Back to Journals » Infection and Drug Resistance » Volume 13
Emergence of Methicillin-Resistant Staphylococcus aureus Belonging to Clonal Complex 15 (CC15-MRSA) in Kuwait Hospitals
Authors Udo EE , Boswihi SS, Mathew B, Noronha B, Verghese T, Al-Jemaz A , Al Saqer F
Received 5 November 2019
Accepted for publication 11 January 2020
Published 21 February 2020 Volume 2020:13 Pages 617—626
DOI https://doi.org/10.2147/IDR.S237319
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Edet E Udo, Samar S Boswihi, Bindu Mathew, Bobby Noronha, Tina Verghese, Aisha Al-Jemaz, Fatma Al Saqer
Department of Microbiology, Faculty of Medicine, Kuwait University, Safat, Kuwait
Correspondence: Edet E Udo
Department of Microbiology, Faculty of Medicine, Kuwait University, P. O. Box 24923, Safat 13110, Kuwait
Tel +965 498 36773
Fax +965 2533 2719
Email [email protected]
Purpose: Methicillin-resistant S. aureus (MRSA) belonging to clonal complex 15 (CC15-MRSA) is rare among clinical isolates with few reports from retail camel meat and human patients. This study investigated the genetic relatedness of CC15-MRSA isolated for the first time from patients in Kuwait hospitals.
Methods: Antibiotic susceptibility was tested by the disk diffusion method. Minimum inhibitory concentration was determined using Etest strips. Molecular typing was performed using spa tying, multilocus sequence tying and DNA microarray.
Results: Of 1327 MRSA isolates, 42 (3.1%) were identified as CC15-MRSA. The 42 isolates belonged to sequence type ST1535-harbored SCCmec type V and spa types t084 (36 isolates), t346 (3 isolates) and one of t114, t228 and t7583. All 42 isolates were resistant to gentamicin, kanamycin, fusidic acid and cadmium acetate; 38 isolates were resistant to tetracycline. The isolates harbored aacA-aphD and fusC that codes for gentamicin and fusidic acid resistance, respectively. Tet(K) was present in the tetracycline-resistant isolates. In addition, the 42 isolates carried inu(A) (lincosamide nucleotidyltransferase) that confers resistance to lincomycin and clindamycin although phenotypically susceptible to these antibiotics. The isolates belonged to accessory gene regulator type II and capsular polysaccharide group 8 but lacked genes for Staphylococcus enterotoxins, toxic shock syndrome toxin, collagen-binding adhesins and Panton–Valentine leukocidin.
Conclusion: This study revealed the emergence and transmission of a previously rare MRSA clone among human patients in Kuwait hospitals and highlights the increasing infiltration of rare MRSA into the human population.
Keywords: DNA microarray, MRSA, antibiotic resistance, MLST
Introduction
Antibiotic-resistant pathogens continue to threaten the effective delivery of healthcare globally due to the associated limited choices of antibiotics for therapy of serious infections caused by these organisms, prolonged illness, prolonged hospitalization leading to increased cost of treatment and mortality.1
Methicillin-resistant Staphylococcus aureus (MRSA) remains an important antibiotic-resistant pathogen known to cause a wide range of infections including superficial skin and soft tissue infections and serious invasive infections such as pneumonia, osteomyelitis, and septicemia.2 Since the initial report in the United Kingdom in the early 1960,3 MRSA strains have been reported from healthcare facilities and in communities worldwide.2 Furthermore, the epidemiology of MRSA has changed substantially since the initial report in the UK. MRSA changed from being primarily associated with infections among elderly patients in healthcare facilities (healthcare-acquired MRSA) to colonizing or infecting young and apparently healthy individuals with no previous history of hospitalization in the community (Community-associated MRSA).1,4,5 The emergence of community-associated MRSA (CA-MRSA) was soon followed by reports of MRSA that were associated with livestock or humans associated with livestock which have been described as livestock-associated MRSA (LA-MRSA).6–11
Since their description in the early 1990s, different CA-MRSA clones have emerged in different geographical regions with different capacities for global transmission. Whereas strains belonging to ST30-MRSA-IV and ST80-MRSA-IV have been widely reported in different countries, others such as ST59-MRSA and ST93-MRSA-IV have restricted geographical distribution.1,2 In addition, novel MRSA clones as well as variants of the established CA-MRSA clones continue to emerge.12,13,15,18 The constant changes in the distribution of MRSA clones warrants regular surveillance of MRSA in local healthcare facilities to detect the newly emerging clones so as to institute effective control measures and prevent their transmission.14,15
MRSA strains belonging to clonal complex 15 (CC15-MRSA) are rare in the literature with only small numbers reported in retail camel meat in Saudi Arabia12 and in a small number of human patients in Saudi Arabia,13,16 Iran17,18 and Italy.19 Whole genome sequencing of four CC15-MRSA isolates cultured from human patients (N=2), and retail meat (N=2) in Saudi Arabia revealed that all four isolates belonged to the same sequence type, ST1535 (13-13-1-1-81-11-13), which is a single locus variant of ST15 and harbored a novel SCCmec V/SCCfus composite genetic element.20
Significant changes in the distribution of MRSA clones have been reported in Kuwait public hospitals in recent years. The number of MRSA clones in Kuwait hospitals increased from one dominant clone in 1992 to 30 diverse clones in 2010.21 In addition, novel MRSA clones and variants of well-known clones emerged in the country.14 This paper reports on the isolation and molecular characteristics of a newly emerging CC15-MRSA clone cultured from clinical samples of patients in Kuwait hospitals.
