Winterhalter M. Understanding carbapenem translocation through OccD3 (OpdP) of Pseudomonas aeruginosa. ACS Chem Biol. 2017;12(6):1656–1664. | ||
Chalhoub H, Pletzer D, Weingart H, et al. Mechanisms of intrinsic resistance and acquired susceptibility of Pseudomonas aeruginosa isolated from cystic fibrosis patients to temocillin, a revived antibiotic. Sci Rep. 2017;7:40208. | ||
Cheneke BR, Indic M, van den Berg B, Movileanu L. An outer membrane protein undergoes enthalpy- and entropy-driven transitions. Biochemistry. 2012;51(26):5348–5358. | ||
Cheneke BR, van den Berg B, Movileanu L. Analysis of gating transitions among the three major open states of the OpdK channel. Biochemistry. 2011;50(22):4987–4997. | ||
Pothula KR, Dhanasekar NN, Lamichhane U, et al. Single residue acts as gate in OccK channels. J Phys Chem B. 2017;121(12):2614–2621. | ||
Pothula KR, Kleinekathofer U. Theoretical analysis of ion conductance and gating transitions in the OpdK (OccK1) channel. Analyst. 2015;140(14):4855–4864. | ||
Samanta S, D’Agostino T, Ghai I, et al. How to get large drugs through small pores? Exploiting the porins pathway in Pseudomonas aeruginosa. Biophys J. 2017;112(3 Suppl 1):416a. | ||
Tamber S, Maier E, Benz R, Hancock RE. Characterization of OpdH, a Pseudomonas aeruginosa porin involved in the uptake of tricarboxylates. J Bacteriol. 2007;189(3):929–939. | ||
Modi N, Ganguly S, Barcena-Uribarri I, et al. Structure, dynamics, and substrate specificity of the OprO porin from Pseudomonas aeruginosa. Biophys J. 2015;109(7):1429–1438. | ||
Modi N, Barcena-Uribarri I, Bains M, Benz R, Hancock RE, Kleinekathofer U. Role of the central arginine R133 toward the ion selectivity of the phosphate specific channel OprP: effects of charge and solvation. Biochemistry. 2013;52(33):5522–5532. | ||
Modi N, Barcena-Uribarri I, Bains M, Benz R, Hancock RE, Kleinekathofer U. Tuning the affinity of anion binding sites in porin channels with negatively charged residues: molecular details for OprP. ACS Chem Biol. 2015;10(2):441–451. | ||
Pongprayoon P, Beckstein O, Wee CL, Sansom MS. Simulations of anion transport through OprP reveal the molecular basis for high affinity and selectivity for phosphate. Proc Natl Acad Sci U S A. 2009;106(51):21614–21618. | ||
Catel-Ferreira M, Nehme R, Molle V, et al. Deciphering the function of the outer membrane protein OprD homologue of Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56(7):3826–3832. | ||
Catel-Ferreira M, Coadou G, Molle V, et al. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J Antimicrob Chemother. 2011;66(9):2053–2056. | ||
Zahn M, D’Agostino T, Eren E, Basle A, Ceccarelli M, van den Berg B. Small-molecule transport by CarO, an abundant eight-stranded beta-barrel outer membrane protein from Acinetobacter baumannii. J Mol Biol. 2015;427(14):2329–2339. | ||
Iyer R, Delcour AH. Complex inhibition of OmpF and OmpC bacterial porins by polyamines. J Biol Chem. 1997;272(30):18595–18601. | ||
Saint N, Lou KL, Widmer C, Luckey M, Schirmer T, Rosenbusch JP. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J Biol Chem. 1996;271(34):20676–20680. | ||
Lou KL, Saint N, Prilipov A, et al. Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis. J Biol Chem. 1996;271(34):20669–20675. | ||
Phale PS, Schirmer T, Prilipov A, Lou K-L, Hardmeyer A, Rosenbusch JP. Voltage gating of Escherichia coli porin channels: role of the constriction loop. Proc Natl Acad Sci U S A. 1997;94(13):6741–6745. | ||
Stock JB, Rauch B, Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977;252(21):7850–7861. | ||
