Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Evaluation of Personalized Skincare Through in-silico Gene Interactive Networks and Cellular Responses to UVR and Oxidative Stress
Authors Markiewicz E , Idowu OC
Received 26 July 2022
Accepted for publication 13 October 2022
Published 19 October 2022 Volume 2022:15 Pages 2221—2243
DOI https://doi.org/10.2147/CCID.S383790
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Ewa Markiewicz, Olusola C Idowu
Hexis Lab, The Catalyst, Newcastle Helix, Newcastle upon Tyne, UK
Correspondence: Olusola C Idowu, HexisLab Limited, The Catalyst, Newcastle Helix, Newcastle upon Tyne, NE4 5TG, UK, Tel +44 1394 825487, Email [email protected]
Purpose: Personalized approaches in dermatology are designed to match the specific requirements based on the individual genetic makeup. One major factor accounting for the differences in skin phenotypes is single nucleotide polymorphism (SNP) within several genes with diverse roles that extend beyond skin tone and pigmentation. Therefore, the cellular sensitivities to the environmental stress and damage linked to extrinsic aging could also underlie the individual characteristics of the skin and dictate the unique skin care requirements. This study aimed to identify the likely biomarkers and molecular signatures expressed in skin cells of different ethnic backgrounds, which could aid further the design of personalized skin products based on specific demands.
Methods: Using data mining and in-silico modeling, the association of SNP-affected genes with three major skin types of European, Asian and African origin was analyzed and compared within the structure-function gene interaction networks. Cultured dermal fibroblasts were subsequently subjected to ultraviolet radiation and oxidative stress and analyzed for DNA damage and senescent markers. The protective applications of two cosmetic ingredients, Resveratrol and Quercetin, were validated in both cellular and in-silico models.
Results: Each skin type was characterized by the presence of SNPs in the genes controlling facultative and constitutive pigmentation, which could also underlie the major differences in responses to photodamage, such as oxidative stress, inflammation, and barrier homeostasis. Skin-type-specific dermal fibroblasts cultured in-vitro demonstrated distinctive sensitivities to ultraviolet radiation and oxidative stress, which could be modulated further by the bioactive compounds with the predicted capacities to interact with some of the genes in the in-silico models.
Conclusion: Evaluation of the SNP-affected gene networks and likely sensitivities of skin cells, defined as low threshold levels to extrinsic stress factors, can provide a valuable tool for the design and formulation of personalized skin products that match more accurately diverse ethnic backgrounds.
Keywords: ethnic skin types, gene networks, single nucleotide polymorphism, aging, cosmetics
Introduction
Individual characteristics of the skin are governed by genetic signatures that are increasingly recognized as a basis of personalized skincare.1 Variation of skin pigmentation in populations worldwide is attributed to the expression and activity of more than 125 genes involved in the synthesis of melanin, which correlates with the level of protection against ultraviolet radiation (UVR) and photo-damage.2,3 Lighter skin types are more prone to increased production of reactive oxygen species (ROS), loss of elastic fibers and thinning of the epidermis, which is considered a sign of extrinsic aging induced by the environment. The defensive capacities of the skin, such as the ability to counteract ROS and sustain the barrier function, are moreover significantly reduced in aged skin. The major factors responsible for this decline are associated with processes of DNA repair, cell proliferation, mitochondrial activity and metabolic functions of the cells. This process, referred to as genetically programmed or intrinsic aging, occurs in all skin types and can be accelerated by lifestyle and environmental factors such as UVR exposure. It is believed that oxidative stress, caused by an imbalance between the production of ROS and their sufficient detoxification through antioxidants plays a major role in skin aging.4,5 Recent studies have uncovered several gene clusters affected by single nucleotide polymorphism (SNP) and associated with specific traits and aging patterns of the European skin.1 More detailed studies are necessary to address the gene networks behind the phenotypes evident in darker skin types on diverse ethnic backgrounds, for example, highlighting the hyper-pigmentation and weakened barrier function as predominant traits linked to environmental stress and skin damage. SNP is caused by deleting or replacing a nucleotide, resulting in altered alleles in the gene’s locus.6 SNPs occur throughout both coding and non-coding fragments of the entire genome; an estimated ~25% alters gene product function and could have clinical impacts.7,8 SNPs have been also identified in several of the genes responsible for the maintenance of structural, biochemical and metabolic properties of the skin, estimated to be at a frequency higher than 10% within a population.1,9–11 It is likely that the genetic variants and multiple combinations of them can translate into significant differences between skin phenotypes. One major and best-characterized trait of human skin is pigmentation, with several genes responsible for melanogenesis and UVR protection also involved in oxidative stress, DNA damage and repair, and inflammation.12–15 These factors also play important roles in dermal fibroblast physiology and secretory activity and are the major contributors to skin aging including extrinsic aging caused by UVR and oxidative stress within the dermis.16 It is therefore likely that the degree of pigmentation is closely linked to not only the sun sensitivity and the protection of genome integrity but also the factors involved in the accumulation of macromolecular damage and aging characteristics. Such patterns could moreover vary between individuals and ethnic groups.1 The differences in both skin pigmentation and the sensitivities to environmental stress, including genotoxic stress affecting both the epidermis and dermis, can translate to the broad patterns of damage between individual skin types, ranging from the loss of tensile strength and wrinkles, dry and rough skin, to the alterations in pigmentation.
In this study, we identify a group of 37 biomarkers affected by the SNPs and discuss their likely biological activities within a pattern of gene network based on data mining and in-silico modeling algorithms. The genes are clustered within three major groups of ethnic origin; European, Asian and African and contain the factors not only responsible for constitutive and facultative pigmentation but also a range of biological activities that could define likely patterns linked to skin phenotypes, sensitivities to the environmental stress and the mechanisms of defense. Such patterns are centered around DNA damage response, oxidative stress and inflammation, and remodeling of the extracellular matrix (ECM) and barrier, which are linked directly or indirectly to melanogenic factors and emphasized in each skin type. Subsequent in-vitro analysis of dermal fibroblasts from the skin of European, Asian and African origin reveals important differences in the cellular responses to UVR and oxidative stress. The cells were exposed to a single dose of UVR or H2O2 and analyzed for the accumulation of enlarged nuclei and changes in cellular densities based on the images stained with fluorescent intercalating agent DAPI. Further measurements involved the relative expression levels of proliferation marker Ki67, as well as the DNA damage marker γH2AX and senescence marker cyclin-dependent kinase (CDK) inhibitor p16INK4a from the corresponding signal intensity of immunofluorescence images. The level of oxidative stress was validated by quantification of intracellular ROS production and total antioxidant capacity of the cells based on the fluorescent and colorimetric probes. The assays were also performed after pre-treatment of the cells with natural bioactive compounds and cosmetic ingredients, Resveratrol and Quercetin, to evaluate their protective effect against cellular damage and oxidative stress. Based on the cell growth and the markers of DNA damage, oxidative stress and senescence, Asian fibroblasts are the most sensitive to UVR exposure. Exposure to oxidative stress alone causes the most significant production of intracellular ROS in European fibroblasts; in contrast, whilst African fibroblasts have robust antioxidant capacities to reduce ROS, they accumulate DNA damage in a pro-oxidant environment. Treatment of the cells with Resveratrol and Quercetin during UVR exposure has noticeably different effects on the oxidative stress response and cell protection against damage, which appears to be most beneficial in Asian fibroblasts. In-silico models of the interaction networks between the compounds and the genes affected by SNPs reveal the likely effect on the clusters of oxidative stress and inflammatory factors that are emphasized mostly in Asian and European skin types. An additional comparison of the Resveratrol and Quercetin targets could also suggest the differences in the mechanisms of action centered around pigmentation and the enhanced interactions between the factors involved in inflammation and barrier permeability.
Based on these data and in agreement with the current trends, we propose that SNP sampling and database modeling through next-generation platform technologies such as artificial intelligence (AI) combined with in-vitro cellular assays will provide a practical reference for the design of novel solutions in personalized skincare. This approach can now be advanced and expanded to incorporate both the individual genetic makeup and skin ethnicity for a more accurate impact on dermatology and the discovery screening of novel active ingredients for cosmetic applications.
Materials and Methods
Study Design
The study combined the applications of in-silico screening and gene network analysis with the in-vitro cellular assays utilizing a quantitative analytical approach. The data were collected computationally as well as experimentally based on image analysis combined with quantification of fluorescence and immunofluorescence signal, particle size, and molecular colorimetric assays. Variation between three different cellular populations in response to each external factor was statistically validated using one-way ANOVA with post-hoc Tukey HSD test.
Construction of the in-silico Gene Interactive Models
For the gene interactive models, a literature search and data mining were performed and the genes harboring the SNPs, together with the skin types the genes originated from, were organized in a database.
The gene networks associated with the European, Asian and African skin types were subsequently constructed computationally. The analysis of the interactions between the biomarkers was performed using Hexis Lab Pro.X® in-silico screening platform based on deep learning algorithms. The models were subsequently used to build interactive networks with Resveratrol and Quercetin based on data mining. Hexis Lab Pro.X® enabled validation and prototyping of bio-based factors; the genes were systematically screened against the integrated databases for known and predicted bioactivities using machine learning. The platform was used to identify biological targets, mine biomedical literature and access the polypharmacology and interactions between the genes across biological networks.