Materials and Methods
Bacterial Strains
The CC15-MRSA isolates used in this study were collected as part of routine microbiology diagnostic services in Kuwait public hospitals. It is mandatory to submit all MRSA isolates obtained from clinical samples (infection and colonization sites) in public hospitals in Kuwait to the MRSA Reference Laboratory for molecular typing following primary isolation and identification in the diagnostic microbiology laboratories using standard bacteriological methods including growth on mannitol salt agar, gram stain, tube coagulase test and DNase testing. The isolates used in this study were part of the MRSA isolates obtained from 1 January to 31 December, 2016 in all 14 public hospitals. The CC15-MRSA were isolated only in eight diagnostic microbiology laboratories. The isolates were preserved in 40% glycerol (v/v) in brain heart infusion broth at −80 ᴼC. Isolates were subcultured twice on brain-heart infusion agar before processing.
Antibiotic Susceptibility Testing
Antibiotic susceptibility profiles of the isolates were tested by disk diffusion method according to the Clinical Laboratory Standards Institute (CLSI)22 against the following antimicrobial disks (Oxoid, Basingstoke, UK): benzyl penicillin (2U), cefoxitin (30 µg), kanamycin (30 µg), mupirocin (200 µg and 5 µg), gentamicin (10 µg), erythromycin (15 µg), clindamycin (2 µg), chloramphenicol (30 µg), tetracycline (10 µg), trimethoprim (2.5 µg), fusidic acid (10 µg), rifampicin (5 µg), ciprofloxacin (5 µg), teicoplanin (30 µg), and linezolid (30 µg). Susceptibility to the antibiotics was interpreted according to the CLSI. Minimum inhibitory concentration (MIC) of cefoxitin, vancomycin, teicoplanin, mupirocin, erythromycin and clindamycin were determined with Etest strips (bioMerieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. S. aureus strains ATCC25923 and ATCC29213 were used as quality control strains for disk diffusion and MIC determination, respectively. Susceptibility to fusidic acid was interpreted according to guidelines of the British Society to Antimicrobial Chemotherapy.23 Susceptibility to cadmium acetate (50 µg) mercuric chloride (109 µg) and ethidium bromide (50 µg) was performed as described previously.24
Molecular Typing of Isolates
DNA microarray analysis was performed to determine antibiotic resistance genotypes and virulence-related genes, and to assign the isolates to clonal complex (CC) using the Identibac S. aureus genotyping Kit 2.0 (Alere Technology, Jena, Germany) and protocols provided by the manufacturer and described previously.2
DNA for PCR amplification was isolated and purified as described previously.14 Spa typing was performed using protocol and primers published previously.25 Multilocus sequence typing (MLST) was performed using the protocol described by Enright et al.26
Pulsed-field gel electrophoresis (PFGE) was performed as described previously.27 Contour-clamped homogenous electric field (CHEF) electrophoresis of SmaI-digested genomic DNA was performed in CHEF-DRIII system (BioRad, Hercules, CA, USA). The run was for 22 h with 5-s initial pulse and 40-s final pulse. Sma-I digested S. aureus strain NCTC 8325 was used as molecular size marker. Following electrophoresis, the gel was stained in ethidium bromide (0.5 mg/L) and photographed under ultra violet illumination. Strain relatedness was determined using BioNumerics software (version 7.0; Applied Maths, Kortrijk, Belgium). The software established similarity between isolates using band-based dice coefficient method with 1.5% optimization and 1.5% band position tolerance. They were separated into similarity clusters by the unweighted-pair group method using average linkages (UPGMA). Relatedness of PFGE profiles was established at a cutoff level of 100% and 90%. The PFGE profiles of MRSA isolates that showed 100% similarity were considered identical and clustered in a single group, while the PFGE profiles of MRSA isolates that showed less than 100% similarity were considered a subtype of that group. Isolates of PFGE profiles that showed similarity at a cutoff level less than 90% were clustered into different groups.