Testa CA, Johnson LJ. A whole-cell phenotypic screening platform for identifying methylerythritol phosphate pathway-selective inhibitors as novel antibacterial agents. Antimicrob Agents Chemother. 2012;56(9):4906–4913. | ||
Mortimer PGS, Piddock LJV. A comparison of methods used for measuring the accumulation of quinolones by enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother. 1991;28(5):639–653. | ||
Goessens WH, van der Bij AK, van Boxtel R, et al. Antibiotic trapping by plasmid-encoded CMY-2 beta-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob Agents Chemother. 2013;57(8):3941–3949. | ||
Shin JR, Lim KJ, Kim DJ, Cho JH, Kim SC. Display of multimeric antimicrobial peptides on the Escherichia coli cell surface and its application as whole-cell antibiotics. PLoS One. 2013;8(3):e58997. | ||
Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26(4):822–880. | ||
Fox DT, Schmidt EN, Tian H, et al. Sub-inhibitory fosmidomycin exposures elicits oxidative stress in Salmonella enterica serovar Typhimurium LT2. PLoS One. 2014;9(4):e95271. | ||
Davis TD, Gerry CJ, Tan DS. General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem Biol. 2014;9(11):2535–2544. | ||
Zhou Y, Joubran C, Miller-Vedam L, et al. Thinking outside the “bug”: a unique assay to measure intracellular drug penetration in gram-negative bacteria. Anal Chem. 2015;87(7):3579–3584. | ||
Bezrukov SM, Berezhkovskii AM, Szabo A. Diffusion model of solute dynamics in a membrane channel: mapping onto the two-site model and optimizing the flux. J Chem Phys. 2007;127(11):115101. | ||
O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem. 2008;51(10):2871–2878. | ||
Scorciapino MA, Acosta-Gutierrez S, Benkerrou D, et al. Rationalizing the permeation of polar antibiotics into Gram-negative bacteria. J Phys Condens Matter. 2017;29(11):113001. | ||
Krasilnikov OV, Sabirov RZ, Ternovsky VI, Merzliak PG, Muratkhodjaev JN. A simple method for the determination of the pore radius of ion channels in planar lipid bilayer membranes. FEMS Microbiol Immunol. 1992;5(1–3):93–100. | ||
Barcena-Uribarri I, Thein M, Maier E, Bonde M, Bergstrom S, Benz R. Use of nonelectrolytes reveals the channel size and oligomeric constitution of the Borrelia burgdorferi P66 porin. PLoS One. 2013;8(11): e78272. | ||
Krasilnikov OV, Da Cruz JB, Yuldasheva LN, Varanda WA, Nogueira RA. A novel approach to study the geometry of the water lumen of ion channels: colicin Ia channels in planar lipid bilayers. J Membr Biol. 1998;161(1):83–92. | ||
Krasilnikov OV. Sizing channels with neutral polymers. In: Kasianowicz JJ, Kellermayer M, Deamer DW, editors. Structure and Dynamics of Confined Polymers. Dordrecht: Springer; 2002:97–115. | ||
Bajaj H, Acosta Gutierrez S, Bodrenko I, et al. Bacterial outer membrane porins as electrostatic nanosieves: exploring transport rules of small polar molecules. ACS Nano. 2017;11(6):5465–5473. | ||
Schwarz G, Danelon C, Winterhalter M. On translocation through a membrane channel via an internal binding site: kinetics and voltage dependence. Biophys J. 2003;84(5):2990–2998. | ||
Tran QT, Williams S, Farid R, Erdemli G, Pearlstein R. The translocation kinetics of antibiotics through porin OmpC: insights from structure-based solvation mapping using WaterMap. Proteins. 2013;81(2):291–299. | ||
Fontana JM, Alexander E, Salvatore M. Translational research in infectious disease: current paradigms and challenges ahead. Transl Res. 2012;159(6):430–453. | ||
Goldman M. The innovative medicines initiative: a European response to the innovation challenge. Clin Pharmacol Ther. 2012;91(3):418–425. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.