Dermal Fibroblasts Cultures
Human dermal fibroblasts (HDF) of European and African skin type origin were purchased from PromoCell (adult, C-12302). Adult dermal fibroblasts of Asian origin were obtained from the Asian Skin Biobank (ASB) at the Skin Research Institute of Singapore (SRIS). The cell cultures were established at young passages in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% Fetal Bovine Serum (FBS, ThermoFisher) and maintained in a 5% CO2, 95% humidified incubator at 37°C. For cell subculture, the cells were treated with Trypsin-EDTA buffer (Gibco) and the cultures were seeded at the same initial density of 6000 cells/cm2 area. For UVR treatment, the cells were exposed to a germicidal lamp (Philips TUV G30T8 30 W bulb) providing predominantly 254-nm light, for 2 minutes and maintained for 72 hours before analysis. UV dose (mJ/cm2) was calculated from lamp specification 125 μW/cm2 at distance 1m x exposure time in seconds, 125 μW/cm2 x 120 sec at 0.75 m distance = 20 mJ/cm2. For H2O2 treatment, the cells were incubated with 250 μM H2O2 for 24 hours, followed by incubation in fresh media for 72 hours. For the treatment with Resveratrol and Quercetin, the cells were pre-incubated with 0.1% compound (concentrated stock dissolved at 10% in DMSO) for 24 hours before UV irradiation. The 0.1% compound concentration was selected after a dose-response validation, ranging from 1% to 0.01% where the toxic high concentrations (particularly Quercetin) and the low non-effective concentrations were discarded. The assay samples included control cells alongside the UVR, H2O2 and Resveratrol/Quercetin-treated cells, in triplicates.
Immunofluorescence and Image Analysis
HDF cultures growing on glass coverslips at the same initial cell density of 6000 cells/cm2 were fixed in 4% formaldehyde/phosphate-buffered saline (PBS) for 15 minutes at room temperature (RT), permeabilized with 0.5% TRITON X-100 for 5 minutes and washed 3 x PBS. For immunostaining, the samples were incubated with primary antibodies diluted in the blocking buffer (PBS + 1% FBS) at 4°C overnight; washed for 30×10 minutes in PBS; incubated with secondary antibodies diluted in the blocking buffer (PBS+1% FBS) at RT for 1 hour; washed for 30×10 minutes in PBS and mounted in the ProLong Gold Antifade Mountant with DAPI (ThermoFisher). The primary antibodies were anti-Ki67 (D3B5, Cell Signaling, 1:500), anti-γH2AX (20E3, Cell Signaling, 1:500), anti-p16INK4a (E6N8P, Cell Signaling, 1:500). Secondary antibodies were FITC anti-rabbit (Jackson ImmunoResearch; 1:1000). The fluorescent images were captured using Leica DM IL LED and photographed using CCD DFC3000G Camera and LAS X 3.6.0.20104 software (Leica Microsystems) with lens magnification 10x and 20x. For the measurements of nuclear size and cell densities, three consecutive images of DAPI-stained nuclei were used for each sample to analyze the size and number of the nuclei using ImageJ software. The cell nuclei from control and test samples were counted manually from each micrograph, and the numbers were recorded. The nuclear size, in microns, was measured in ImageJ as particle size, based on the numbers of image pixels corresponding to microns on the scale bar, with 20 pixels corresponding to 10 microns on the 1047×781 pixels micrograph. The total numbers of cell nuclei obtained per given sample were plotted against the values of nuclear sizes in this sample. The population of enlarged nuclei was subsequently defined as nuclei with a size >20 microns. The percentage of enlarged cell nuclei in control, UVR- and UVR compound-treated samples was calculated based on 100 cells for each treatment. For the measurements of cell densities in the UVR- and UVR + compound-treated samples, the percentage of counted nuclei was calculated in relation to the numbers of nuclei in the control. For each UVR- and UVR + compound-treated sample, the number of Ki67-positive nuclei was recorded and calculated as a percentage of the total number of nuclei in the sample. To quantify the expression levels of γH2AX and p16INK4a, the relative signal intensity was measured for control and treated samples in ImageJ, adjusted to the equal number of cells, with the signal in the control sample equalized to 1. All figures were assembled using GraphPad Prism 9 and PowerPoint software. For statistical analysis, the data comparing variation between skin types for each applied factor were validated using one-way Analysis Of Variance (ANOVA) with post-hoc Tukey HSD test, n = 3, experimental replicates. The graphs represent Mean ± SEM, with statistically significant outputs * p < 0.05, ** p < 0.01, *** p < 0.001.
Intracellular ROS Production Assay
For the measurements of intracellular ROS production, HDFs were seeded onto 96 well plates at the density of 2×103 per well and analyzed 72 hours post-treatment. The media was removed, replaced with PBS and the cells incubated for 30 minutes with a 10 μM 2’7’-dichlorofluorescein diacetate (DCF-DA) probe (Abcam). Conversion into fluorescent DCF was induced by further incubation of the cells in DMEM/10% FBS in a CO2 incubator for 15 minutes. The cells were fixed with 4% formaldehyde/PBS, washed with PBS and relative fluorescence intensity was recorded in a microplate reader (SpectraMax iD5 Microplate Reader, Molecular Devices) at Ex/Em 485/535 nm and adjusted to an equal number of cells. For statistical analysis, the data comparing variation between skin types for each applied factor were validated using one-way Analysis Of Variance (ANOVA) with post-hoc Tukey HSD test, n = 3, experimental replicates. The graphs represent Mean ± SEM, with statistically significant outputs *p < 0.05, **p < 0.01, ***p < 0.001.
Total Antioxidant Capacity Assay
The total antioxidant capacity of the HDF cultures was measured according to the manufacturer’s protocol (Abcam, ab65329). Briefly, the cells were washed with cold PBS, homogenized in 100 μL of ddH2O, incubated for 10 minutes on ice and centrifuged. Trolox standard (1mM) was prepared for several dilutions containing 0–200 nmol Trolox/well. The cell supernatants and Trolox standards were transferred to new wells and incubated with 100 μL Cu2+ Working Solution for 90 minutes at room temperature in dark. The colorimetric output was measured in a microplate reader (SpectraMax iD5 Microplate Reader, Molecular Devices) at OD 570 nm and adjusted to an equal number of cells. Sample total antioxidant capacity (TAC) was calculated as TAC = (Ts/Sn)*D, where Ts = TAC amount in the sample well calculated from a standard curve (nmol), Sn = sample volume added in the sample wells (μL), D = sample dilution factor. For statistical analysis, the data comparing variation between skin types for each applied factor were validated using one-way Analysis Of Variance (ANOVA) with post-hoc Tukey HSD test, n = 3, experimental replicates. The graphs represent Mean ± SEM, with statistically significant outputs *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Single Nucleotide Polymorphism in the Context of Skin Ethnicity
Single nucleotide polymorphism (SNP) is present in several genes involved in the maintenance of skin structure and biological activity. The SNPs can be located both within coding and non-coding regions of the gene and often affect its activity; with the skin types also likely to have different combinations of the modified genes. The biomarkers could additionally interact differently with the genes involved in the melanogenesis pathway in each skin type, creating patterns of structural features and physiology linked to subtle changes in pigmentation. To expand on this further, we performed a literature search of the genes affected by SNPs uncovered in genome-wide association studies (GWAS) across three different ethnic skin types: European, Asian and African (Figure 1 and Table 1).
 |
Table 1 Summary of the Genes Affected by SNPs in European, Asian and African Skin Types |
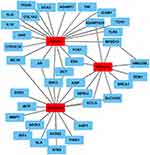 |
Figure 1 Network of the genes affected by the SNPs across three different ethnic skin types based on published data. |
Genes affected by the SNPs in European skin are predominantly linked to melanogenesis and variation of pigmentation: androgen receptor, AR and attractin, ATRN;15 agouti-signaling protein, ASIP;8,17,22 dopachrome tautomerase, DCT;18 GATA binding protein 3, GATA3;6,19 HECT and RLD domain containing E3 ubiquitin protein ligase 2; HERC2;8 interferon regulatory factor 4, IRF4;7 ligand for the receptor-type protein-tyrosine kinase KIT, KITLG;20 melanocortin 1 receptor, MC1R;9,10 microphthalmia-associated transcription factor, MITF;11 solute carrier family 24 member 5, SLC24A521 and tyrosinase-related protein 1, TYRP1.11,22 Lighter skin reflectance is frequently accompanied by sensitivity to the sun and decreased tanning abilities linked to ASIP, HERC2, IRF4, MC1R and TYRP1.7–10,22 Other SNPs could predispose to changes in pigmentation, ie, MC1R and ASIP to freckles;10,22 basonculin-2, BNC2 to pigment spots;19 KITLG and human leukocyte antigen, HLA to solar lentigines.23,24 SNPs in melanogenic genes such as MITF or TYRP1 could have an additional impact on vitamin D deficiency.11 European skin also harbors SNPs in the genes involved in redox homeostasis: superoxide dismutase 2, SOD21 and acetyl-coenzyme A synthetase 2, ACSS2;25 hydration: aquaporin-3, AQP3 and skin elasticity: matrix metalloproteinase 1, MMP1.1
Asian skin harbors SNPs in the genes regulating constitutive pigmentation, including AR, ectodysplasin A, EDA;15 DCT;18 MC1R;26 major facilitator superfamily domain-containing 12, MFSD12;27 oculocutaneous albinism II, OCA228 and SLC24A5,29 usually resulting in lighter skin appearance. Some of the factors associated with darker phenotypes in Asian skin are the fixed alleles of the melanogenic genes involved in UVR response such as damage-specific DNA binding protein 1, DDB1 and MFSD12.27 Skin color variation is also associated with SNPs in the genes linked to hyperpigmentation, for example, BNC2 resulting in facial pigmented spots.30 A significant cluster of the genes is likely centered around the innate immune responses and inflammatory processes, involving intercellular adhesion molecule 1, ICAM1 and tumor necrosis factor, TNF;31 interleukin 10, IL10;32 integrin alpha E, ITGAE and Toll-like receptor 6, TLR6.33 SNPs are also present in the genes involved in xenobiotic metabolism and oxidative stress: aryl hydrocarbon receptor, AHR;30 cytochrome P450 family 2 subfamily C member 19; CYP2C19;34 and high mobility group 20B, HMG20.35 Additional cluster is comprised of the biomarkers regulating the structure and barrier function of the epidermis: filaggrin, FLG and trichohyalin, TCHH;33 skin barrier permeability and trans-epidermal water loss (TEWL), disintegrin and metalloproteinase domain-containing protein 17, ADAM1715 and ficolin-1, FCN136 as well as ECM integrity, ADAM metallopeptidase with thrombospondin type 1 motif 20, ADAMTS2015 and collagen type 1 alpha 2 chain, COL1A2.30
High melanin index in African skin is linked to the fixed alleles within the genes responsible for constitutive pigmentation and UVR responses: DDB1, HERC2, HMG20, MFSD12;27 ASIP37 and KITLG.38 Skin color variation is also determined by other specific SNPs in EDA,15 KITLG,20 DDB1, HERC2 and SLC24A5,27 resulting in lighter pigmentation. SNPs are additionally present in the genes involved in epidermal homeostasis, barrier permeability and hydration: BRCA1 DNA repair associated; BRCA115 and FCN136 as well as remodeling of the ECM, ADAMTS20.15
Skin Biomarkers Affected by Gene Polymorphism Assemble into a Structure-Function Interaction Network
The genes harboring SNPs could be aligned with biological functions relevant to multiple cellular activities and skin homeostasis based on published data (Table 2). We have subsequently captured the molecular hierarchy of the genes through in-silico modeling and interactive network based on the structure-function and protein–protein interactions machine learning (ML) algorithms (Figure 2).