Plasmid analysis was performed by the CTAB method as described previously.27 Curing, mixed-culture transfer and conjugation experiments were performed as described previously.28
Results
Molecular Typing of Isolates
The results of DNA microarray analysis performed on 1327 MRSA isolates obtained from different clinical samples in 2016 identified 42 (3.1%) isolates as CC15-MRSA-V+SCCfus. The distribution of other MRSA clones obtained in 2016 was reported elsewhere.14 The 42 isolates were cultured from wound swabs (N=14), nasal swabs (N=10), high vaginal swabs (N=5), groin swabs (N=3) blood (N=2), urine (N=2), tracheal aspirate (N=1), eye swab (N=1) with no clinical sources specified for two isolates in eight hospitals including Mubarak hospital, MUB (N=12), Adan hospital, ADH (N=5), Jahra hospital, JH (N=3), Razi hospital, RAZ (N=3), Maternity hospital, MAT (N=11), Sabah hospital, SAB (N=4), Chest-Disease hospital, CDH (N=1), and Farwaniya hospital, FAW (N=3). The isolates were obtained from 25 males and 15 females. The gender of two of the patients was not provided.
Spa typing of the 42 isolates revealed five spa types. These were t084 (N=36), t346 (N=3), t228 (N=1), t144 (N=1) and t7583 (N=1). MLST was performed on 20 representative isolates selected on the basis of their spa types as follows: 14 of t084 isolates, all three of t346 and one each of t228, t144 and t7583 isolate. All 20 isolates belonged to the same sequence type, ST1535 (13-13-1-1-81-11-13). ST1535 is a single locus variant of ST15 at the pta locus. These isolates carried pta 81 instead of pta12 in traditional ST15 isolates.20
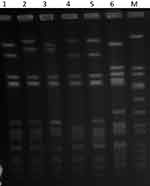
PFGE of the 42 isolates yielded four closely related patterns designated types A-D with the majority of the isolates belonging to PFGE type A and its subtypes (Figure 1). The t084 isolates were clustered among PFGE type A (N=28) and its subtypes A1 (N=9), type A2 (N=1) and type D (N=1). Isolates of PFGE types B (N-1) and C (N=3) corresponded to spa types t228 and t346, respectively. The t114 isolate belonged to PFGE type A1.
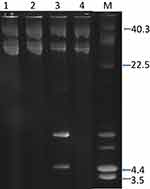
Plasmid analysis of the 42 isolates revealed the presence of a single c.28kb plasmid in 39 isolates. Three isolates harbored a single plasmid of c18kb and one isolate contained a 4.4kb plasmid (Figure 2).
Four isolates representing three plasmid patterns and spa types, t084, t114, t346, t228 and t7583 were selected for curing and transfer experiments to determine the resistance phenotypes of the plasmids. Colonies were screened for the loss of resistance to gentamicin, kanamycin, trimethoprim, tetracycline, and cadmium acetate. A minimum of 600 colonies were screened for loss of resistance to the selected antibiotics. None of the resistance screened was lost in any of the four isolates. Similarly, no resistance was transferred from any of the four isolates when used as donors in conjugation and mixed-culture transfer experiments.
Antibiotic Resistance Phenotypes and Genotypes
All 42 isolates were susceptible to vancomycin (MIC: ≤2 mg/L), teicoplanin (MIC: ≤2 mg/L), linezolid, rifampicin, trimethoprim, chloramphenicol, erythromycin (MIC: ≤0.5 mg/L) and clindamycin (MIC: 0.19–0.38 mg/L) but were resistant to penicillin G and cefoxitin (N=42), gentamicin (N=42), Kanamycin (N=42), fusidic acid (N=42), tetracycline (N=36) and ciprofloxacin (N=2). All isolates were resistant to cadmium acetate.