 |
Table 2 Gene Function Relevant to Cellular Homeostasis and Skin Health |
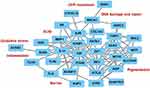 |
Figure 2 An interactive in-silico network of the biomarkers affected by the SNPs based on the structure-function and protein-protein binding algorithms. |
Most of the analyzed genes form defined clusters corresponding to their biological activity whilst engaged in multi-target molecular interactions or synergistic processes related to pigmentation, DNA repair, inflammation and immunity, oxidative stress, ECM remodeling and barrier maintenance. The most prominent gene-enriched or central clusters with a predominant number of potential binding interactions are responsible for the control of pigmentation as well as oxidative stress and inflammation. The main cluster of pigmentation genes is comprised of ASIP, BNC2, MC1R, MITF, OCA2, SLC24A5 and TYRP1. The genes in this cluster are involved in the regulation of pigmentation through transcriptional and enzymatic control of the melanin synthesis pathway and melanosome structure and maturation.12,39–44 This cluster aligns closely with other biomarkers including ATRN, DCT, IRF4 and KITLG, which are involved in the control of melanogenesis, proliferation and survival of melanocytes, and melanin polymerization.24,45–47 Within the melanogenic genes cluster, several, including ASIP, ATRN, IRF4 and MITF are directly involved in cellular responses to UVR, sun sensitivity, DNA repair and inflammation.12,39,45,47 The genes, except for SLC4A5 and KITGL, are predominantly affected by the SNPs in the European and Asian skin.
The genes involved in facultative pigmentation, melanin polymerization and sun sensitivity, including ASIP, DCT, IRF4, KITLG, MC1R and MITF, interact with the biomarkers associated with immune responses and inflammation: ADAM17, GATA3, ICAM1, IL10, ITGAE, TLR6 and TNF.48–54 These genes, except for GATA3, have the SNPs identified in Asian skin. The cluster is also involved in multiple interactions with the factors implicated in skin barrier function: AQP3, EDA, FLG and TCHH;55–58 antioxidant capacities and oxidative stress: ACSS2 and SOD2;59,60 xenobiotic metabolism: AHR, AR and CYP2C1961–63 as well as ECM remodeling: ADAMTS20, COL1A2 and MMP1.16,64,65 SNPs in these genes have been identified predominantly in European and Asian skin, with an emphasis on oxidative stress and ECM remodeling attributed to European skin.
A small cluster of the DNA damage and repair genes, BRCA1, DDB1 and HERC2,14,66,67 which interact with the markers of pigmentation and xenobiotic metabolism, could also be identified through this analysis. This specific group of genes bears the SNPs identified in African skin. Finally, several biomarkers, such as FCN1, HLA1, HMG20B and MFSD12, involved in immunity and inflammation, genome integrity and pigmentation,68–71 were not gathered in this network, indicative of the potential novel interactions that should be investigated further (Figure 2 and Table 2).
This analysis indicated that the three examined major skin types could differ in the patterns of the genes modified by the SNPs. The biomarkers form distinctive clusters, which are also involved in the major interactions with the melanogenic factors responsible for skin pigmentation. This points to the functional association between skin color and its structure and physiology that dictates homeostatic responses to the environment at the molecular and cellular levels.
Dermal Fibroblasts of Different Skin Type Origin Demonstrate Distinct Responses to UVR and Oxidative Stress
Skin color is directly associated with not only the ability to regulate pigmentation, ie, tanning, in response to the environment but also with the individual sensitivities to UVR and UVR-induced damage. It is therefore likely that different skin types would be characterized by diverse cellular responses to several extrinsic factors, including genotoxic stress.
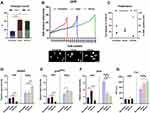
To expand on this further, we cultured dermal fibroblasts isolated from the European, Asian and African skin types, corresponding, respectively, to the phototypes I–II, II–III and IV–V on the Fitzpatrick scale, and analyzed the response of the cells to UVR and oxidative stress. The cells were cultured at the same initial density in monolayers, irradiated with a single dose of 0 mJ/cm2 (control) and 20 mJ/cm2 UVR and the cultures were maintained for an additional 72 hours. Since the UVR causes DNA damage, cell cycle arrest and growth inhibition,72 in the initial approach, we measured the size of the nuclei from the images of the cultures stained with fluorescent probe DAPI, with particular focus on enlarged, >20 microns in diameter nuclei. An enlarged cell nucleus is an indicator of the increase in cellular size and appearance of cell cycle-arrested senescent cells in the culture.73 In control cultures, the percentage of enlarged nuclei was initially higher in both Asian and African fibroblasts compared to the European fibroblasts, as a general characteristic of the cells. The proportion of enlarged nuclei in the UVR-treated cultures was 10% in the European, 25% in Asian and 21% in African fibroblasts, with a respective increase of 4%, 7% and 2% compared to untreated control (Figure 3A). Distribution analysis of the nuclear size versus cell numbers revealed that these changes were also accompanied by significant differences in cell densities in the UV-irradiated cultures. Whilst Asian fibroblasts responded to UVR with a significant reduction in cell densities, the decrease in the densities was less pronounced in European and African fibroblasts (Figure 3B and C). Differences in cell densities were closely associated with the expression of proliferation marker Ki67, with 7.5% of African fibroblasts and 21–28% of European and Asian fibroblasts counted as Ki67 positive in the UVR-treated cultures, indicative of likely prolonged cell cycle and delayed contact inhibition in the latter (Figure 3C). Changes in cellular growth were also reflected in the expression levels of DNA damage marker γH2AX and cyclin-dependent kinase (CDK) inhibitor p16INK4a. Both proteins have been implicated as the markers of persistent DNA damage response and cell senescence.74,75 Expression of both γH2AX and p16INK4a was significantly increased in UV-irradiated European and Asian fibroblasts but remained low in the African fibroblasts (Figure 3D and E, UVR, left panel). The expression of both markers was accompanied by an increase in intracellular ROS production, which was most pronounced in Asian fibroblasts and lowest in African fibroblasts (Figure 3F, UVR, left panel). The differences in the levels of ROS were not reflected in the non-enzymatic, total antioxidant capacities (TAC) of the cells, which remained constant across all three cell types (Figure 3G, UVR, left panel).
Exposure to UVR is a major trigger for oxidative stress, resulting in ROS production and macromolecular damage. To gain more insight into the possible differences in the patterns of the UVR effect versus the effect of oxidative stress alone, the fibroblasts were treated with 0 μM H2O2 (control) and 250 μM H2O2 for 24 hours, and the cultures were maintained for the additional 72 hours. Treatment with H2O2 also triggered the increase in expression of γH2AX and p16INK4a, with the levels of both proteins significantly higher compared to the effect of UVR. Interestingly, expression of γH2AX was visibly increased in the African fibroblasts, indicative of persistent DNA lesions induced by oxidative stress (Figure 3D and E, H2O2, right panel). Similar to UVR, H2O2 treatment also resulted in intracellular ROS production, which remained at similar levels in Asian and African fibroblasts. In contrast, the ROS level induced by H2O2 was ¬3-fold higher in the European fibroblasts (Figure 3F, H2O2, right panel). Oxidative stress induced by H2O2 also triggered an increase in TAC across all cell types, which was ¬3-4-fold higher compared to TAC measured in the UV-irradiated cells. Consistent with the intracellular ROS levels, TAC was lowest in the European fibroblasts and highest in the African fibroblasts, likely reflecting the strongest capacity to neutralize internal ROS (Figure 3G, H2O2, right panel). These data demonstrated that UVR-induced damage had the most pronounced effect on dermal fibroblasts of Asian skin type origin, causing inhibition of cell growth and the expression of senescence and oxidative stress markers. In contrast, African fibroblasts were relatively most resistant to the effects of UVR; however, they did accumulate increased levels of DNA damage upon treatment with H2O2. This type of damage nevertheless occurred in the absence of increased ROS production, which also correlated with the high antioxidant capacities of the African fibroblasts. The sensitivity to oxidative stress triggered by H2O2, based on intracellular ROS and TAC levels, was highest in the European fibroblasts, indicative of a relatively weak capacity to neutralize ROS.
Diverse Effects of Photo-Protective and Antioxidant Cosmetic Ingredients on Ethnic Skin Fibroblasts
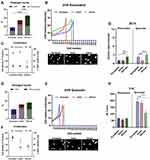
To validate the effects of UVR and oxidative stress on the skin fibroblasts further, we applied two natural compounds, Resveratrol and Quercetin, which are also used in cosmetic formulations as protective and anti-aging ingredients. Similar to the previous approach, the fibroblasts were cultured at the same initial density in monolayers, pre-treated for 24 hours with 0% (control) and 0.1% compound, irradiated with a single dose of 0 mJ/cm2 (control) and 20 mJ/cm2 UVR and the cultures were analyzed after 72 hours. Resveratrol is a stilbene compound with a strong UVR absorbance capacity that can offer a photo-protection.76 The proportion of enlarged nuclei in the Resveratrol/UV-irradiated cultures compared to UVR-control decreased from 10% to 7% in European and from 25% to 9.5% in Asian fibroblasts, whilst the African fibroblasts remained largely unaffected (Figure 4A). Distribution analysis of the nuclear size versus cell numbers and expression of Ki67 revealed that the presence of Resveratrol did slow down the growth of the UV-irradiated African fibroblasts. This was consistent with a ¬50% reduction in cell densities despite the presence of Ki67-positive cells, indicative of a likely prolonged cell cycle and delayed contact inhibition (Figure 4B and C).
To expand on these observations further, we next performed the analysis in the presence of Quercetin. Quercetin is a flavonoid with a powerful antioxidant and anti-inflammatory capacity, which has been also implicated in anti-aging applications as a senolytic ingredient.77 The proportion of enlarged nuclei in the Quercetin/UV-irradiated cultures compared to UVR-control decreased from 25% to 7% in Asian and from 21% to 15% in African fibroblasts, whilst in the European fibroblasts it remained constant (Figure 4D). Distribution analysis of the nuclear size versus cell numbers revealed that a reduction in the numbers of cells with enlarged nuclei was associated with a decrease in cell densities, suggesting that Quercetin could stimulate their removal. Ki67 expression was strongly enhanced by Quercetin in European fibroblasts whilst having a moderate effect on cycling Asian and African fibroblasts (Figure 4E and F).
The presence of Resveratrol during UV irradiation also prevented intracellular ROS production in the European and Asian fibroblasts; it had a minimal effect on African fibroblasts where the level of ROS was already low (Figures 3F and 4G, Resveratrol, left panel). The presence of Quercetin during UV irradiation also inhibited intracellular ROS production, which was most pronounced in Asian fibroblasts (Figure 4G, Quercetin, right panel). UV irradiation in the presence of Resveratrol was accompanied by only a small increase in TAC of the cells, from ¬40 nM Trolox power in UVR control to ¬60 nM Trolox power in the Resveratrol pre-treated cultures (Figures 3G and 4H, Resveratrol, left panel). Quercetin, on the other hand, had a simultaneous strong antioxidant effect on the cells, increasing TAC levels across all three cell types; however, the values were significantly higher in European and Asian fibroblasts (¬190 nM Trolox power) compared to African fibroblasts (¬100 nM Trolox power, Figure 4H, Quercetin, right panel).
These data demonstrated that Resveratrol protected skin dermal fibroblasts from UVR-induced damage, which was the most significant in Asian and European fibroblasts. Inhibition of the intracellular ROS production was not associated with substantial enhancement of the cellular TAC; rather it was likely a direct UVR-protective effect of Resveratrol, which had the weakest impact in African fibroblasts. Quercetin could have a possible senolytic effect targeting the senescent cells whilst enhancing the antioxidant defense in the tissue. The two mechanisms would overlap most strongly in Asian fibroblasts.
The Bioactive Compounds Demonstrate Subtle Differences in the in-silico Skin Gene Target Interactive Networks
The in-vitro analysis revealed that both Resveratrol and Quercetin had protective effects on skin fibroblasts during UV irradiation; however, the impact of both compounds was slightly different across skin types suggesting the possible differences in the mechanisms of action. To expand on this observation further, we performed an in-silico analysis of the known interactions between Resveratrol and Quercetin with the SNP-affected skin gene targets based on data mining and Hexis Lab Pro.X® in-silico platform utilizing deep learning algorithms (Figure 5). Both compounds demonstrated the interactions with five biomarkers, SOD2, IL10, AHR, BRCA1 and FLG, indicative of the strong activation of the antioxidant and anti-inflammatory axis. The antioxidant and anti-inflammatory mechanisms of Resveratrol and Quercetin were also indicated by inhibiting interactions with five other biomarkers: TNF, ICAM1, MMP1, AR and CYP2C19. All biomarkers, except for BRCA1, are linked with Asian and European skin types, which also strongly responded to the protective effects of Resveratrol and Quercetin against UVR in the in-vitro assays.
Resveratrol had additional interaction sites with two melanogenic genes, DCT and MITF, indicative of their inhibition (Figure 5A). In the skin, both biomarkers are involved in responses to UVR and control of pigmentation; dermal fibroblasts isolated from darkly pigmented, ie, African skin could have a decreased sensitivity to Resveratrol.
The interactive target gene network for Quercetin indicated an enhanced activity towards oxidative stress and inflammation. The axis modulated by Quercetin involves GATA3 which forms additional links with TNF, ICAM1, AHR as well as AHR and BRCA1 through up-regulated COL1A2. Enhanced interactions are also present between the inflammatory and epidermal barrier integrity factors involving ADAM17, TNF, IL10 and MMP1 (Figure 5B). The anti-inflammatory and antioxidant capacity of Quercetin could be responsible for a strong protective effect on Asian fibroblasts during UVR exposure.
Discussion
In this study, we highlighted a panel of the genes affected by small nucleotide polymorphisms (SNPs) in three major skin types. In-silico analysis of the interactive network between the biomarkers revealed further functional clusters and synergistic patterns with relevance to the biological activities of the skin. Presently, the most established approaches in personalized skincare are focused on the validation of genetic variations such as SNPs between individuals within a specific ethnic group. Similar analysis across different ethnic groups could uncover further complex patterns with significant potential toward a more stratified characterization of human skin. Although many SNPs studied so far do not always result in the altered expression of the corresponding genes, this argues for a more validated approach correlating such measurements linked to a particular trait and searching for a mechanistic association between interacting genes. Any specific trait that is determined by the combination of several genes forming functional clusters or networks would involve alterations in one gene triggering downstream responses in other activities, for example, reciprocal interactions between melanogenesis and UVR-induced DNA repair or maintenance of skin barrier and control of inflammation.
Whilst the SNPs will not contribute entirely to the different skin types, they are likely to affect both the individual signatures and the traits prevalent within given skin ethnicity. One characteristic associated with genotype–phenotype correlations could be significant compartmentalization affecting the distribution of specific traits across skin ethnicity. Based on the genes involved, these characteristics would likely concentrate around pigmentation in European skin; pigmentation, inflammation, barrier and oxidative stress in Asian skin and epidermal integrity and barrier homeostasis in African skin. These diverse patterns could reflect the specific sensitivity and protection requirements of each skin type; uncovering these requirements would also facilitate a better design and application of effective skin products.
To gain an insight into these patterns, we cultured dermal fibroblasts obtained from European, Asian and African skin and challenged them with extrinsic factors, UVR and H2O2, known to induce DNA damage and oxidative stress. Both these factors also play a central role in signal transduction linked to skin pigmentation, inflammation and barrier homeostasis. European fibroblasts responded to UVR by increased expression of DNA damage marker γH2AX and senescence marker, cyclin-dependent kinase (CDK) inhibitor p16INK4a. This was accompanied by intracellular production of ROS, which could be reduced in the presence of Resveratrol without significant changes to the total antioxidant capacity (TAC) of the cells. TAC of European fibroblasts was enhanced most profoundly by Quercetin; the cells also demonstrated very high sensitivity to oxidative stress induced by H2O2. These results suggest that European fibroblasts could be particularly prone to damage caused by free radicals in an oxidative environment; such an environment could be also more pronounced in lightly pigmented skin that is weakly protected by melanin. The cells would also benefit from strong antioxidants such as Quercetin or molecules with a photo-protective capacity due to UVR absorbance such as Resveratrol.
Asian fibroblasts were characterized by pronounced sensitivity to UVR. UVR responses were associated with an increased percentage of the cells with enlarged nuclei, reduced cell densities and expression of γH2AX and p16INK4a, suggesting induction of senescent phenotypes. Asian fibroblasts also responded to UVR by significant production of intracellular ROS, which could be reduced by both Resveratrol and Quercetin. Both ingredients were also capable of reducing the percentage of the cells with enlarged nuclei and increasing cell densities. Compared to European fibroblasts, Asian fibroblasts produced less ROS and had higher TAC values when challenged with H2O2. These data suggest that Asian fibroblasts could be particularly prone to damage caused directly by UVR but show stronger defenses in a purely pro-oxidant environment. Resveratrol and Quercetin analyzed in this study could be particularly beneficial to Asian fibroblasts for anti-aging and rejuvenating potential.
African fibroblasts demonstrated the most prominent resistance to UVR-induced cellular damage. It was evidenced by the preservation of high cell densities and significantly low levels of γH2AX, p16INK4a and intracellular ROS expression. The levels of ROS remained unaffected or slightly higher in the presence of Resveratrol and Quercetin. Response of African fibroblasts to Quercetin also involved a moderate increase in TAC together with a significant reduction in cell densities, suggestive of potential clearance of UVR-damaged cells. One additional striking phenotype of African fibroblasts was a low production of intracellular ROS and high TAC when the cells were challenged with H2O2, indicative of efficient mobilization of antioxidant defenses in a pro-oxidant environment. However, treatment of African fibroblasts with H2O2 also resulted in unusually high levels of γH2AX expression. These results suggest that African fibroblasts could have efficient mechanisms for counteracting the oxidative stress and damage caused by UVR but are more susceptible to DNA lesions in a pro-oxidant environment. Whilst the protective effect of Resveratrol and Quercetin might be less beneficial for African fibroblasts, other active ingredients able to shield from such damage could be identified in the future. Unraveling the molecular differences in biological pathways, such as those associated with DNA damage or oxidative stress, can also offer a better insight into a range of traits responsible for the individual characteristics of the skin.