The resistance genotypes of the isolates are summarized in Table 1. The resistance genotypes matched the corresponding resistance phenotypes with the exception of the presence of inu(A). inu(A) (lincosamide nucleotidyltransferase), which confers resistance to lincomycin and clindamycin, was detected in all 42 isolates although they were phenotypically susceptible to clindamycin (MIC: 0.19–0.38 mg/L).
 |
Table 1 Characteristics of CC15-MRSA-V+SCCfus Isolates |
All isolates carried fusC which mediates fusidic acid resistance. In addition to aacA-aphD which mediates resistance to gentamicin, kanamycin and tobramycin; 39 of the isolates were positive for aadD which encodes resistance to kanamycin, neomycin and tobramycin. None of the isolates was positive for the aphA3, which mediates resistance to neomycin and kanamycin. One isolate was positive for erm(C).
Prevalence of Virulence-Related Genes
Results of DNA microarray analysis revealed that, with few exceptions, the isolates were remarkably homogeneous in the carriage of virulence-associated genes as presented in Table 1.
All 42 isolates were positive for genes for accessory gene regulator (agr) type II, and capsular polysaccharide type 8 (cap8), clumping factor A (clfA), clumping factor B (clfB), cell-wall-associated fibronectin-binding proteins (ebh), fibronectin-binding protein A and B (fnbA/B), biofilm-associated genes, icaA, icaC and icaD, and genes encoding microbial surface compounds recognizing adhesive matrix molecules, enolase, and hyaluronate lyase (hysA1). The isolates were also positive for transferrin binding protein, isdA and the type 1 site-specific deoxyribonuclease subunit, hsdSx (CC15) and hsdSx (CC25) but not hsdS1, hsdS2 and hsdS3.
All isolates lacked genes for Panton–Valentine leucocidin (PVL), staphylococcal enterotoxins, toxic-shock syndrome toxin (TSST), exfoliative toxins, and epidermal cell differentiation inhibitors, edinA, edinB and edinC, arginine catabolic mobile element (ACME) and collagen-binding adhesins (cna).
The isolates were variable in the carriage of genes for leukocidins, haemolysins; Immune evasion clusters and proteases as summarized in Table 1. Whereas all isolates were negative for lukM components, they were all positive for lukF, lukS, lukD, but differed in the carriage of luKE, lukX and hlgA. For the Immune evasion cluster genes, all 42 isolates were positive for Chp (chemotaxis-inhibiting proteins) and scn (staphylococcal complement inhibitor) and were negative for sak (staphylokinase). The haemolysins encoding gene hlb (haemolysin B) was absent in one t084 and in the t346 isolates (Table 1).
Distribution of the CC15-MRSA–V+SCCfus Isolates in the Hospitals
The time line for the isolation of the CC15-MRSA-V isolates is summarized in Table 2. The isolates were obtained from eight hospitals with most of them identified in two hospitals, namely, MUB (N=13) and MAT (N=11). The first isolate was obtained in January from a High Vaginal Swab sample of a patient at the MAT. The numbers that were isolated in subsequent months varied with higher numbers isolated in November (N=8), August (N=7), April (N=5) and December (N=5).
 |
Table 2 Isolation and Distribution of CC15-MRSA-V+SCCfus Isolates |
Table 3 shows the distribution of the 36 spa type t084 isolates. The t084 isolates were widely distributed with 19 of the 36 isolates originating from two of the eight hospitals. The isolates were obtained from different clinical samples representing colonization and infection sites with most of the isolates cultured from nasal (N=9) and wound (N=8) samples.
 |
Table 3 Distribution of t084 in Clinical Samples in Different Hospitals |
Discussion
This study reports the isolation of CC15-MRSA-V+SCCfus isolates from human patients in Kuwait hospitals. Prior to this report, CC15-MRSA-V+SCCfus isolates were isolated sporadically from retail camel meat12 and human patients in Saudi Arabia.13,20 The report of 42 CC15-MRSA-V+SCCfus isolates in this study represents the largest collection of CC15-MRSA-V+SCCfus isolates reported in the literature among human patients and represents a significant change in the epidemiology of this clone and its expansion into the human patient population.