Phenotypic Traits Related to the Skin Ethnicity
Pigmentation
Variation of skin color is determined by the quantity of melanin and the proportion of eumelanin to pheomelanin in the epidermis.78 Melanin contributes to natural protection against UVR, which manifests as light absorption and dissipation as well as re-distribution or de novo synthesis of the pigment in tanning responses.79,80 In lightly pigmented skin, particularly of the European type, UVR can penetrate the epidermis and dermis coupled with diminished capacity to repair DNA damage.81 Decreased ability to tan frequently leads to increased sun sensitivity in European skin, due to a decrease in density and activity of melanocytes.79 Relatively higher content of melanin in Asian and African skin typically correlates with better sun protection; however, all skin types demonstrate a considerable degree of constitutive pigmentation variation on the individual level.82,83
Within the Fitzpatrick skin type classification, which was originally developed to evaluate the susceptibility of the skin to erythema after exposure to UV radiation, degree of pigmentation and sun sensitivity would correspond to type I–IV for European skin, type II–III for East Asian skin, type III–V for South Asian skin and type V–VI for African skin.84,85 Whereas types I to IV on the Fitzpatrick scale describe lighter skin more accurately, types V and VI have relatively limited capacity to categorize darker pigmentation in skin types that do not burn easily, mostly representing an Asian and African heritage.86
Skin characterization based on genetic biomarkers combined with melanin index could offer a better representation of all skin types. The pigmentation genes bearing SNPs and responsible for skin tone and color are mostly present in the European skin type. Both constitutive and facultative pigmentation are affected by the SNPs in major genes including ASIP, DCT, IRF4, KITLG, MC1R, MITF and TYRP1, predominantly resulting in lighter skin reflectance, decreased tanning abilities and increased sun sensitivity. Lighter skin appearance in both Asian and African skin is controlled by the genes of constitutive pigmentation, including EDA and SLC24A5. Constitutive pigmentation is also responsible for high melanin index and darker skin appearance, with the fixed alleles of the genes involved in UVR response, DDB1 and MFSD12, present in both Asian and African skin.
Elasticity
Photodamage resulting from prolonged sun exposure is frequently linked to the significant loss of elasticity and deep wrinkles characterizing European skin.87 These changes arise from decreased synthesis and fragmentation of collagen, elastin and hyaluronic acid leading to the impaired organization of the epidermis.88,89 In comparison to European skin, Asian skin types are relatively resistant to wrinkles, with the decline in skin elasticity most likely due to an increase in cross-linked collagen and advanced glycation end products (AGE).90,91 One of the most prominent features of African skin is the increased elasticity and relative resistance to wrinkles due to abundant collagen and relatively thicker dermis compared to European skin.92 African skin is also resistant to degradation of collagen through photo-damage, contains a higher amount of tropoelastin, fibrillin and fibulin, and has enhanced profiles of growth factors responsible for cell migration and proliferation of cells in the papillary dermis.93,94 Genes involved in the control of skin elasticity and remodeling of ECM are present in European skin, including MMP1, as well as Asian skin, including ADAMTS20 and COL1A2. SNPs in MMP1 and COL1A2 could be responsible for alterations of collagen remodeling or susceptibility for modifications such as glycation. Changes in the activity of ADAMTS20, which also harbors SNP in African skin, could have an additional effect on the processing of proteoglycans in the dermis.
Skin Barrier
European skin is characterized by a relatively thin epidermis that undergoes further degeneration with time.95 The thinner epidermis, with a more fragile system of barrier corneocytes and lipids, is also characteristic of East Asian skin.90,95 Consequently, the skin is more sensitive to environmental pollutants causing inflammation and an increase in oxidative stress.96,97 This can further facilitate enhanced microbial invasion with subsequent colonization of sebaceous glands and acne.98
South Asian skin has relatively low levels of surface-free fatty acids and increased accumulation of sebum.99,100 This in turn can lead to increased sensitivity to allergens, activation of immune cells, overproduction of cytokines and skin inflammation.101 African skin is characterized by a relatively thicker epidermis and an abundance of the outermost layer of lipids.92,100 Because of this, skin hydration is likely enhanced and TEWL decreased in African skin.102,103 However, significantly increased TEWL has been also recorded as a likely thermoregulatory mechanism in both African and South Asian skin.104 Despite a higher amount of lipids and enhanced barrier, African skin appears to contain decreased levels of the cornified envelope proteins such as filaggrin in the epidermis.105 This could render the epidermis more susceptible to xerosis in cold temperatures and low humidity, which can deplete the natural lipid layer.106 The biomarkers with SNPs involved in the control of the structure and barrier function are predominant in Asian skin, including FLG, TCHH, ADAM17 and FCN1. SNPs in the genes involved in barrier permeability, hydration and immunity are also present in the European skin, including AQP3, and African skin, including BRCA1 and FCN1.
Hyper-Pigmentation
Hyper-pigmentation induced by UVR, such as solar lentigines, is frequently present in European skin and characterized by an increase in the density and activity of melanocytes.107 Hyper-pigmentation caused by excess synthesis or local re-distribution of melanin in the epidermis is one of the major problems evident in Asian skin. Uneven pigmentation in East Asian skin is accompanied by increased sensitivity to air pollution and a predisposition to acne.108,109 Hyper-pigmentation present in South Asian skin is frequently linked to sun damage, inflammatory conditions, acne and injuries.110,111
The genes harboring SNPs that could predispose to changes in pigmentation are characteristic of European skin, including MC1R, ASIP, BNC2, KITLG and HLA as well as Asian skin, including BNC2 and the genes of xenobiotic metabolism and oxidative stress AHR, CYP2C19 and HMG20.
Inflammation
Inflammation is often present in European skin as a consequence of excess UVR exposure, resulting in sunburns, solar lentigines or tumorigenesis.112 Inflammation in Asian skin types is frequently accompanied by hyperactive melanocytes and hyperpigmentation problems. Accumulation of pigmented cells and melanin in the dermis is also detected in hormone-induced melasma and post-inflammatory hyperpigmentation.111,113 Inflammation associated with acne breakout and post-injury in Asian skin is a frequent cause of thinning of the dermis and the formation of scars in the affected area.114 A cluster of the innate immune responses and inflammation genes affected by the SNPs involves ICAM1, TNF, IL10, ITGAE and TLR6, which have been detected in Asian skin.
Compartmentalization of Aging Patterns Across Skin Ethnicity
Skin aging is influenced by genetic factors that are responsible for individual variation in appearance, both intrinsic and extrinsic aging factors combine to drive a progressive deterioration of the skin over time. Extrinsic skin aging is driven by gene expression and signal transduction pathways upon exposure to environmental stress. Cumulative molecular damage caused by UVR is linked to altered expression of the factors involved in remodeling of the elastic fibers in the dermis and is responsible for faster-aging phenotypes predominantly in European skin. In contrast, Asian and African skin demonstrate greater elasticity accounting for fewer wrinkles with age, most likely because of the presence of thicker dermis containing more collagen and elastin. Aging is associated with an increase in oxidative stress caused by altered expression of ROS scavenging factors and antioxidant enzymes relevant in response to air pollution and in detoxification of the skin, which could be affecting more the European and Asian skin. Dermal fibroblasts originated from the skin of European and Asian heritage and analyzed in this study demonstrated a marked sensitivity to UVR, which was significantly higher compared to the fibroblasts from African skin. The UVR sensitivity was associated with the accumulation of the cells with enlarged nuclei, inhibition of cell growth, expression of γH2AX and p16INK4a, and intracellular production of ROS. Such responses, in particular the levels of ROS, were moreover more pronounced in Asian than in European fibroblasts. It is likely that the accumulation of the senescent cells with persistent DNA damage and ROS production caused by UVR has a more deteriorating effect on collagen and elastic fibers in European than Asian skin. European fibroblasts also responded to oxidative stress, induced by H2O2 exposure, by significantly higher production of ROS. Asian fibroblasts could have a better capacity to neutralize ROS caused by oxidative stress, given the relatively high total antioxidant capacity detected in this study. Pre-treatment of the cells with Resveratrol, the natural compound with photoprotective and antioxidant capacities, during UVR exposure had a beneficial effect on both European and Asian fibroblasts in terms of inhibition of cell senescence and ROS production. In contrast, the presence of Quercetin during UVR exposure increased cellular TAC to significantly higher levels compared to Resveratrol. Both ROS production and accumulation of senescent cells were inhibited most efficiently by Quercetin in Asian fibroblasts.
One aspect of skin aging caused by photo-damage is its association with constitutive pigmentation and tanning responses, which are thought to have direct protective effects against photo-damage caused by UVR. Darker pigmentation is frequently linked to fixed gene polymorphism across skin types of Asian and African origin, likely such genes as dominant traits could also contribute to guiding the photo-aging patterns in persons of mixed ethnic backgrounds. Due to naturally higher melanin content, which can be increased further upon sun exposure, aging of darker skin is frequently accompanied by hyperpigmentation caused by age spots, solar lentigines or melasma. Hyperpigmentation linked to inflammation is prevalent in Asian skin. Premature aging of East Asian skin could be also associated with hyperpigmentation coupled with increased sensitivity and inflammation triggered by environmental pollutants. Secretion of the inflammatory molecules has been moreover described as part of the skin aging process. The presence of H2O2 in the cellular environment is a long-established factor causing inflammation; Asian fibroblasts responded to both UVR and H2O2 treatment with a significant increase in the expression of p16INK4a. Accumulation of senescent cells in the dermis could be one factor responsible for the establishment of a pro-inflammatory environment triggering hyperpigmentation in Asian skin.
Skin aging is also accompanied by loss of hydration; whilst increased TEWL has been associated with an impaired barrier in European skin, decreased hydration and xerosis can be also one of the major problems of African skin, most likely linked to the factors regulating water-binding rather than the thickness of the epidermis. Changes in the barrier homeostasis are also frequently linked to inflammation and increased sensitivity to pro-inflammatory factors exasperating skin hydration further. Dermal fibroblasts from African skin analyzed in this study were relatively resistant to UVR and ROS production induced by H2O2. The cells did however respond to H2O2 treatment with a significant accumulation of persistent DNA lesions as determined by γH2AX expression. This could suggest a likely increased sensitivity to genotoxic stress linked to inflammation, which could also translate to sensitivity and cell renewal in the epidermis.
Given these patterns, the discoveries made through in-silico analysis combined with cellular assays could guide a better understanding of the genetic traits, sensitivities to environmental factors and aging differences across all skin types. Making such connections can be translated into the more rapid development of effective anti-aging therapeutic products.