The 42 CC15-MRSA-V+SCCfus in this study belonged to ST1535, harbored SCCmec V, agr II, and the novel SCCfus composite genetic element similar to the isolates obtained from retail camel meat12 and human patients reported previously in Saudi Arabia20 and may be related to them. However, although the isolates obtained from Kuwait and Saudi Arabia belonged to the same genotype, CC15-MRSA-V+SCCfus, they were not homogeneous. Although the majority of the isolates from Kuwait belonged to spa type t084, few isolates of spa types, t114, t228, t346 and t7583 were also detected. Similarly, in addition to spa type t084, the CC15-MRSA-V+SCCfus identified in Saudi Arabia also included isolates with different spa types, t328, t393 and t38520 suggesting that the isolates from both countries may have evolved differently. The detection of CC5-MRSA-V+SCCfus isolates with diverse spa types provides evidence of an ongoing diversification of the clone. The report of CC15-MSSA belonging to spa type t346 among S. aureus isolated in different European countries29 supports the independent acquisition of the SCCmec V and fusC, as seen in this study, by CC15-MSSA isolates. Therefore, it is important to continue to monitor the evolution of this clone in the region.
Prior to the report of CC15-MRSA-V+SCCfus isolates in Saudi Arabia, CC15-MRSA-IV isolates belonging to ST15 and accessory gene regulator group II (agr II), were reported in a small number of human patients in Saudi Arabia16 and Iran,17,18 and an isolate of ST15-MRSA-I was reported in a patient in Italy.19 Therefore, the CC15/ST1535-MRSA-V+SCCfus isolates in this study and those obtained in Saudi Arabia12,13 represent newly emerging variants of the CC15-MRSA lineage. Further support for the new emergence of the CC15-MRSA-V+SCCfus isolates is the report of the presence of hsdM/hsdS variants of the type 1 site-specific deoxyribonuclease subunit in the isolates from Saudi Arabia.20 The hsdM/hsdS variants were suggested to facilitate the uptake of new foreign DNA that promoted the emergence of CC15-MRSA.20 The CC15/ST1535-MRSA-V+SCCfus isolates carried one SCCfus composite genetic element which has recently been reported in fusidic acid-resistant MRSA isolates belonging to different genetic backgrounds including CC1, CC5, CC8, CC22, CC30, CC45, CC152, CC97 and CC91314,21,30–33 and appears to have contributed to the recent increases in the prevalence of fusidic acid resistance in MRSA isolates.
Surprisingly, although phenotypically susceptible to erythromycin (MIC: ≤0.5 mg/L) and clindamycin (MIC: MIC: 0.19–0.38 mg/L), the isolates were positive for inu(A) that confers resistance to clindamycin and lincomycin, and one of the isolates was positive for erm(C) that encodes erythromycin resistance suggestive of the presence of unexpressed resistance genes. Similarly, Raji et al12 reported the presence of mecA in CC15-MRSA-V+SCCfus isolates that were phenotypically susceptible to cefoxitin.
Our results, obtained with DNA microarray, mirrored the results obtained with whole genome sequencing of the isolates from Saudi Arabia with regards to the presence of some virulence-related genes.20 Similar to the results with isolates from Saudi Arabia,20 the isolates in this study harbored a variety of virulence genes except genes for staphylococcal enterotoxins, toxic shock syndrome toxin and exfoliative toxins (Table 1). Similarly, non-human clones of Livestock-associated MRSA isolates such as CC3988,34,35 and CC9736 also lack genes for staphylococcal enterotoxins and toxic shock syndrome toxin suggesting that the CC15-MRSA-V+SCCfus may be related to LA-MRSA lineage.
The results also revealed the presence of the Immune evasion cluster genes, scn and chp, and the absence of sak. Since sak is thought to be associated with tissue invasion, its absence in the CC15 lineage has been used to explain their association with carriage rather than invasive infections.20 However, a few of the isolates in this study were obtained from blood cultures and other invasive clinical samples suggesting that at least some of the CC15-MRSA isolates can also cause invasive infections. Unfortunately, no information was available to indicate whether these patients had catheter or IV lines which could explain their presence in the bloodstreams. It is possible that because only few CC15-MRSA isolates have been reported till date, the range of infections they are capable of causing has not been fully appreciated. As more CC15-MRSA-V+SCCfus are isolated from human patients and studied, more will be known about their capacity to cause infections.