Applications of AI Technologies to Genetic Traits for the Design of Personalized Cosmetic Products
The SNP-based DNA analysis offers a unique and novel approach to personalized skincare. The development of genetically guided, individual skincare products relies on innovation defining specific traits to achieve optimal long-term skin health. The approaches involving genetic tests could enable subsequent comparison with the information available in future databases. Assembly of a personal profile based on genetic signatures can highlight the complex bio-molecular networks and specific requirements to enhance individual characteristics and ameliorate undesirable problems.
The future of skin analysis for cosmetic and therapeutic interventions will ultimately be dependent on rapid methods for collecting data for larger cohort studies and validating in-silico models. Such screening methods likely require a more accurate analysis and prediction using Machine Learning (ML) and Artificial Intelligence (AI) platform technologies. In this study, we applied the data mining and Hexis Lab Pro.X® in-silico platform to analyze the interactions of two cosmetic ingredients, Resveratrol and Quercetin, with the molecular targets of the skin cell. This approach allowed us to visualize and interpret the likely axes of the biomarkers affected by the compounds. The interactive networks demonstrated the strong antioxidant and anti-inflammatory capacities of the compounds and allowed the identification of specific gene clusters enhanced through additional reciprocal connections. The biomarkers were mostly associated with European and Asian skin types and the strong capacity of Resveratrol and Quercetin to reduce oxidative stress could be validated in the in-vitro cellular assays.
The approach based on in-silico analyses can facilitate the search for new personalized skincare cosmetic ingredients. The AI algorithms can be applied to the biomarker characteristics for better resolution of their dynamic interactions with particular emphasis on skin ethnicity. Such application could also capture specific changes that can occur as a result of environmental damage and might represent an adaptive response to stress that constitutes one of the targets for rejuvenation potential.
Conclusion
In this study, we investigated the molecular bases and cellular characteristics behind the likely differences between ethnic skin types, with a view to contributing to the emerging theme of personalized skincare and the ways to advance its applications. Using the in-silico approach, we constructed networks of the genes that are described in the literature as affected by single nucleotide polymorphism (SNP) and uncovered clusters across both skin types and functional categories. Since the genes are involved in defined activities in the skin, playing roles in oxidative stress and inflammation, UVR responses, DNA damage and repair, pigmentation, ECM and barrier, it indicates that the cells present in different skin types would have diverse characteristics and responses triggered by some of these factors. Using skin dermal fibroblasts from the skin of European, Asian and African origin, we uncovered a number of signatures manifested in response to UVR and oxidative stress. Two active ingredients used in cosmetic products, Resveratrol and Quercetin, affect in a specific manner some of the genes identified in our networks; both ingredients also have a slightly different effect on three types of fibroblasts included in this study. The findings are discussed in the broader context of known characteristics and aging patterns of human skin, with the proposal that uncovering genetic traits and specific sensitivities to environmental stress factors could contribute to applications in cosmetic science.
Abbreviations
ACSS2, acetyl-coenzyme A synthetase 2; ADAM17, disintegrin and metalloproteinase domain-containing protein 17; ADAMTS20, ADAM metallopeptidase with thrombospondin type 1 motif 20; AGE, advanced glycation end products; AHR, aryl hydrocarbon receptor; AI, artificial intelligence; AQP3, Aquaporin-3; AR, androgen receptor; ASIP, agouti-signaling protein; ATRN, Attractin; BNC2, basonculin-2; BRCA1, BRCA1 DNA repair associated; COL1A2, collagen type 1 alpha 2 chain; CYP2C19, cytochrome P450 family 2 subfamily C member 19; DAPI, 4’,6-diamidino-2-phenylindole; DCF-DA, 2’7’-dichlorofluorescein diacetate; DCT, dopachrome tautomerase; DDB1, damage specific DNA binding protein 1; DMEM, Dulbecco’s modified Eagle’s medium; DMSO, dimethylsulfoxide; ECM, extracellular matrix; EDA, ectodysplasin A; EDTA, ethylenediamine tetraacetic acid; FCN1, ficolin-1; FITC, fluorescein isothiocyanate; FLG, Filaggrin; GATA3, GATA binding protein 3; GWAS, genome-wide association studies; H2O2, hydrogen peroxide; γH2AX, H2A histone family member X, phospho-isoform; HDF, human dermal fibroblasts; HERC2, HECT and RLD domain containing E3 ubiquitin protein ligase 2; HLA, human leukocyte antigen; HMG20, high mobility group 20B; ICAM1, intercellular adhesion molecule 1; IL10, interleukin 10; IRF4, interferon regulatory factor 4; ITGAE, integrin alpha E; Ki67, marker of proliferation Ki67; KITLG, ligand for the receptor-type protein-tyrosine kinase KIT; MC1R, melanocortin 1 receptor; MFSD12, major facilitator superfamily domain-containing 12; MITF, microphthalmia-associated transcription factor; ML, Machine Learning; MMP1, matrix metalloproteinase 1; OCA2, oculocutaneous albinism II; p16INK4a, cyclin-dependent kinase (CDK) inhibitor p16; PBS, phosphate buffered saline; ROS, reactive oxygen species; SLC24A5, solute carrier family 24 member 5; SNP, single nucleotide polymorphism; SOD2, superoxide dismutase 2; TAC, total antioxidant capacity; TCHH, Trichohyalin; TEWL, trans-epidermal water loss; TLR6, Toll-like receptor 6; TNF, tumor necrosis factor; TYRP1, tyrosinase-related protein 1; UVR, ultraviolet radiation.
Acknowledgments
The authors would like to thank the Asian Skin Biobank (ASB) at the Skin Research Institute of Singapore (SRIS), for generating and providing human primary dermal fibroblasts. The ASB is funded by Singapore’s Agency for Science, Technology & Research (A*STAR) through core fund and under the IAF-PP Project (H1701a0004).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Naval J, Alonso V, Herranz MA. Genetic polymorphisms and skin aging: the identification of population genotypic groups holds potential for personalized treatments. Clin Cosmet Investig Dermatol. 2014;7:207–214. doi:10.2147/CCID.S55669
2. Brenner M, Hearing VJ. The protective role of melanin against UV damage in human skin. Photochem Photobiol. 2008;84(3):539–549. doi:10.1111/j.1751-1097.2007.00226.x
3. Solano F. Melanins: skin pigments and much more- types, structural models, biological functions and formation routes. New J Sci. 2014;2014:1–28. doi:10.1155/2014/498276
4. McCullough JL, Kelly KM. Prevention and treatment of skin aging. Ann N Y Acad Sci. 2006;1067:323–331. doi:10.1196/annals.1354.044
5. Zouboulis CC, Makrantonaki E. Clinical aspects and molecular diagnostics of skin aging. Clin Dermatol. 2011;29:3–14. doi:10.1016/j.clindermatol.2010.07.001
6. Jacobs LC, Wollstein A, Lao O, et al. Comprehensive candidate gene study highlights UGT1A and BNC2 as new genes determining continuous skin color variation in Europeans. Hum Genet. 2013;132:147–158. doi:10.1007/s00439-012-1232-9
7. Han J, Kraft P, Nan H, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4(5):e1000074. doi:10.1371/journal.pgen.1000074
8. Zhang M, Song F, Liang L, et al. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum Mol Genet. 2013;22(14):2948–2959. doi:10.1093/hmg/ddt142
9. Han J, Kraft P, Colditz GA, Wong J, Hunter DJ. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer. 2006;119(8):1976–1984. doi:10.1002/ijc.22074
10. Candille SI, Absher DM, Beleza S, et al. Genome-wide association studies of quantitatively measured skin, hair and eye pigmentation in four European populations. PLoS One. 2012;7(10):e48294. doi:10.1371/journal.pone.0048294
11. Saternus R, Pilz S, Graber S, et al. A closer look at evolution: variants (SNPs) of genes involved in skin pigmentation, including EXOC2, TYR, TYRP1 and DCT are associated with 25(OH)D serum concentration. Endocrinology. 2015;156(1):39–47. doi:10.1210/en.2014-1238
12. Malcov-Brog H, Alpert A, Golan T, et al. UV-protection timer controls linkage between stress and pigmentation skin protection systems. Mol Cell. 2018;72:444–456. doi:10.1016/j.molcel.2018.09.022
13. Bajana S, Roach K, Turner S, Paul J, Kovats S. Interferon regulatory factor 4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J Immunol. 2012;189(7):3368–3377. doi:10.4049/jimmunol.1102613
14. Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nature Cell Biol. 2010;12:80–86. doi:10.1038/ncb0410-412
15. Izagirre N, Garcia I, Junquera C, de la Rua C, Alonso S. A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol Biol Evol. 2006;23(9):1697–1706. doi:10.1093/molbev/msl030
16. Lago JC, Puzzi MB. The effect of aging in primary human dermal fibroblasts. PLoS One. 2019;14(7):e0219165. doi:10.1371/journal.pone.0219165