Although isolated in eight hospitals, most of the isolates were obtained from two hospitals. However, the pattern of isolation shown in Table 2 suggests that the isolates were not associated with outbreaks. This is supported by the fact that the isolates were obtained from single patients except in November where two isolates were obtained from two different sites in the same patient. Nevertheless, the isolation of spa type t084 in each of the eight hospitals suggests that, given the right conditions, patient-to-patient transmission can occur. It will be interesting to see how this clone will evolve in the future.
Limitations of this study include lack of information regarding travel history of the patients who harbored these strains which would help establish any epidemiological relationship with the isolates from Saudi Arabia. We also do not have information regarding animals contact or consumption of camel meat. Information is also not available regarding whether the patients were colonized or infected at the time of admission to the hospital. Furthermore, it is not known whether patients whose blood cultures yielded CC15-MRSA-V+SCCfus had catheters or IV lines which could aid the survival of these strains in the blood stream.
Conclusion
We report the isolation and characterization of an emerging CC15-MRSA-V+SCCfus clone from patients in eight hospitals in Kuwait. The isolates belonged to sequence type, ST1535 and closely related pulsotypes. The isolates belonged to different spa types with t084 as the dominant spa types. The isolates expressed resistance to multiple antibiotics including fusidic acid, gentamicin, and tetracycline, harbored genes for multiple virulence factors but lacked genes for staphylococcal enterotoxins, toxic shock syndrome toxins and exfoliative toxins. Future surveillance is required to monitor the isolation of this clone in other hospitals in the country.
Ethical Approval
This study does not require ethical approval.
Data Sharing Statement
All data are available in the text.
Acknowledgment
The authors are grateful for the technical staff in the MRSA Reference Laboratory located in Microbiology department located in Faculty of Medicine for their technical assistance.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the final version for publication, and agreed to be accountable for all aspects of the work.
Funding
This study was not funded by any external organization or institution.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31:4.
2. Monecke S, Coombs G, Shore AC, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(4):e17936. doi:10.1371/journal.pone.0017936
3. Jevons MP. “Celbenin”-resistant staphylococci. Br Med J. 1961;1(5219):124–125. doi:10.1136/bmj.1.5219.124-a
4. Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25(2):97–108. doi:10.1016/0195-6701(93)90100-E
5. David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–687. doi:10.1128/CMR.00081-09
6. Armand-Lefevre L, Ruimy R, Andremont A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis. 2005;11(5):711–714. doi:10.3201/eid1105.040866
7. Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11(12):1965–1966. doi:10.3201/eid1112.050428
8. Fluit AC. Livestock-associated Staphylococcus aureus. Clin Microbiol Infect. 2012;18(8):735–744. doi:10.1111/j.1469-0691.2012.03846.x
9. Fitzgerald JR. Human origin for livestock-associated methicillin-resistant Staphylococcus aureus. MBio. 2012;3(2):e00082–e00012. doi:10.1128/mBio.00082-12
10. Liu CM, Price LB, Hungate BA, et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 2015;1(5):e1400216. doi:10.1126/sciadv.1400216
11. Larsen J, Stegger M, Andersen PS, et al. Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2016;63(10):1349–1352. doi:10.1093/cid/ciw532
12. Raji MA, Garaween G, Ehricht R, Monecke S, Shibl AM, Senok A. Genetic characterization of Staphylococcus aureus isolated from retail meat in Riyadh, Saudi Arabia. Front Microbiol. 2016;7:911. doi:10.3389/fmicb.2016.00911
13. Senok A, Ehricht R, Monecke S, Al-Saedan R, Somily A. Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect. 2016;14:13–18. doi:10.1016/j.nmni.2016.07.009
14. Boswihi SS, Udo EE, Monecke S, et al. Emerging variants of methicillin-resistant Staphylococcus aureus genotypes in Kuwait hospitals. PLoS One. 2018;13(4):e0195933. doi:10.1371/journal.pone.0195933
15. Senok A, Somily AM, Nassar R, et al. Emergence of novel methicillin-resistant Staphylococcus aureus strains in a tertiary care facility in Riyadh, Saudi Arabia. Infect Drug Resist. 2019;12:2739–2746. doi:10.2147/IDR.S218870
16. Abou Shady HM, Bakr AE, Hashad ME, Alzohairy MA. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz J Infect Dis. 2015;19(1):68–76. doi:10.1016/j.bjid.2014.09.005
17. Japoni-Nejad A, Rezazadeh M, Kazemian H, Fardmousavi N, van Belkum A, Ghaznavi-Rad E. Molecular characterization of the first community-acquired methicillin-resistant Staphylococcus aureus strains from Central Iran. Int J Infect Dis. 2013;17(11):e949–e954.
18. Goudarzi M, Seyedjavadi SS, Nasiri MJ, Goudarzi H, Sajadi Nia R, Dabiri H. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from patients with bacteremia based on MLST, SCCmec, spa, and agr locus types analysis. Microb Pathog. 2017;104:328–335. doi:10.1016/j.micpath.2017.01.055
19. Campanile F, Bongiorno D, Borbone S, Stefani S. Hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) in Italy. Ann Clin Microbiol Antimicrob. 2009;8:22. doi:10.1186/1476-0711-8-22
20. Senok AC, Somily AM, Slickers P, et al. Investigating a rare methicillin-resistant Staphylococcus aureus strain: first description of genome sequencing and molecular characterization of CC15-MRSA. Infect Drug Resist. 2017;10:307–315. doi:10.2147/IDR.S145394
21. Boswihi SS, Udo EE, Al-Sweih N. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait Hospitals: 1992–2010. PLoS One. 2016;11(9):e0162744. doi:10.1371/journal.pone.0162744
22. Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement M100-S25. Wayne, PA, USA: CLSI; 2015.
23. British Society to Antimicrobial Chemotherapy (BSAC) (BSAC, 2013). Available from: http://bsac.org.uk/susceptibility.
24. Udo EE, Jacob LE, Mathew B. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J Med Microbiol. 2001;50(10):909–915. doi:10.1099/0022-1317-50-10-909
25. Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi:10.1128/JCM.41.12.5442-5448.2003
26. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi:10.1128/JCM.38.3.1008-1015.2000
27. Udo EE, Al-Bustan MA, Jacob LE, Chugh TD. Enterotoxin production by coagulase-negative staphylococci in restaurant workers from Kuwait City may be a potential cause of food poisoning. J Med Microbiol. 1999;48(9):819–823. doi:10.1099/00222615-48-9-819
28. Udo EE, Jacob LE. Conjugative transfer of high-level mupirocin resistance and the mobilization of non-conjugative plasmids in Staphylococcus aureus. Microb Drug Resist. 1998;4(3):185–193.
29. Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7(1):e1000215. doi:10.1371/journal.pmed.1000215
30. Oliveira DC, Milheirico C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006;50(10):3457–3459. doi:10.1128/AAC.00629-06
31. Williamson DA, Monecke S, Heffernan H, et al. High usage of topical fusidic acid and rapid clonal expansion of fusidic acid-resistant Staphylococcus aureus: a cautionary tale. Clin Infect Dis. 2014;59(10):1451–1454. doi:10.1093/cid/ciu658
32. Ellington MJ, Reuter S, Harris SR, et al. Emergent and evolving antimicrobial resistance cassettes in community-associated fusidic acid and meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2015;45(5):477–484. doi:10.1016/j.ijantimicag.2015.01.009
33. Monecke S, Jatzwauk L, Muller E, et al. Diversity of SCCmec elements in Staphylococcus aureus as observed in South-Eastern Germany. PLoS One. 2016;11(9):e0162654. doi:10.1371/journal.pone.0162654
34. Heikinheimo A, Johler S, Karvonen L, et al. New dominant spa type t2741 in livestock-associated MRSA (CC398-MRSA-V) in finnish fattening pigs at slaughter. Antimicrob Resist Infect Control. 2016;5:6. doi:10.1186/s13756-016-0105-8
35. Cuny C, Abdelbary MMH, Kock R, et al. Methicillin-resistant Staphylococcus aureus from infections in horses in Germany are frequent colonizers of veterinarians but rare among MRSA from infections in humans. One Health. 2016;2:11–17. doi:10.1016/j.onehlt.2015.11.004
36. Monecke S, Kuhnert P, Hotzel H, Slickers P, Ehricht R. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet Microbiol. 2007;125(1–2):128–140. doi:10.1016/j.vetmic.2007.05.016
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.