17. Valenzuela RK, Henderson BS, Walsh MH, et al. Predicting phenotype from genotype: normal pigmentation. J Forensic Sci. 2010;55(2):315–322. doi:10.1111/j.1556-4029.2009.01317.x
18. Alonso S, Izagirre N, Smith-Zubiaga I, et al. Complex signatures of selection for the melanogenic loci TYR, TYRP and DCT in humans. BMC Evol Biol. 2008;8:74. doi:10.1186/1471-2148-8-74
19. Jacobs LC, Hamer MA, Gunn DA, et al. A genome-wide association study identifies the skin color genes IRF4, MC1R, ASIP and BNC2 influencing facial pigmented spots. J Invest Dermatol. 2015;135(7):1735–1742. doi:10.1038/jid.2015.62
20. Martin AR, Lin M, Granka JM, et al. An unexpectedly complex architecture for skin pigmentation in Africans. Cell. 2017;171(6):1340–1353. doi:10.1016/j.cell.2017.11.015
21. Beleza S, Johnson NA, Candille SI, et al. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 2013;9(3):e1003372. doi:10.1371/journal.pgen.1003372
22. Sulem P, Gudbjartsson DF, Stacey SN, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40(7):835–837. doi:10.1038/ng.160
23. Moskvina V, Smith M, Ivanov D, et al. Genetic differences between five European populations. Hum Hered. 2010;70(2):141–149. doi:10.1159/000313854
24. Amyere M, Vogt T, Hoo J, et al. KITLG mutations cause familial progressive hyper- and hypopigmentation. J Investig Dermatol. 2011;131(6):1234–1239. doi:10.1038/jid.2011.29
25. Brown KM, MacGregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40(7):838–840. doi:10.1038/ng.163
26. Rana BK, Hewett-Emmett D, Jin L, et al. High polymorphism at the human melanocortin 1 receptor locus. Genetics. 1999;151(4):1547–1557. doi:10.1093/genetics/151.4.1547
27. Crawford NG, Kelly DE, Hansen MEB, et al. Loci associated with skin pigmentation identified in African populations. Science. 2017;358(6365):eaan8433. doi:10.1126/science.aan8433
28. Edwards M, Bigham A, Tan J, et al. Association of the OCA2 polymorphism His615Arg with melanin content in east Asian populations: further evidence of convergent evolution of skin pigmentation. PLoS Genet. 2010;6(3):e1000867. doi:10.1371/journal.pgen.1000867
29. Mishra A, Nizammuddin S, Mallick CB, et al. Genotype-phenotype study of the middle gangetic plain in India shows association of rs2470102 with skin pigmentation. J Invest Dermatol. 2017;137(3):670–677. doi:10.1016/j.jid.2016.10.043
30. Gao W, Tan J, Huls A, et al. Genetic variants associated with skin aging in the Chinese Han Population. J Dermatol Sci. 2017;86(1):21–29. doi:10.1016/j.jdermsci.2016.12.017
31. Sengupta S, Farheen S, Mukherjee N. DNA sequence variation and haplotype structure of the ICAM1 and TNF genes in 12 ethnic groups of India reveal patterns of importance in designing association studies. Ann Hum Genet. 2004;68:574–587. doi:10.1046/j.1529-8817.2003.00126.x
32. Yelmen B, Mondal M, Marnetto D, et al. Ancestry-specific analyses reveal differential demographic histories and opposite selective pressures in modern South Asian populations. Mol Biol Evol. 2019;36(8):1628–1642. doi:10.1093/molbev/msz037
33. Chambers JC, Abbott J, Zhang W, et al. The South Asian genome. PLoS One. 2014;9(8):e102645. doi:10.1371/journal.pone.0102645
34. Jose R, Chandrasekaran A, Sam SS, et al. CYP2C9 and CYP2C19 genetic polymorphisms: frequencies in the south Indian population. Fund Clin Pharmacol. 2005;19(1):101–105. doi:10.1111/j.1472-8206.2004.00307.x
35. Kooner JS, Saleheen D, Sim X, et al. Genome-wide association study in people of South Asian ancestry identifies six novel susceptibility loci for type 2 diabetes. Nat Genet. 2011;43(10):984–989. doi:10.1038/ng.921
36. Zhang M, Li B, Wu S, et al. A genome-wide association study of basal transepidermal water loss finds that variants at 9q34.3 are associated with skin barrier function. J Invest Dermatol. 2017;137(4):979–982. doi:10.1016/j.jid.2016.11.030
37. Bonilla C, Boxill LA, McDonald SA, et al. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum Genet. 2005;116(5):402–406. doi:10.1007/s00439-004-1251-2
38. Miller CT, Beleza S, Pollen AA. Cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131(6):1179–1189. doi:10.1016/j.cell.2007.10.055
39. Ollmann MM, Lamoreux ML, Wilson BD, Barsh GS. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998;12:316–330. doi:10.1101/gad.12.3.316
40. Visser M, Palstra RJ, Kayser M. Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum Mol Genet. 2014;23:5750–5762. doi:10.1093/hmg/ddu289
41. Wolf Horell EM, Boulanger MC, D’Orazio JA. Melanocortin 1 receptor: structure, function and regulation. Front Genet. 2016;7:95. doi:10.3389/fgene.2016.00095
42. Johanson HC, Chen W, Wicking C, Sturm RA. Inheritance of a novel mutated allele of the OCA2 gene associated with high incidence of oculocutaneous albinism in a Polynesian community. J Hum Genet. 2010;55:103–111. doi:10.1038/jhg.2009.130
43. Ginger R, Askew SE, Ogborne RM, Green MR. SLC24A5 encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem. 2008;283(9):5486–5495. doi:10.1074/jbc.M707521200
44. Sarangarajan R, Boissy RE. Tyrp1 and oculocutaneous albinism type 3. Pigment Cell Res. 2001;14(6):437–444. doi:10.1034/j.1600-0749.2001.140603.x
45. Gunn TM, Miller KA, He L, et al. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature. 1999;398:152–156. doi:10.1038/18217
46. Aroca P, Solano F, Salina C, Garcia-Borron JC, Lozano JA. Regulation of the final phase of mammalian melanogenesis. The role of dopachrome tautomerase and the ratio between 5,6-dihydroxyindole-2-carboxylic acid and 5,6-dihydroxyindole. Eur J Biochem. 1992;208(1):155–163. doi:10.1111/j.1432-1033.1992.tb17169.x
47. Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155(5):1022–1033. doi:10.1016/j.cell.2013.10.022
48. Claus-Werner F, Cobzaru C, Triantafyllopoulou A, et al. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand-dependent terminal keratinocyte differentiation. J Exp Med. 2012;209(6):1105–1119. doi:10.1084/jem.20112258
49. Zeitvogel J, Jokmin N, Rieker S, Klug I, Brandenberger C, Werfel T. GATA3 regulates FLG and FLG2 expression in human primary keratinocytes. Sci Rep. 2017;7:11847. doi:10.1038/s41598-017-10252-x
50. Caughman SW, Li LJ, Degitz K. Human intercellular adhesion molecule-1 gene and its expression in the skin. J Invest Dermatol. 1992;98(6):S61–S65. doi:10.1111/1523-1747.ep12462226
51. Kang K, Gilliam AC, Chen G, Tootell E, Cooper KD. In human skin, UVB initiates early induction of IL-10 over IL-12 preferentially in the expanding dermal monocytic/macrophagic population. J Invest Dermatol. 1998;111(1):31–38. doi:10.1046/j.1523-1747.1998.00121.x
52. Pauls K, Schon M, Kubitza RC, Homey B, Wiesenborn A, Lehmann P. Role of integrin alphaE(CD103)beta7 for tissue-specific epidermal localization of CD8+ T lymphocytes. J Invest Dermatol. 2001;117(3):569–575. doi:10.1046/j.0022-202x.2001.01481.x
53. Skabytska Y, Wolbing F, Gunther C, et al. Cutaneous innate immune sensing of Toll-like receptor 2-6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity. 2014;41:762–775. doi:10.1016/j.immuni.2014.10.009
54. Bashir MM, Sharma MR, Werth VP. TNF-α production in the skin. Arch Dermatol Res. 2009;301:87–91. doi:10.1007/s00403-008-0893-7
55. Bollag WB, Aitkens L, White J, Hyndman KA. Aquaporin-3 in the epidermis: more than skin deep. AJP-Cell. 2020;318(6):1144–1153. doi:10.1152/ajpcell.00075.2020
56. Li S, Zhou J, Zhang L, Li J, Yu J, Ning K. Ectodysplasin A regulates epithelial barrier function through sonic hedgehog signalling pathway. J Cell Mol Med. 2018;22(1):230–240. doi:10.1111/jcmm.13311
57. Kezic S, Jakasa I. Filaggrin and skin barrier function. Curr Probl Dermatol. 2016;49:1–7. doi:10.1159/000441539
58. Medland SE, Nyholt DR, Painter JN, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85:750–755. doi:10.1016/j.ajhg.2009.10.009
59. Chen R, Xu M, Nagati J, Garcia JA. Coordinate regulation of stress signaling and epigenetic events by Acss2 and HIF-2 in cancer cells. J Exp Med. 2017;12(12):e0190241. doi:10.1371/journal.pone.0190241
60. Velarde MC, Flynn JM, Day NU, Melov S, Campisi J. Mitochondrial oxidative stress caused by Sod2 deficiency promotes cellular senescence and aging phenotypes in the skin. Aging. 2012;4(1):3–12. doi:10.18632/aging.100423
61. Esser C, Bargen I, Weighardt HST, Krutmann J, Krutmann J. Functions of the aryl hydrocarbon receptor in the skin. Semin Immunopathol. 2013;35(6):677–691. doi:10.1007/s00281-013-0394-4
62. Lai JJ, Chang P, Lai KP, Chen L, Chang C. The role of androgen and androgen receptor in the skin-related disorders. Arch Dermatol Res. 2012;304(7):499–510. doi:10.1007/s00403-012-1265-x
63. Veith A, Moorthy B. Role of cytochrome P450S in the generation and metabolism of reactive oxygen species. Curr Opin Toxicol. 2018;7:44–51. doi:10.1016/j.cotox.2017.10.003
64. Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. BBA Mol Basis Dis. 2011;1812(12):1616–1629. doi:10.1016/j.bbadis.2011.08.009
65. Pittayapruek P, Meephansan J, Prapapan O, Komine M, Ohtsuki M. Role of matrix metalloproteinases in photoaging and photocarcinogenesis. Int J Mol Sci. 2016;17(6):868. doi:10.3390/ijms17060868
66. Sotiropoulou PA, Karambelas AE, Debaugnies M, et al. BRCA1 deficiency in skin epidermis leads to selective loss of hair follicle stem cells and their progeny. Genes Dev. 2013;27(1):39–51. doi:10.1101/gad.206573.112
67. Cang Y, Zhang J, Nicholas SA, Kim AL, Zhou P, Goff SP. DDB1 is essential for genomic stability in developing epidermis. PNAS. 2007;104(8):2733–2737. doi:10.1073/pnas.0611311104
68. Kilpatrick DC, Chalmers JD. Human L-Ficolin (Ficolin-2) and its clinical significance. J Biomed Biotechnol. 2012;2012:138797. doi:10.1155/2012/138797
69. Richmond JM, Harris JE. Immunology and skin in health and disease. Cold Spring Harb Perspect Med. 2014;4(12):a015339. doi:10.1101/cshperspect.a015339
70. Lee M, Daniels MJ, Garnett MJ, Venkitaraman AR. A mitotic function for the high-mobility group protein HMG20b regulated by its interaction with the BRC repeats of the BRCA2 tumor suppressor. Oncogene. 2011;30:3360–3369. doi:10.1038/onc.2011.55
71. Wei CY, Zhu MX, Lu NH, et al. Bioinformatics-based analysis reveals elevated MFSD12 as a key promoter of cell proliferation and a potential therapeutic target in melanoma. Oncogene. 2019;38:1876–1891. doi:10.1038/s41388-018-0531-6
72. Gentile M, Latonen L, Laiho M. Cell cycle arrest and apoptosis provoked by UV radiation-induced DNA damage are transcriptionally highly divergent responses. Nucleic Acids Res. 2003;31(16):4779–4790. doi:10.1093/nar/gkg675
73. Zhao H, Darzynkiewicz Z. Biomarkers of cell senescence assessed by imaging cytometry. Methods Mol Biol. 2013;965:83–92. doi:10.1007/978-1-62703-239-1_5
74. Bernadotte A, Mikhelson VM, Spivak IM. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging. 2016;8(1):3–11. doi:10.18632/aging.100871
75. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130(8):1715–1725. doi:10.1002/ijc.27316
76. Reagan-Shaw S, Mukhtar H, Ahmad N. Resveratrol imparts photoprotection of normal cells and enhances the efficacy of radiation therapy in cancer cells. Photochem Photobiol. 2008;84(2):415–421. doi:10.1111/j.1751-1097.2007.00279.x
77. Lesjak M, Beara I, Simin N, et al. Antioxidant and anti-inflammatory activities of quercetin and its derivatives. J Funct Foods. 2018;40:68–75. doi:10.1016/j.jff.2017.10.047
78. Ito S, Wakamatsu K, Sarna T. Photodegradation of eumelanin and pheomelanin and its pathophysiological implications. Photochem Photobiol. 2018;94:409–420. doi:10.1111/php.12837
79. Tadokoro T, Yamaguchi Y, Batzer J, et al. Mechanisms of skin tanning in different racial/ethnic groups in response to ultraviolet radiation. J Invest Dermatol. 2005;124(6):1326–1332. doi:10.1111/j.0022-202X.2005.23760.x
80. Nielsen KP, Zhao L, Stamnes JJ, Stamnes K, Moan J. The importance of the depth distribution of melanin in skin for DNA protection and other photobiological processes. J Photochem Photobiol B Biol. 2006;82(3):194–198. doi:10.1016/j.jphotobiol.2005.11.008
81. Rees JL. The genetics of sun sensitivity in humans. AJHG. 2004;75(5):739–751. doi:10.1086/425285
82. Sturm RA, Duffy DL. Human pigmentation genes under environmental selection. Genome Biol. 2012;13:248. doi:10.1186/gb-2012-13-9-248
83. Del Bino S, Duval C, Bernerd F. Clinical and biological characterization of skin pigmentation diversity and its consequences on UV impact. Int J Mol Sci. 2018;19:2668. doi:10.3390/ijms19092668
84. Fitzpatrick TB. The validity and practicality of sun-reactive skin types I through VI. Arch Dermatol. 1988;124:869–871. doi:10.1001/archderm.124.6.869
85. Sachdeva S. Fitzpatrick skin typing: applications in dermatology. Indian J Dermatol Venereol Leprol. 2009;75:93–96. doi:10.4103/0378-6323.45238
86. Ware OR, Dawson JE, Shinohara MM, Taylor SC. Racial limitations of Fitzpatrick skin type. Cutis. 2020;105(2):77–80.
87. Naylor EC, Watson RE, Sherratt MJ. Molecular aspects of skin ageing. Maturitas. 2011;69(3):249–256. doi:10.1016/j.maturitas.2011.04.011
88. Fisher GJ, Quan T, Purohit T, et al. Collagen fragmentation promotes oxidative stress and elevates matrix metalloproteinase-1 in fibroblasts in aged human skin. Am J Pathol. 2009;174:101–114. doi:10.2353/ajpath.2009.080599
89. Flament F, Bazin R, Laquieze S, Rubert V, Simonpietri E, Piot B. Effect of the sun on visible clinical signs of aging in Caucasian skin. Clin Cosmet Investig Dermatol. 2013;6:221–232. doi:10.2147/CCID.S44686
90. Galzote C, Estanislao R, Suero MO, et al. Characterization of facial skin of various Asian populations through visual and noninvasive instrumental evaluations: influence of age and skincare habits. Skin Res Technol. 2013;19(4):454–465. doi:10.1111/srt.12069
91. Vashi NA, Maymone MBC, Kundu RV. Aging differences in ethnic skin. J Clin Aesthet Dermatol. 2016;9(1):31–38.
92. Brissett AE, Naylor MC. The aging African-American face. Facial Plast Surg. 2010;26(2):154–163. doi:10.1055/s-0030-1253501
93. Girardeau-Hubert S, Pageon H, Asselineau D. In vivo and in vitro approaches in understanding the differences between Caucasian and African skin types: specific involvement of the papillary dermis. Int J Dermatol. 2012;51(Suppl.1):1–4. doi:10.1111/j.1365-4632.2012.05553.x
94. Langton AK, Alessi S, Hann M, et al. Aging in skin of color: disruption to elastic fiber organization is detrimental to skin’s biomechanical function. J Invest Dermatol. 2019;139(4):779–788. doi:10.1016/j.jid.2018.10.026
95. Rawlings AV. Ethnic skin types: are there differences in skin structure and function? Int J Cosmet Sci. 2006;28(2):79–93. doi:10.1111/j.1467-2494.2006.00302.x
96. Rinnerthaler M, Bischof J, Streubel MK, Trost A, Richter K. Oxidative stress in aging human skin. Biomolecules. 2015;5(2):545–589. doi:10.3390/biom5020545
97. Araviiskaia E, Berardesca E, Bieber T, et al. The impact of airborne pollution on skin. J Eur Acad Dermatol Venereol. 2019;33(8):1496–1505. doi:10.1111/jdv.15583
98. Goh CL, Noppakun N, Micali G. Meeting the challenges of acne treatment in Asian patients: a review of the role of dermocosmetics as adjunctive therapy. J Cutan Aesthet Surg. 2016;9(2):85–92. doi:10.4103/0974-2077.184043
99. Mukherjee S, Mitra R, Maitra A, et al. Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci Rep. 2016;6:36062. doi:10.1038/srep36062
100. Shetage SS, Traynor MJ, Brown MB, Chilcott RP. Sebomic identification of sex- and ethnicity-specific variations in residual skin surface components (RSSC) for bio-monitoring or forensic applications. Lipids Health Dis. 2018;17:194. doi:10.1186/s12944-018-0844-z
101. Makrantonaki E, Ganceviciene R, Zouboulis C. An update on the role of the sebaceous gland in the pathogenesis of acne. Dermatoendocrinol. 2011;3(1):41–49. doi:10.4161/derm.3.1.13900
102. Diridollou S, de Rigal J, Querleux B, Leroy F, Holloway Barbosa V. Comparative study of the hydration of the stratum corneum between four ethnic groups: influence of age. Int J Dermatol. 2007;46(Suppl 1):11–14. doi:10.1111/j.1365-4632.2007.03455.x
103. Pappas A, Fantasia J, Chen T. Age and ethnic variations in sebaceous lipids. Dermato-Endocrinology. 2013;5(2):319–324. doi:10.4161/derm.25366
104. Voegeli R, Gierschendorf J, Summers B, Rawlings AV. Facial skin mapping: from single point bio-instrumental evaluation to continuous visualization of skin hydration, barrier function, skin surface pH, and sebum in different ethnic skin types. Int J Cosmet Sci. 2019;41:411–424. doi:10.1111/ics.12562
105. Girardeau-Hubert S, Deneuville C, Pageon H, et al. Reconstructed skin models revealed unexpected differences in epidermal African and Caucasian skin. Sci Rep. 2019;9:7456. doi:10.1038/s41598-019-43128-3
106. Wan DC, Wong VW, Longaker MT, Yang GP, Wei FC. Moisturizing different racial skin types. J Clin Aesthet Dermatol. 2014;7(6):25–32.
107. Choi W, Yin L, Smuda C, Batzer J, Hearing VJ, Kolbe L. Molecular and histological characterization of age spots. Exp Dermatol. 2017;26(3):242–248. doi:10.1111/exd.13203
108. Nouveau-Richard S, Yang Z, Mac-Mary S, et al. Skin aging: a comparison between Chinese and European populations. A pilot study. J Dermatol Sci. 2005;40(3):187–193. doi:10.1016/j.jdermsci.2005.06.006
109. Krutmann J, Moyal D, Liu W, et al. Pollution and acne: is there a link? Clin Cosmet Investig Dermatol. 2017;10:199–204. doi:10.2147/CCID.S131323
110. Vashi N, Kundu R. Facial hyperpigmentation: causes and treatment. Br J Dermatol. 2013;169:41–56. doi:10.1111/bjd.12536
111. Nouveau S, Agrawal D, Kohli M, Bernerd F, Misra N, Nayak CS. Skin hyperpigmentation in Indian population: insights and best practice. Indian J Dermatol. 2016;61(5):487–495. doi:10.4103/0019-5154.190103
112. Neagu M, Constantin C, Caruntu C, Dumitru C, Surcel M, Zurac S. Inflammation: a key process in skin tumorigenesis. Oncol Lett. 2019;17:4068–4084. doi:10.3892/ol.2018.9735
113. Davis EC, Callender VD. Postinflammatory hyperpigmentation. A review of the epidemiology, clinical features and treatment options in skin of color. J Clin Aesthet Dermatol. 2010;3(7):20–31.
114. Handog EB, Datuin MSL, Singzon IA. Chemical peels for acne and acne scars in Asians: evidence based review. J Cutan Aesthet Surg. 2012;5(4):239–246. doi:10.4103/0974-2077.104911
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.